Highlights
-
•
Swine coronaviruses responsible for significant economic losses to the swine industry.
-
•
Vaccines available only for TGEV and PEDV.
-
•
Types of vaccines include inactivated, live attenuated, recombinant, vectored and DNA vaccines.
-
•
Most vaccines aim to induce lactogenic immunity by immunizing sows at the end of gestation.
Keywords: Review, Pigs, Coronaviruses, Vaccines
Abstract
The recent introduction of the porcine epidemic diarrhea virus (PEDV) into the North American swine herd has highlighted again the need for effective vaccines for swine coronaviruses. While vaccines for transmissible gastroenteritis virus (TGEV) have been available to producers around the world for a long time, effective vaccines for PEDV and deltacoronaviruses were only recently developed or are still in development. Here, we review existing vaccine technologies for swine coronaviruses and highlight promising technologies which may help to control these important viruses in the future.
1. Swine Coronaviruses
Coronaviruses were first described in the mid-1960s and subsequently isolated from a number of species including man, mice, swine and chicken. These viruses share a common morphological characteristic, a fringe of club-shaped projections, 12–24 nm long, around a pleomorphic 60–220 nm viral particle, having a resemblance to a solar corona (Masters, 2013). Coronaviruses infect humans and various animal species, causing respiratory, gastrointestinal and neurological diseases as well as hepatitis. Prominent examples include the severe acute respiratory syndrome virus (SARS-CoV), middle-eastern respiratory syndrome virus (MERS-CoV) and the feline infectious peritonitis virus (FIPV), to name a few.
Swine coronaviruses can be divided into respiratory (PRCoV) and enteropathogenic coronaviruses such as transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV) and porcine deltacoronavirus (PDCoV). The latter have similar epidemiological, clinical and pathological features. The family Coronaviridae is currently divided into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. TGEV and PEDV belong to the Alphacoronavirus genus, whereas PDCoV belongs to genus of Deltacoronaviruses.
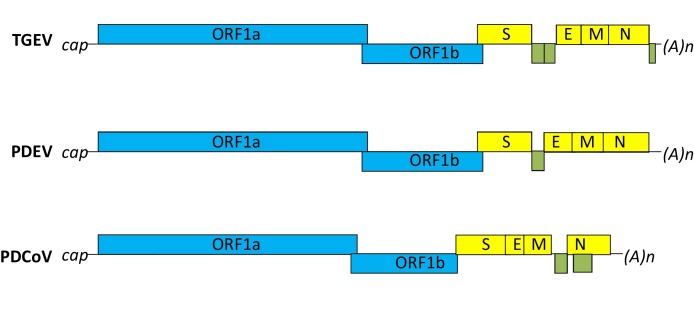
Coronaviruses are enveloped, single-stranded, positive-sense RNA viruses with the largest RNA genome of approximately 30 kb reported to date. The genomic RNA includes 5′ and 3′ untranslated regions (UTR), and it is capped and polyadenylated. Open reading frame (ORF) 1a and ORF1ab occupy the 5′ two-thirds of the genome and encode two replicase polyproteins (pp1a and pp1ab). Expression of pp1ab protein requires a ribosomal frameshift during translation of the genomic RNA. Produced polyproteins are proteolytically cleaved into 16 nonstructural proteins, nsp1 through nsp16 by the proteinase activity of nsp3 and nsp5. The 3′-proximal one-third of the genome encodes four structural proteins, including spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. Some betacoronaviruses have an additional membrane protein, hemagglutinin esterase (HE). Interspersed between these genes are genes encoding accessory proteins. The number of these genes varies between different coronaviruses. For instance, TGEV has 3 accessory genes, PDCoV has 2, whereas PEDV has only one (Fig. 1 ).
Fig. 1.
Genome organization of swine enteric coronaviruses TGEV, PEDV, and PDCoV. Genes for structural proteins are presented in yellow. Putative accessory genes are shown in green. Nonstructural proteins encoded by ORF1a/b are presented in blue. TGEV, transmissible gastroenteritis coronavirus; PEDV, porcine epidemic diarrhea virus; PDCoV, porcine deltacoronavirus; S, spike; E, envelope; M, membrane; N, nucleocapsid. Genomes have 5′ cap and 3′ poly A tail.
The viral RNA genome is packaged by the N protein into a helical nucleocapsid. In addition to the structural role, the N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, up-regulates interleukine-8 expression and antagonizes type I interferon production (Ding et al., 2014, Xu et al., 2013b). The S protein, which forms peplomers on the virion surface, mediates binding to host receptors and membrane fusion. It can be divided into S1 and S2 domains. In some coronaviruses the S protein is processed into S1 and S2 fragments by cellular proteases or trypsin (Belouzard et al., 2012, Wicht et al., 2014). The S protein is a major target for virus neutralizing antibodies (Chang et al., 2002, Reguera et al., 2012). The M protein is the most abundant virion component and also contains conserved linear B-cell epitopes (Zhang et al., 2012). The E protein is responsible for the assembly of virion, and it causes endoplasmic reticulum stress and interleukin-8 expression up-regulation (Xu et al., 2013a). Accessory genes are dispensable for virus growth in vitro, but they may play an important role in the virus survival in the infected host. Indeed, the product of the TGEV accessory gene ORF7 reduces the expression of genes involved in the antiviral defense of the immune system, e.g. the interferon response, and inflammation (Cruz et al., 2011). The ORF3 protein of PEDV functions as an ion channel, and it is thought to be related with virulence of PEDV (Song et al., 2003, Wang et al., 2012). One of the PEDV non-structural proteins, nsp1, was shown to be a type I interferon suppressor (Zhang et al., 2016a). Interestingly, PDCoV lacks the nsp1 gene.
