Highlights
-
•
IPI-2I cells are susceptible to TGEV, PDCoV, and PEAV.
-
•
IPI-2I cells can be infected with PEDV, but with low efficiency.
-
•
A homogeneous cell line IPI-FX is obtained from IPI-2I cells by sub-cloning.
-
•
IPI-FX cells are highly susceptible to PEDV, TGEV, PDCoV, and PEAV.
Keywords: Swine enteric coronavirus, Porcine ileum epithelial cell (IPI-2I), Susceptibility
Abstract
Swine enteric coronavirus (CoV) is an important group of pathogens causing diarrhea in piglets. At least four kinds of swine enteric CoVs have been identified, including transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), porcine deltacoronavirus (PDCoV), and the emerging HKU2-like porcine enteric alphacoronavirus (PEAV). The small intestines, particularly the jejunum and ileum, are the most common targets of these four CoVs in vivo, and co-infections by these CoVs are frequently observed in clinically infected pigs. This study was conducted to investigate the susceptibility of the porcine ileum epithelial cell line, IPI-2I, to different swine enteric CoVs. We found that IPI-2I cells are highly susceptible to TGEV, PDCoV, and PEAV, as demonstrated by cytopathic effect and virus multiplication. However, only a small number of cells could be infected by PEDV, possibly due to the heterogeneity of IPI-2I cells. A homogeneous cell line, designated IPI-FX, obtained from IPI-2I cells by sub-cloning with limited serial dilutions, was found to be highly susceptible to PEDV. Furthermore, IPI-FX cells were also highly susceptible to TGEV, PDCoV, as well as PEAV. Thus, this sub-cloned IPI-FX cell line is an ideal cell model to study the mechanisms of infection, particularly co-infections of swine enteric CoVs.
1. Introduction
Coronaviruses (CoVs) are enveloped, single-stranded, positive sense RNA viruses that can be divided into four genera: Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV), and Deltacoronavirus (δ-CoV) (Su et al., 2016). CoVs can cause respiratory, enteric, hepatic, or neurological diseases of varying severity in a variety of animals (Gerdts and Zakhartchouk, 2017). Swine enteric CoV is one of the main causative pathogens of viral diarrhea in piglets. To date, at least four kinds of swine enteric CoVs have been identified, including transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), porcine deltacoronavirus (PDCoV), and HKU2-like porcine enteric alphacoronavirus (PEAV). TGEV, PEDV and PEAV belong to the α-CoVs, while PDCoV is a member of the genus δ-CoV (Pan et al., 2017; Pensaert and de Bouck, 1978; Woo et al., 2012). Among the four swine enteric CoVs, TGEV was the first to be identified in the United States as early as 1946 (Doyle and Hutchings, 1946). PEDV was discovered in the United Kingdom in the early 1970s and was subsequently identified in many European and Asian countries (Lee, 2015; Pensaert and de Bouck, 1978). Since late 2010, variant PEDV strains have reemerged in China and have now become the dominant global strain (Huang et al., 2013; Li et al., 2012). PDCoV was originally found in pig manure in Hong Kong in 2009, and the first outbreak was reported in 2014 in the United States (Wang et al., 2014; Woo et al., 2012). The newly identified PEAV is a bat-HUK2-like CoV in swine which was found in a diarrhea outbreak in swine herds in Guangdong, China in 2017 (Gong et al., 2017; Pan et al., 2017; Zhou et al., 2018). All four swine enteric CoVs cause severe diarrhea, vomiting, and mortality in piglets, resulting in great economic losses and posing a continuing threat to the development of the pig industry.
Regarding in vitro culture of these four swine enteric CoVs, ST (swine testicle) and PK-15 (porcine kidney) cells are widely used for TGEV isolation and propagation (Ding et al., 2017); Vero (African green monkey kidney) cells are highly susceptible to PEDV and PEAV infection (Hofmann and Wyler, 1988; Pan et al., 2017); ST and LLC-PK1 (porcine kidney) cells are permissive for PDCoV infection (Hu et al., 2015). It is well known that porcine intestinal epithelial cells are the primary target cells in vivo for swine enteric CoVs. However, none of the cells mentioned above (Vero, ST, PK-15, and LLC-PK1) are derived from the porcine intestinal tract. Biological experiments with enteric CoVs on non-intestinal epithelial cells often do not mimic real infections and are unsuitable for studying cell-virus interactions. IPEC-J2 is a line of porcine intestinal epithelial cells derived from neonatal pig jejunum (Brosnahan and Brown, 2012). Some studies have shown that IPEC-J2 cells were susceptible to PEDV infection, while others reported the opposite (Zhang et al., 2018; Zhao et al., 2014). However, a subclone of IPEC-J2 cells, IPEC-DQ, supports efficient PEDV propagation (Zhang et al., 2018). Recently, Jung et al. also tested the susceptibility of IPEC-J2 cells to PDCoV infection and found that IPEC-J2 cells supported PDCoV propagation but cytopathic effect (CPE) could only be observed after the 3rd serial passage of PDCoV in this cell type (Jung et al., 2018).