2. Pathogenesis and clinical disease
Coronaviruses target predominantly type I and II pneumocytes (PRCoV) or villous- and crypt enterocytes in the intestine (TGEV, PEDV and PDCoV). PEDV also infection Goblet cells in the small intestine (Jung and Saif, 2015). In addition, infection of alveolar macrophages and lamina propria macrophages has been shown for some but not all swine coronaviruses (Laude et al., 1984, Park and Shin, 2014). Entrance of the virus into the target cells is mediated by a series of receptor ligand interactions including heparin sulfate (Huan et al., 2015) and aminopeptidase N (APN) (Chen et al., 1996, Li et al., 2007). Importantly, the expression levels of aminopeptidase N appear to correlate with the level of infection, at least for PEDV. The higher the expression levels the more severe is the infection (Li et al., 2007, Zhang and Yoo, 2016). As a result, it may be perceivable that piglets born with lower APN levels in the brush border may be more resistant to PEDV than piglets with higher levels.
Enteric infections with TGEV and PEDV are characterized by severe diarrhea, vomiting and dehydration with high morbidity and mortality especially in piglets less than two weeks of age. In contrast, infections with respiratory coronaviruses cause very mild and transient disease in pigs of all ages, which often get unnoticed by the producer. Unless complicated by concurrent infections, PRCoV infections are only short lasting with temporary phases of coughing and respiratory distress. However, PRCoV can become a more significant problem during co-infections with other pathogens such as the porcine reproductive and respiratory syndrome virus PRRSV (Jung et al., 2009).
Infection of enterocytes with PEDV results in villous atrophy which can lead to malabsorption, diarrhea and anorexia. Within 24–48 h post infection, vomiting may occur, which typically does not last longer than 2–3 days post infection. Diarrhea can be found within 24 to 36 h post infection depending on the dose of the virus and the age of the piglets. Diarrhea typically lasts for about 5-8 days, but can last longer, and results in severe weight loss that often cannot be made up during the normal production cycle. Viral shedding is highest between days 3–5, but can last for days to weeks post infection. Surviving piglets start to recover around 6–8 days post infection, typically around the same time when proliferation of the crypt epithelium and regeneration of the villi occurs. Similarly, TGEV infects villous enterocytes and causes disease that clinically is indistinguishable from PEDV. Mortality rates are highest in young piglets, often reaching about 100%. In contrast, infection with PDCoV causes milder infections in piglets between 3 and 5 weeks of age. Diarrhea, vomiting and anorexia can be found in infected animals. In general, infected animals display much milder signs compared to infections with PEDV and TGEV.
3. Immunity to swine coronaviruses
The innate immune response to enteric coronaviruses in pigs is characterized by a rapid antiviral response in the intestine, including the release of interferons, nuclear factor кB and other antiviral molecules (Chattha et al., 2015, Jung and Saif, 2015, Sang et al., 2010). Pigs can produce three types of interferons (Sang et al., 2014): Type I interferons include well known interferons such as interferon α and β (IFN-α/β) and in pigs are encoded by as many as 17 different genes. The only type II interferon in pigs is IFN-ɣ. Type III interferons include IFN-λ1 (interleukin 29; IL-29), IFN-λ2 (IL-28A), IFN-λ3 (IL-28B) and IFN-λ4 (Kotenko et al., 2003, Park et al., 2012, Prokunina-Olsson et al., 2013, Sheppard et al., 2003, Zhang and Yoo, 2016). Their functions are unknown in pigs. Especially type I and III interferons are used by the host to counteract viral infections. In response, most viruses including PEDV and TGEV have developed strategies to evade and interfere with the interferon response. Several viral proteins, including structural and non-structural proteins have been identified for PEDV and TGEV that can suppress the interferon response. For an excellent review on the evasion of immunity by porcine coronaviruses please see (Zhang and Yoo, 2016).
The adaptive immune response to swine enteric coronaviruses is based on secretory antibodies and cytotoxic T cells. These include secretory IgA antibodies (SIgA) that are produced by antibody-secreting cells in the lamina propria of the mucosal tissues and systemic antibodies such as IgG and IgM are found in serum and interstitial tissues and some isotypes can be transsudated across the mucosal epithelium into the lumen (Chattha et al., 2015, Horton and Vidarsson, 2013). The cellular response to swine coronaviruses is characterized by T helper cells that are supporting the production of antibodies and cytotoxic T cells that are targeting virus infected epithelial cells. In pigs, these are predominantly ɣɗ-cells, most of which can be found within the intraepithelial layer (Bonneville et al., 2010). The majority of T cell epitopes are located in the Spike and nucleoprotein of coronaviruses (Channappanavar et al., 2014, Saif, 2004, Sestak et al., 1999). Additionally, CD8 T cell epitopes have been found in the membrane protein of the human SARS-CoV (Yang et al., 2006)
In neonatal piglets, the main mechanism of protection is mediated by lactogenic immunity. During lactation, SIgA, IgG and IgM are passively transferred to the piglet via colostrum and milk (Bohl and Saif, 1975, Saif and Bohl, 1979, Saif and Bohl, 1983, Salmon et al., 2009). Colostrum contains predominantly IgG, which is transudated from sow serum and absorbed by the piglet within the first 24–48 h of life. Secretory IgA is predominantly found in milk, after transitioning from colostrum to milk around 3–4 days of age (Langel et al., 2016). SIgA is produced by antibody secreting cells in the mammary gland, and it was shown many years ago by Bohl and Saif (Bohl et al., 1972a) that these cells migrate from the gut to the mammary gland at the end of pregnancy. This was confirmed by others in a variety of species and chemokines such as CCL28 and others have been found responsible for recruiting these antibody secreting cells to the mammary gland (Bourges et al., 2008, Lazarus et al., 2003, Meurens et al., 2006, Meurens et al., 2007, Wilson and Butcher, 2004). Thus, in order to enhance the level of maternal immunity the oral route seems to be the most obvious choice for vaccinating the sow. Indeed, most vaccines for enteric coronaviruses are designed to induce lactogenic immunity by vaccinating the sow, however, most of them are administered via systemic injection. In the absence of effective vaccines for PEDV, many producers are currently using a lock-down of the barn combined with feeding back infectious live virus to pregnant sows. However, the duration of immunity often does not extent more than a few years (Table 1 ), depending on the type of vaccine being used with live vaccines typically providing longer lasting immunity. Even after feed-back, immunity starts to wane after a relatively short period of time, often even less than a few months. For an excellent review of the role of lactogenic immunity for PEDV see (Langel et al., 2016). In addition to antibodies, the colostrum also contains innate effector molecules such as defensins and antimicrobial peptides, interleukins and cytokines (Bandrick et al., 2014, Hlavova et al., 2014, Mair et al., 2014, Nechvatalova et al., 2011, Salmon et al., 2009).
Table 1.
Vaccine strategies for swine enteric Coronaviruses.
| Vaccine type | Development | Advantages | Disadvantages | Coronavirus antigen |
|---|---|---|---|---|
| Inactivated virus (Baek et al., 2016, Frederickson et al., 2014, Berube et al., 2015) |
Virions are inactivated with chemicals. | Easy to prepare; cannot cause disease if properly inactivated. | Can induce Th2-skewed immune response; needs adjuvant. | Whole virus |
| Live-attenuated virus (de Arriba et al., 2002, Sato et al., 2011) |
Genomes are mutated using multiple passages in Vero cells. | Inexpensive; strong cellular and humoral immune responses; can be given orally. | Reversion to virulence; can still cause some disease; protection is dose dependent. | Whole virus |
| Viral vectored (Yuan et al., 2015, Tuboly and Nagy, 2001). |
Unrelated viral genome (Poxvirus, Adenovirus) engineered to express the gene of interest. | Strong cellular and humoral immune responses; intrinsic adjuvant properties; can be given orally. | Preexisting immunity against vector virus. | Spike protein |
| Subunit (Oh et al., 2014, Makadiya et al., 2016, Bae et al., 2003) |
Antigen is expressed in mammalian, baculovirus, yeast or plant cells. | Cannot cause disease from viral infection; can generate high-titer neutralizing antibodies. | Expensive; needs adjuvant; protection can be incomplete. | Spike protein |
| DNA vaccines (Zhang et al., 2016b, Meng et al., 2013) |
Genes encoding antigens are cloned into plasmid expression vector. | Cannot cause disease from viral infection; can be given orally when introduced into Lactobacillus or Salmonella. | Th1-skewed immune response when used alone. | Spike, nucleocapsid or membrane proteins |
| Viral replicating particles vaccine (Mogler et al., 2014a, Mogler et al., 2014b) |
Replicon RNA containing gene of interest is packaged into alphavirus virion particles. | Cannot cause disease from viral infection; intrinsic adjuvant properties; high level of antigen expression. | Oral delivery has not been demonstrated. | Spike protein |
The level of cross-protection is somewhat unclear for coronaviruses. For PEDV, Goede et al. reported that 3-day old piglets born to sows that had been infected with a mild strain of PEDV seven months previously, were protected against infection with a more virulent strain of PEDV (Goede et al., 2015). In this experiment the sows were challenged with a more virulent PEDV virus at day 109 of gestation, and orally re-challenged when the piglets were three days old. None of the sows displayed significant clinical symptoms. The piglets were orally challenged with 1 ml of mucus scrapings of the more virulent PEDV. While mortality and morbidity rates varied significantly amongst the piglets in each group, the overall morbidity and mortality was reduced in piglets born to sows that had been pre-exposed to PEDV.
4. Vaccines for TGEV and other coronaviruses
In the 90s, TGEV was responsible for severe economic losses around the globe. Several vaccine technologies were developed and commercialised. By administration to sows, the importance of lactogenic immunity was established (Bohl et al., 1972b, Saif and Bohl, 1983). However, with a disappearance of the disease in many parts of the world, fewer vaccines are now commercially available in North America and Europe (Table 2 ). Most current commercial TGEV vaccines are live attenuated vaccines that are given to the sow during gestation in order to provide lactogenic immunity to the newborn piglet. These vaccines are often available as bi- or trivalent vaccines combined with rotavirus, PEDV and/or Escherichia coli. Experimental vaccines include novel DNA vaccines, vectored vaccines and recombinant vaccines (Table 1). For example, the porcine adenovirus was used to deliver the TGEV spike protein (Tuboly and Nagy, 2001). Yuan et al. used the swine pox virus to express the A epitope of the spike protein (Yuan et al., 2015). DNA plasmids were generated for both PEDV and TGEV for the development of a DNA vaccine (Meng et al., 2013). Recombinant proteins (spike and nucleocapsid) have been extensively evaluated as recombinant vaccine following expression in bacteria, yeast and plants. Many of these are being assessed for their potential to mucosal immunity after oral administration.
Table 2.
Vaccines for TGEV and PEDV.
| Virus | Region/country | Vaccines in development | Commercial vaccines |
|---|---|---|---|
| TGEV | North America | Recombinant proteins expressed in baculovirus, yeast, and plants; live attenuated vaccine; DNA vaccine | Live attenuated vaccines (mono, bi- and trivalent for TGEV, rota and E.coli) |
| Europe | Recombinant proteins expressed in baculovirus, yeast, and plants; live attenuated vaccine; DNA vaccine | Live attenuated vaccines (mono, bi- and trivalent for TGEV, rota and E.coli) | |
| Asia | Recombinant proteins expressed in baculovirus, yeast, and plants; live attenuated vaccine | Inactivated vaccines (mono, bi- and trivalent for TGEV, rota, PEDV and/or E.coli); live attenuated trivalent TGEV, PEDV and porcine rotavirus (China) | |
| PEDV | North America | Recombinant proteins expressed in yeast and baculovirus; DNA vaccine; infectious clone for live-attenuated vaccine | Inactivated vaccine; Recombinant alphavirus-based vaccine |
| Europe | DNA vaccine | Inactivated vaccines | |
| Asia | Recombinant vaccines expressed in baculovirus, yeast, plants, Lactobaccilus casei, Salmonella Typhimurium and others | Inactivated bi-valent TGEV and PEDV vaccine (China; strain CV777); Live attenuated tri-valent TGEV, PEDV and porcine rotavirus (China; PEDV strain CV777); Live attenuated vaccine (Japan; PEDV strain 83P-5; South Korea; PEDV strains SM98-1 and DR-13, Philippines, PEDV strain DR13); Inactivated vaccine (South Korea, PEDV strain SM98-1) |
5. Vaccines for PEDV in North America
The first vaccine for PEDV in the US was developed by Harrisvaccines™ in Iowa, and conditionally licensed in 2013. The vaccine, initially called iPED vaccine, was based on a truncated version of the PEDV spike gene produced in the SirraVax℠ RNA Particle Technology platform (Mogler et al., 2014b), which is based on the a pVEK replicon vector derived from the Venezuelan equine encephalitis virus. The technology is a propagation defective, single cycle RNA particle technology that is believed to target dendritic cells. A longer version of the spike genes was codon optimized and used in the second-generation vaccine called iPED plus, which now is commercially available as Porcine Epidemic Diarrhea Vaccine, RNA (PED RNA). The vaccine induced immunity in young pigs after two doses given intramuscularly in a three-week interval. Vaccinated weaned pigs when challenged with homogenized gut tissue from a clinical isolate displayed significantly reduced severity of clinical signs (diarrhea) and reduced viral shedding for the first 72 h (Mogler et al., 2014a). Vaccine efficacy was further tested in naïve sows. After three vaccinations at 8, 5, and 1 weeks pre-farrowing piglets from both groups were challenged between 2 and 6 days of age with 103 TCID50 (PEDV/CO/2013). Average litter mortality in the control group was 91%, while average mortality in the vaccinated groups was 69% (Crawford et al., 2015). Similar results were found in sows previously exposed to PEDV. After oral challenge of piglets within the first week average litter mortality was reduced from 59% in the control group to 45% in the vaccinated group. Finally, Greiner et al. (2015) evaluated the vaccine in 80 sows that had been previously exposed to PEDV. Vaccination induced higher titers against the S1 protein in the colostrum of vaccinated sows and reduced overall pig mortality by 3% (Greiner et al., 2015).