Jejunum and ileum are the most common in vivo targets of swine enteric CoVs. In addition to IPEC-J2, another porcine intestinal epithelial cell line derived from pig ileum, IPI-2I (Kaeffer et al., 1993), is a candidate cell line that may support swine enteric CoVs infection. However, whether IPI-2I cells are susceptible to swine enteric CoVs has not been characterized. In this study, we investigated the susceptibility of IPI-2I cells to four different swine enteric CoVs and established a sub-cloned homogeneous cell population (designated IPI-FX), which can be efficiently infected by all four swine enteric CoVs.
2. Materials and methods
2.1. Cells, viruses and reagents
IPI-2I, Vero, and ST cells were obtained from the China Center for Type Culture Collection (Wuhan, China) and cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, USA), 100 U/mL penicillin and 10 μg/mL streptomycin sulfates at 37 ℃ with 5% CO2 in a humidified incubator. LLC-PK1 cells were acquired from the American Type Culture Collection (ATCC number CL-101; Manassas, VA) and cultured under the conditions described above. TGEV strain WH1 (GenBank accession no. HQ462571), PEDV strain AJ1102 (GenBank accession no. JX188454.1), PDCoV strain CHN-HN-2014 (GenBank accession no. KT336560), and PEAV strain CHN-GD-2017 (GenBank accession no. MH539766) were isolated from piglets with severe diarrhea in China in 2010, 2011, 2014 and 2017, respectively (Bi et al., 2012; Ding et al., 2017; Dong et al., 2016). Mouse monoclonal antibodies (mAbs) against TGEV nucleocapsid (N) protein, PEDV nucleocapsid (N) protein, PDCoV spike (S) protein were described previously (An et al., 2014; Ding et al., 2014; Zhu et al., 2018). The mAb against PEAV S protein was produced from hybridoma cells derived from SP2/0 myeloma cells and spleen cells of BALB/c mice immunized with recombinant S1 protein of PEAV strain CHN-GD-2017.
2.2. Virus inoculation, CPE and growth curve
IPI-2I cells seeded in 24-well plates were inoculated with PDCoV, TGEV or PEAV at a multiplicity of infection (MOI) of 1 or infected with PEDV at MOI 5. At 6, 12, 18, 24 and 30 h post-infection (hpi), CPE was examined to compare with mock-infected cells. Similarly, IPI-FX cells were inoculated with PDCoV, TGEV, PEAV at MOI 1 or infected with PEDV at MOI 5. At 24 hpi, CPE was examined. To get the multi-step growth kinetics curves, IPI-2I or IPI-FX cells in 24-well plates were inoculated with PDCoV, TGEV, PEAV or PEDV (MOI = 0.1). LLC-PK1 cells were infected with PDCoV (MOI = 0.1), ST cells were infected with TGEV (MOI = 0.1) and Vero cells were infected with PEAV (MOI = 0.1). Whole cell samples were collected at 6, 12, 18, 24 or 30hpi followed by freezing and thawing three times, and centrifugation at 3000 r/min for 10 min to collect the supernatant. Viral titers were determined by 50% tissue culture infectious dose (TCID50) assay.
2.3. Indirect immunofluorescence assay (IFA)
IPI-2I cells in 24-well plates were mock-infected or infected with PDCoV, TGEV, PEAV at MOI 1 or infected with PEDV at MOI 5. At different time-points after inoculation, the cells were washed thrice with phosphate-buffered saline (PBS), then fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.2% Triton X-100 for 10 min at room temperature. After three washes with PBS, the cells were blocked with PBS containing 5% bovine serum albumin for 1 h and then incubated with mAbs against PDCoV S protein, PEAV S protein, TGEV N protein or PEDV N protein, respectively, for 1 h. The cells were rinsed with PBS and then treated with Alexa Fluor 488-conjugated anti-mouse secondary antibody (Santa Cruz Biotechnology, USA) for 1 h, followed by 0.01% 4′,6-diamidino-2-phenylindole (DAPI) staining for 15 min to detect nuclei. Fluorescent images were visualized using an inverted fluorescence microscope (Olympus IX73).