A second vaccine for PEDV in the US was developed by Zoetis and made commercially available in 2014 under a conditional license. The vaccine consists of an inactivated whole virus formulated with an adjuvant. The vaccine was tested in PEDV negative sows, which were vaccinated twice, 3 weeks apart, with the vaccine (n = 23) or an adjuvant placebo (n = 3). The vaccine was safe and immunogenic and induced neutralizing antibodies to both the whole virus and the spike protein (Frederickson et al., 2014). The vaccine was also tested under field conditions in a PEDV positive commercial herd. Sows (n = 120) were vaccinated at 5- and 2 weeks pre-farrowing, control sows (n = 120) received placebo. Vaccination resulted in reduction of pre-weaning mortality due to PEDV from 6.3% in piglets born to control sows versus 0.6% in piglets born to vaccinated sows. Vaccination resulted in about 3 times higher neutralization antibody titers compared to the control group, and an additional 1.8 pigs per sows survived (Rapp-Gabrielson et al., 2014).
A third vaccine was recently developed by the Vaccine and Infectious Disease Organization-InterVac in Canada. The vaccine is based on inactivated virus formulated with an adjuvant. When administered to seronegative sows 4 and 2 weeks prior to farrowing, high levels of neutralizing antibodies against PEDV were found in colostrum and milk, as well as in the serum of piglets born to vaccinated sows. Piglets were orally infected at 5 days of life with 300 pfu of PEDV isolate CO 025. It was found that 95% of all piglets (n = 83) born to vaccinated sows survived the infection and showed significantly reduced clinical symptoms, weight loss and viral shedding. In contrast, all piglets from unvaccinated sows displayed severe clinical symptoms including weight loss and dehydration, and 50% of these piglets died within 6 days post infection. These results were confirmed in two additional clinical trials. A large field trial involving > 600 sows was performed in three commercial units in Saskatchewan, Canada to assess the vaccine in different genetics, health statuses and management systems. The vaccine demonstrated to be completely safe to use; no adverse events including injection site reactions and reproductive complications were observed. Vaccine efficacy was evaluated in 8% of these animals by transporting the pregnant sows a week before farrowing to the VIDO-InterVac high containment facility. Following oral challenge at 5 days of age, survival was significantly higher in piglets born to vaccinated sows than those from control sows (Berube et al., 2015). The assessment of duration of immunity is currently ongoing. In addition, an affinity tagged PEDV S1 protein was expressed in the HEK293 system to be used as subunit vaccine. When administered to pregnant sows the vaccine partially protected newborn piglets against infection with PEDV (Makadiya et al., 2016).
6. PEDV vaccines in Asia
Since early 1980s, PEDV outbreaks have been reported in several Asian countries, including China, Japan, Thailand, Taiwan, the Philippines, South Korea and Vietnam. In October 2010, a large-scale outbreak of severe PEDV was reported in China (Wang et al., 2013). In late 2013, PEDV outbreaks occurred in Japan, South Korea and Taiwan (Lee and Lee, 2014). Phylogenetic analysis of PEDV full-length genomic sequences reveals that PEDV can be genetically divided into 2 groups: G1 (classical) and G2 (field epidemic or pandemic). Each group can be further divided into two subgroups: 1a and 1b, 2a and 2b, respectively. It is also possible that the low to moderate effectiveness of current PEDV vaccines are due to genetic differences between vaccine and field epidemic strains (Kim et al., 2015).
For disease control, an inactivated bivalent TGEV and PEDV vaccine was introduced in China in 1995. In March 2015, a trivalent attenuated vaccine (PEDV, TGEV and porcine rotavirus) was also approved. All these vaccines were based on the classical CV777 (G1-a) strain that can be grown to high titers in green monkey kidney Vero cells. There are no data published on the efficacy of these vaccines. However, de Arriba et al. (de Arriba et al., 2002) orally inoculated 11-day-old conventionally reared piglets with two different doses of the attenuated CV-777 strain and challenged with the same virulent PEDV strain three weeks later. The vaccinated pigs were partially protected against the challenge, and 25% of the low dose- and 50% of the high dose-exposed pigs did not shed virus after challenge.
Since winter of 2010, China experienced severe PED outbreaks with devastating damage to the swine industry. These outbreaks can be explained by re-emergence of new PEDV strains. To answer industry demand, Chinese researchers have developed a bivalent (PEDV and TGEV) attenuated vaccine that contained the PEDV strain ZJ08 (G1-b) and a bivalent attenuated vaccine based on the AJ1102 strain (G2-b). Inactivated bivalent vaccine based on the G2-b strain has also been developed. These vaccines are currently under clinical evaluation (Wang et al., 2016).
In Japan, PEDV strain 83P-5 (G1-a) was attenuated after 100 passages in Vero cells (Sato et al., 2011). Subsequently, this strain has been employed as an intramuscular (IM) live attenuated vaccine (P–5 V) in Japan and South Korea. In addition, two South Korean virulent PEDV strains (SM98-1 and DR-13) were attenuated by serial cell culture passages. The attenuated SM98-1 strain has been used as an IM live or killed vaccine, whereas DR-13 was used as an oral live vaccine. This oral vaccine was registered and commercialised in the Philippines in 2011. In South Korea, the multiple dose vaccination program (3 or 4 IM vaccinations in the following order: live-killed–killed or live-live-killed-killed, respectively) at 2- or 3-week intervals starting before farrowing is commonly recommended in pregnant sows (Lee, 2015).
According to a South Korean study, the administration of commercial vaccines increased the survival rate of piglets challenged with a virulent wild-type PEDV from 18.2% to over 80%. However, all vaccines did not significantly reduce morbidity rate and virus shedding (Lee, 2015). Also, Song et al. (Song et al., 2007) reported 60% reduction of the mortality rate of the suckling piglets born to the sows that were IM vaccinated with DR-13 PEDV vaccine.
Despite of nationwide use of commercially available vaccines, South Korea experienced a devastated PED epidemic in 2013–2014. PEDV G2-b strains were responsible for recent severe PED epidemics in Asian countries and North America. Considering this fact, South Korean researchers (Baek et al., 2016) tested an inactivated vaccine based on serially cultured G2-b strain KOR/KNU-141112/2014. Pregnant sows were immunized IM with the inactivated adjuvanted vaccine at 6 and 3 weeks prior to farrowing. Six-day old piglets were challenged with the homologous virulent virus. Piglets born to vaccinated sows had reduced morbidity, mortality and quickly recovered daily weight gain. Further studies are needed to evaluate efficacy of this vaccine in 1- or 2-day old piglets under field conditions.
All commercially available vaccines that are in use in Asia are traditional live attenuated or killed vaccines. However, researchers are working on the next-generation vaccines for PEDV (Table 1). For instance, Hou et al. (Hou et al., 2007) expressed the PEDV N protein on the surface of lactobacillus. Oral and intranasal inoculations of recombinant L. casei into pregnant sow and mice resulted in high levels of N-specific serum IgG and mucosal IgA. Similarly, Liu et al. (Liu et al., 2012) expressed S1 and N protein in recombinant L. casei and reported enhanced mucosal and systemic immune responses after oral immunization of mice. Meng et al. (Meng et al., 2013) evaluated the immunogenicity of recombinant DNA plasmids expressing S genes from PEDV and TGEV in mice. The results showed that the recombinant DNA plasmids increased the proliferation of T lymphocytes and the number of CD4+ and CD8+ T lymphocytes. In addition, the DNA vaccines induced a high level of IFN-γ in the immunized mice. At 35 days post-immunization, the recombinant DNA plasmids bearing full-length S genes of TGEV and PEDV stimulated high levels of virus-neutralizing antibodies. Zhang et al. (Zhang et al., 2016b) have also constructed bivalent DNA vaccine co-expressing S genes of TGEV and PEDV. Attenuated Salmonella Typhimurium was used for oral delivery this vaccine into pigs. Vaccinated pigs developed TGEV and PEDV-specific cellular and humoral immune responses; however, challenge experiment was not conducted in this study.
There have also been reports on expressing recombinant PEDV S protein fragments in plants (Bae et al., 2003) and in a cell line (Oh et al., 2014). Feeding mice with transgenic tobacco plants that express the S protein fragment containing the PEDV neutralizing epitope induced PEDV-specific antibody and cell-mediated immune responses. In another study, Oh et al. (Oh et al., 2014) established porcine cell line stably expressing S1 fragment of the PEDV spike protein. Pregnant sows were immunized IM 3 times with the purified and adjuvanted protein 6, 4 and 2 weeks prior to farrowing. The sows developed PEDV-specific neutralizing antibody response in serum and colostrum. One 4- to 5-day-old piglet was selected randomly from each farrowing sow for challenge with virulent PEDV. The piglets born to vaccinated sows showed reduced morbidity, mortality and virus shedding. However, a low number of challenged piglets hamper the author’s conclusion about efficient protection of neonatal piglets.
7. Future perspectives
With the disappearance of TGEV around the world, the need for TGEV vaccines has dropped over the last few years. For North America and Europe, only two major international animal health companies continue to offer TGEV vaccines. In contrast, many parts of Asia including China and Korea are still dealing with TGEV outbreaks, and different types of vaccines are still available. The introduction of PEDV into the North American herd in 2013–2014, however, has reversed this picture and has highlighted the global need for effective vaccines. Coronaviruses represent an important group of animal pathogens that can have devastating impacts in a variety of species. For an industry to rely on biosecurity alone seems somewhat risky and, in case of an emerging disease, can lead to major economic losses, as witnessed in North America over the last two years. Indeed, the phylogeny of coronaviruses demonstrates a great deal of diversity in antigenic variants, which may lead to limited cross-protection against infection with different strains. Thus, it is important to continue to survey novel PEDV variants that may emerge locally or globally through antigenic drift (point mutations) or antigenic shift (recombination events).
Effective prevention and control of PEDV and other coronaviruses can only be achieved through the use of vaccines. An ideal vaccine prevents mortality and clinical disease in newborn piglets, the age group most seriously affected by the disease, as well as viral shedding. As lactogenic immunity is a key mechanism of protection, efforts to enhance the levels of antibodies in the milk through formulation (adjuvants) and delivery (mucosal) are critical. However, time is also of essence when dealing with a new strain, as traditional manufacturing vaccine methods like virus isolation, inactivation or attenuation can be time-consuming. Therefore, research on next-generation vaccines such as RNA particle, DNA, sub-unit and viral-vectored approaches is critical for the prevention of future outbreaks of emerging coronavirus diseases.
Conflict of interest statement
The authors declare no conflict of interest.