2.4. TCID50 assay
Cells (LLC-PK1, Vero, ST, IPI-2I or IPI-FX) were seeded into 96-well plates and cultured for 80% confluence. The cell monolayers were washed twice with maintenance medium contain 7.5 μg/ml trypsin or 2% fetal bovine serum. One hundred microliters of 10-fold dilutions of virus samples were inoculated in eight replicates per dilution. Viral CPE was monitored for 3–5 days, and virus titers were calculated by using the Reed-Muench method (Reed and Muench, 1938) and expressed as TCID50 per milliliter.
2.5. RNA extraction and reverse transcription polymerase chain reaction (RT-PCR)
Total cellular RNA was extracted with TRIzol reagent (Promega, USA) from PEDV-infected IPI-2I cells. After the RNA was reverse-transcribed to cDNA, PCR assays were performed to detect PEDV RNA with the following primers: PEDV-F: 5′-GCAACAACAGGTCCAGAT-3′; PEDV-R: 5′-CTCACGAACAGCCACATT-3′.
2.6. Sub-clonal screening of IPI-2I cells
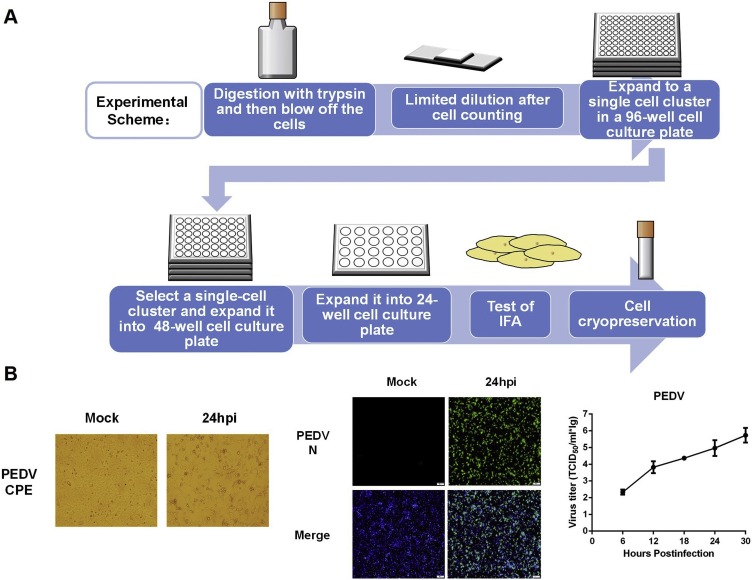
To obtain a homogeneous cell pool, IPI-2I cells were sub-cloned by finite continuous dilution. First, the cells were digested into a single cell suspension by trypsin, then counted and diluted into a 96-well cell culture plate. In principle, only one cell was present in each well. The medium was changed every 3–5 days until the cells grew into single clusters. The clusters with the best growth status in the 96-well cell culture plate were digested and amplified in a larger cell culture plate. When the cell number was sufficient, IFA was used to detect the infection rate.
2.7. Statistical analysis
Data are shown as the mean ± standard deviation of three independent experiments. The results were analyzed for significance by Student’s t test using GraphPad Prism 6 software. Differences between groups were considered statistically significant when P < 0.05.