References
- Bae J.L., Lee J.G., Kang T.J., Jang H.S., Jang Y.S., Yang M.S. Induction of antigen-specific systemic and mucosal immune responses by feeding animals transgenic plants expressing the antigen. Vaccine. 2003;21:4052–4058. doi: 10.1016/s0264-410x(03)00360-8. [DOI] [PubMed] [Google Scholar]
- Baek P.S., Choi H.W., Lee S., Yoon I.J., Lee Y.J., Lee du S., Lee C. Efficacy of an inactivated genotype 2b porcine epidemic diarrhea virus vaccine in neonatal piglets. Vet. Immunol. Immunopathol. 2016;174:45–49. doi: 10.1016/j.vetimm.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandrick M., Ariza-Nieto C., Baidoo S.K., Molitor T.W. Colostral antibody-mediated and cell-mediated immunity contributes to innate and antigen-specific immunity in piglets. Dev. Comp. Immunol. 2014;43:114–120. doi: 10.1016/j.dci.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube N., Hauta Shirley, Popowych Yurij, Connor Wayne, Erickson Jan, Tetland Sherry, Bock Ken, Allan Brenda, van den Hurk Jan, Makadiya Niraj, van Moorlehem Elaine, Brownlie Robert, Zakhartchouk Alexander, Rawlyk Neil, Walker Stew, Wheler Colette, Don Wilson S.K., Tikoo, Yan Zhou, Andrew Potter A., Volker Gerdts . North American PRRS Symposium; Chicago: 2015. Development of a Novel Vaccine for Porcine Epidemic Diarrhea Virus. [Google Scholar]
- Bohl E.H., Saif L.J. Passive immunity in transmissible gastroenteritis of swine: immunoglobulin characteristics of antibodies in milk after inoculating virus by different routes. Infect. Immun. 1975;11:23–32. doi: 10.1128/iai.11.1.23-32.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohl E.H., Gupta R.K., McCloskey L.W., Saif L. Immunology of transmissible gastroenteritis. J. Am. Vet. Med. Assoc. 1972;160:543–549. [PubMed] [Google Scholar]
- Bohl E.H., Gupta R.K., Olquin M.V., Saif L.J. Antibody responses in serum, colostrum, and milk of swine after infection or vaccination with transmissible gastroenteritis virus. Infect. Immun. 1972;6:289–301. doi: 10.1128/iai.6.3.289-301.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneville M., O'Brien R.L., Born W.K. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 2010;10:467–478. doi: 10.1038/nri2781. [DOI] [PubMed] [Google Scholar]
- Bourges D., Meurens F., Berri M., Chevaleyre C., Zanello G., Levast B., Melo S., Gerdts V., Salmon H. New insights into the dual recruitment of IgA+ B cells in the developing mammary gland. Mol. Immunol. 2008;45:3354–3362. doi: 10.1016/j.molimm.2008.04.017. [DOI] [PubMed] [Google Scholar]
- Chang S.H., Bae J.L., Kang T.J., Kim J., Chung G.H., Lim C.W., Laude H., Yang M.S., Jang Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells. 2002;14:295–299. [PubMed] [Google Scholar]
- Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 2014;59:118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattha K.S., Roth J.A., Saif L.J. Strategies for design and application of enteric viral vaccines. Annu. Rev. Anim. Biosci. 2015;3:375–395. doi: 10.1146/annurev-animal-022114-111038. [DOI] [PubMed] [Google Scholar]
- Chen H., Kinzer C.A., Paul W.E. p161, a murine membrane protein expressed on mast cells and some macrophages, is mouse CD13/aminopeptidase N. J. Immunol. 1996;157:2593–2600. [PubMed] [Google Scholar]
- Crawford K., Mogler M., Hicks J., Harris D.L. Protective efficacy of a replicon particle vaccine in both naïve and previously exposed gilts against porcine epidemic diarrhea virus. Ann. Proc. Am. Assoc. Swine Veterinarians. 2015:212–213. [Google Scholar]
- Cruz J.L., Sola I., Becares M., Alberca B., Plana J., Enjuanes L., Zuniga S. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog. 2011;7:e1002090. doi: 10.1371/journal.ppat.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederickson, D., Bandrick, M., Taylor, L.P., Coleman, D.W., Pfeiffer, A., Locke, C.R., Huether, M.J., Zhang, J., Verhelle, R., Hildebrandt, T.K., Hardham, J.M., Rapp-Gabrielson, V.J., 2014. Safety and antibody response of pigs to an experimental Porcine Epidemic Diarrhea Virus (PEDV) vaccine, killed virus. 2014 North American PRRS Symposium, abstract, 69.
- Goede D., Murtaugh M.P., Nerem J., Yeske P., Rossow K., Morrison R. Previous infection of sows with a mild strain of porcine epidemic diarrhea virus confers protection against infection with a severe strain. Vet. Microbiol. 2015;176:161–164. doi: 10.1016/j.vetmic.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Greiner L., Connor J., Graham A., Mellor J., Lowe J. Evaluation of a PED vaccine on piglet mortality and sow immunity. 46th Proceedings of the 2015 Meeting of the American Association of Swine Veterinarians. 2015:361. [Google Scholar]
- Hlavova K., Stepanova H., Faldyna M. The phenotype and activation status of T and NK cells in porcine colostrum suggest these are central/effector memory cells. Vet. J. 2014;202:477–482. doi: 10.1016/j.tvjl.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Horton R.E., Vidarsson G. Antibodies and their receptors: different potential roles in mucosal defense. Front. Immunol. 2013;4:200. doi: 10.3389/fimmu.2013.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X.L., Yu L.Y., Liu J., Wang G.H. Surface-displayed porcine epidemic diarrhea viral (PEDV) antigens on lactic acid bacteria. Vaccine. 2007;26:24–31. doi: 10.1016/j.vaccine.2007.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan C.C., Wang Y., Ni B., Wang R., Huang L., Ren X.F., Tong G.Z., Ding C., Fan H.J., Mao X. Porcine epidemic diarrhea virus uses cell-surface heparan sulfate as an attachment factor. Arch. Virol. 2015;160:1621–1628. doi: 10.1007/s00705-015-2408-0. [DOI] [PubMed] [Google Scholar]
- Jung K., Saif L.J. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Renukaradhya G.J., Alekseev K.P., Fang Y., Tang Y., Saif L.J. Porcine reproductive and respiratory syndrome virus modifies innate immunity and alters disease outcome in pigs subsequently infected with porcine respiratory coronavirus: implications for respiratory viral co-infections. J. Gen. Virol. 2009;90:2713–2723. doi: 10.1099/vir.0.014001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lee J.M., Jung J., Kim I.J., Hyun B.H., Kim H.I., Park C.K., Oem J.K., Kim Y.H., Lee M.H., Lee K.K. Genetic characterization of porcine epidemic diarrhea virus in Korea from 1998 to 2013. Arch. Virol. 2015;160:1055–1064. doi: 10.1007/s00705-015-2353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Langel S.N., Paim F.C., Lager K.M., Vlasova A.N., Saif L.J. Lactogenic immunity and vaccines for porcine epidemic diarrhea virus (PEDV): historical and current concepts. Virus Res. 2016 doi: 10.1016/j.virusres.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Charley B., Gelfi J. Replication of transmissible gastroenteritis coronavirus (TGEV) in swine alveolar macrophages. J. Gen. Virol. 1984;65(Pt. 2):327–332. doi: 10.1099/0022-1317-65-2-327. [DOI] [PubMed] [Google Scholar]
- Lazarus N.H., Kunkel E.J., Johnston B., Wilson E., Youngman K.R., Butcher E.C. A common mucosal chemokine (mucosae-associated epithelial chemokine/CCL28) selectively attracts IgA plasmablasts. J. Immunol. 2003;170:3799–3805. doi: 10.4049/jimmunol.170.7.3799. [DOI] [PubMed] [Google Scholar]
- Lee S., Lee C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg. Infect. Dis. 2014;20:1223–1226. doi: 10.3201/eid2007.140294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.X., Ge J.W., Li Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology. 2007;365:166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.Q., Ge J.W., Qiao X.Y., Jiang Y.P., Liu S.M., Li Y.J. High-level mucosal and systemic immune responses induced by oral administration with Lactobacillus-expressed porcine epidemic diarrhea virus (PEDV) S1 region combined with Lactobacillus-expressed N protein. Appl. Microbiol. Biotechnol. 2012;93:2437–2446. doi: 10.1007/s00253-011-3734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair K.H., Sedlak C., Kaser T., Pasternak A., Levast B., Gerner W., Saalmuller A., Summerfield A., Gerdts V., Wilson H.L., Meurens F. The porcine innate immune system: an update. Dev. Comp. Immunol. 2014;45:321–343. doi: 10.1016/j.dci.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makadiya N., Brownlie R., van den Hurk J., Berube N., Allan B., Gerdts V., Zakhartchouk A. S1 domain of the porcine epidemic diarrhea virus spike protein as a vaccine antigen. Virol. J. 2016;13:57. doi: 10.1186/s12985-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters, P.S.a.P., S. 2013. Coronaviridae, In: Knipe, D.M.a.P.H. (Ed.) Fields Virology. Wolters Kluwer, 825.
- Meng F., Ren Y., Suo S., Sun X., Li X., Li P., Yang W., Li G., Li L., Schwegmann-Wessels C., Herrler G., Ren X. Evaluation on the efficacy and immunogenicity of recombinant DNA plasmids expressing spike genes from porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus. PLoS One. 2013;8:e57468. doi: 10.1371/journal.pone.0057468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurens F., Berri M., Whale J., Dybvig T., Strom S., Thompson D., Brownlie R., Townsend H.G., Salmon H., Gerdts V. Expression of TECK/CCL25 and MEC/CCL28 chemokines and their respective receptors CCR9 and CCR10 in porcine mucosal tissues. Vet. Immunol. Immunopathol. 2006;113:313–327. doi: 10.1016/j.vetimm.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Meurens F., Whale J., Brownlie R., Dybvig T., Thompson D.R., Gerdts V. Expression of mucosal chemokines TECK/CCL25 and MEC/CCL28 during fetal development of the ovine mucosal immune system. Immunology. 2007;120:544–555. doi: 10.1111/j.1365-2567.2006.02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogler, M.A., Gander, J.R., Ray, D.D., Harris, D.L., 2014b. Vaccination of PEDV-naïve dams with replicon RNA particle vaccine protects suckling piglets from challenge. North American PRRS Symposium, abstract 75.
- Mogler M.A., Gander J., Harris D.L.H. Development of an alphavirus RNA particle vaccine against porcine epidemic diarrhea virus. Ann. Proc. Am. Assoc. Swine Veterinarians. 2014:63–64. [Google Scholar]
- Nechvatalova K., Kudlackova H., Leva L., Babickova K., Faldyna M. Transfer of humoral and cell-mediated immunity via colostrum in pigs. Vet. Immunol. Immunopathol. 2011;142:95–100. doi: 10.1016/j.vetimm.2011.03.022. [DOI] [PubMed] [Google Scholar]
- Oh J., Lee K.W., Choi H.W., Lee C. Immunogenicity and protective efficacy of recombinant S1 domain of the porcine epidemic diarrhea virus spike protein. Arch. Virol. 2014;159:2977–2987. doi: 10.1007/s00705-014-2163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Shin H.J. Porcine epidemic diarrhea virus infects and replicates in porcine alveolar macrophages. Virus Res. 2014;191:143–152. doi: 10.1016/j.virusres.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Serti E., Eke O., Muchmore B., Prokunina-Olsson L., Capone S., Folgori A., Rehermann B. IL-29 is the dominant type III interferon produced by hepatocytes during acute hepatitis C virus infection. Hepatology. 2012;56:2060–2070. doi: 10.1002/hep.25897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L., Muchmore B., Tang W., Pfeiffer R.M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I., Chen S., Brand N., Tarway M., Liu L., Sheikh F., Astemborski J., Bonkovsky H.L., Edlin B.R., Howell C.D., Morgan T.R., Thomas D.L., Rehermann B., Donnelly R.P., O'Brien T.R. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp-Gabrielson, Frederickson, D.F., Bandrick, M., Taylor, L.P., Marx, J., Ricker, T., Coleman, D., Pfeiffer, A., Thompson, J.R., Zhang, J., Zager, S., Huether, M., Hardham, J.M., Sornsen, S., 2014. Field efficacy of an experimental Porcine Epidemic Diarrhea (PED) vaccine administered to pregnant sows. 2014 North American PRRS Symposium, abstract 80.
- Reguera J., Santiago C., Mudgal G., Ordono D., Enjuanes L., Casasnovas J.M. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8:e1002859. doi: 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif L.J., Bohl E.H. Passive immunity in transmissible gastroenteritis of swine: immunoglobulin classes of milk antibodies after oral-intranasal inoculation of sows with a live low cell culture-passaged virus. Am. J. Vet. Res. 1979;40:115–117. [PubMed] [Google Scholar]
- Saif L.J., Bohl E.H. Passive immunity to transmissible gastroenteritis virus: intramammary viral inoculation of sows. Ann. N. Y. Acad. Sci. 1983;409:708–723. doi: 10.1111/j.1749-6632.1983.tb26910.x. [DOI] [PubMed] [Google Scholar]
- Saif L.J. Animal coronavirus vaccines: lessons for SARS. Dev. Biol. (Basel) 2004;119:129–140. [PubMed] [Google Scholar]
- Salmon H., Berri M., Gerdts V., Meurens F. Humoral and cellular factors of maternal immunity in swine. Dev. Comp. Immunol. 2009;33:384–393. doi: 10.1016/j.dci.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Sang Y., Rowland R.R., Hesse R.A., Blecha F. Differential expression and activity of the porcine type I interferon family. Physiol. Genomics. 2010;42:248–258. doi: 10.1152/physiolgenomics.00198.2009. [DOI] [PubMed] [Google Scholar]
- Sang Y., Bergkamp J., Blecha F. Molecular evolution of the porcine type I interferon family: subtype-specific expression and antiviral activity. PLoS One. 2014;9:e112378. doi: 10.1371/journal.pone.0112378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Takeyama N., Katsumata A., Tuchiya K., Kodama T., Kusanagi K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Genes. 2011;43:72–78. doi: 10.1007/s11262-011-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestak K., Meister R.K., Hayes J.R., Kim L., Lewis P.A., Myers G., Saif L.J. Active immunity and T-cell populations in pigs intraperitoneally inoculated with baculovirus-expressed transmissible gastroenteritis virus structural proteins. Vet. Immunol. Immunopathol. 1999;70:203–221. doi: 10.1016/S0165-2427(99)00074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J., Ostrander C., Dong D., Shin J., Presnell S., Fox B., Haldeman B., Cooper E., Taft D., Gilbert T., Grant F.J., Tackett M., Krivan W., McKnight G., Clegg C., Foster D., Klucher K.M. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- Song D.S., Yang J.S., Oh J.S., Han J.H., Park B.K. Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3. Vaccine. 2003;21:1833–1842. doi: 10.1016/S0264-410X(03)00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.S., Oh J.S., Kang B.K., Yang J.S., Moon H.J., Yoo H.S., Jang Y.S., Park B.K. Oral efficacy of Vero cell attenuated porcine epidemic diarrhea virus DR13 strain. Res. Vet. Sci. 2007;82:134–140. doi: 10.1016/j.rvsc.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuboly T., Nagy E. Construction and characterization of recombinant porcine adenovirus serotype 5 expressing the transmissible gastroenteritis virus spike gene. J. Gen. Virol. 2001;82:183–190. doi: 10.1099/0022-1317-82-1-183. [DOI] [PubMed] [Google Scholar]
- Wang K., Lu W., Chen J., Xie S., Shi H., Hsu H., Yu W., Xu K., Bian C., Fischer W.B., Schwarz W., Feng L., Sun B. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586:384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhao P., Guo L., Liu Y., Du Y., Ren S., Li J., Zhang Y., Fan Y., Huang B., Liu S., Wu J. Porcine epidemic diarrhea virus variants with high pathogenicity, China. Emerg. Infect. Dis. 2013;19:2048–2049. doi: 10.3201/eid1912.121088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016 doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicht O., Li W., Willems L., Meuleman T.J., Wubbolts R.W., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 2014;88:7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E., Butcher E.C. CCL28 controls immunoglobulin (Ig)A plasma cell accumulation in the lactating mammary gland and IgA antibody transfer to the neonate. J. Exp. Med. 2004;200:805–809. doi: 10.1084/jem.20041069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang H., Zhang Q., Dong J., Liang Y., Huang Y., Liu H.J., Tong D. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013;10:26. doi: 10.1186/1743-422X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang H., Zhang Q., Huang Y., Dong J., Liang Y., Liu H.J., Tong D. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.T., Peng H., Zhu Z.L., Li G., Huang Z.T., Zhao Z.X., Koup R.A., Bailer R.T., Wu C.Y. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin. Immunol. 2006;120:171–178. doi: 10.1016/j.clim.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Lin H., Fan H. Efficacy and immunogenicity of recombinant swinepox virus expressing the A epitope of the TGEV S protein. Vaccine. 2015;33:3900–3906. doi: 10.1016/j.vaccine.2015.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Yoo D. Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling. Virus Res. 2016 doi: 10.1016/j.virusres.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Chen J., Shi H., Chen X., Shi D., Feng L., Yang B. Identification of a conserved linear B-cell epitope in the M protein of porcine epidemic diarrhea virus. Virol. J. 2012;9:225. doi: 10.1186/1743-422X-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang X., Liao X., Huang X., Cao S., Wen X., Wen Y., Wu R., Liu W. Construction of a bivalent DNA vaccine co-expressing S genes of transmissible gastroenteritis virus and porcine epidemic diarrhea virus delivered by attenuated Salmonella typhimurium. Virus Genes. 2016;52:354–364. doi: 10.1007/s11262-016-1316-z. [DOI] [PubMed] [Google Scholar]
- de Arriba M.L., Carvajal A., Pozo J., Rubio P. Mucosal and systemic isotype-specific antibody responses and protection in conventional pigs exposed to virulent or attenuated porcine epidemic diarrhoea virus. Vet. Immunol. Immunopathol. 2002;85:85–97. doi: 10.1016/s0165-2427(01)00417-2. [DOI] [PubMed] [Google Scholar]