3. Results
3.1. IPI-2I cells are susceptible to TGEV, PDCoV, and PEAV
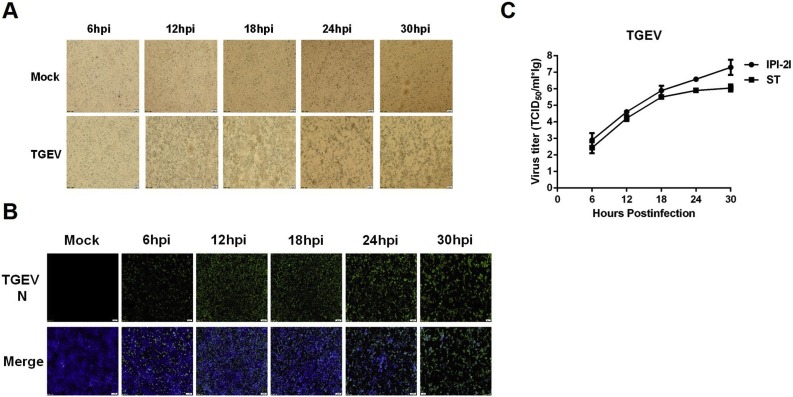
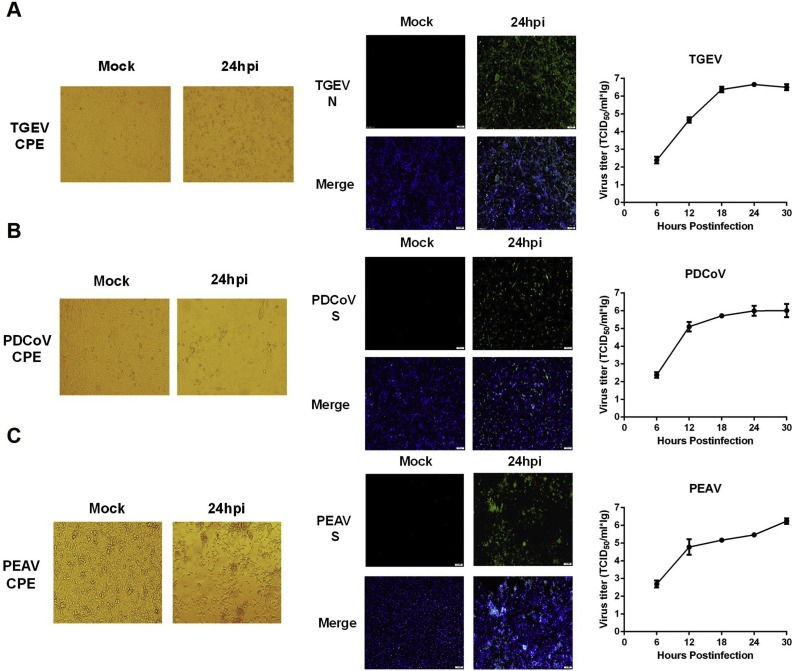
To test the susceptibility of IPI-2I cells to different swine enteric CoVs, we first investigated whether IPI-2I cells support TGEV infection. To this end, IPI-2I cells were infected with TGEV strain WH-1 (MOI = 1) and microscopically monitored for CPE at 6, 12, 18, 24 and 30 hpi. As shown in Fig. 1 A, typical CPE became visible at 12 hpi and was readily apparent as the infection progressed. IFA was performed using mAb against TGEV N protein to further confirm TGEV infection at different time points after inoculation (Fig. 1B). We also compared the growth curves of TGEV in IPI-2I and ST cells (MOI = 0.1). The results showed that the viral titers gradually increased after infection in IPI-2I (Fig. 1C). Furthermore, TGEV achieved higher viral titers in IPI-2I than in ST cells (Fig. 1C), suggesting that IPI-2I cells are highly susceptible to TGEV infection.
Fig. 1.
Cytopathic effect (CPE), immunofluorescence staining, and growth curve of TGEV in IPI-2I cells. (A) IPI-2I cells were mock-infected or infected with TGEV at a multiplicity of infection (MOI) of 1. CPE was observed at 6, 12, 18, 24 and 30 h post-infection (hpi). (B) The infected IPI-2I cells were fixed at different time points post-infection and indirect immunofluorescence assays were performed with a monoclonal antibody against TGEV N protein. (C) IPI-2I cells or ST cells were infected with TGEV (MOI = 0.1). Cells were collected at 6, 12, 18, 24 and 30 hpi to determine the viral titers by TCID50 assay.
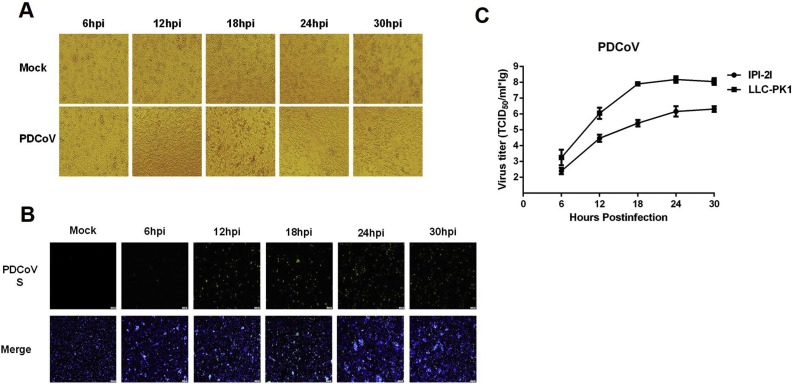
Accumulating evidence has suggested that supplemental trypsin in the cell culture medium contributes to a significant increase in viral titers of PEDV, PDCoV, and PEAV, compared with non-trypsin medium (Li et al., 2015). Before evaluating the susceptibility of IPI-2I cells to infection with these enteric CoVs, we examined the trypsin tolerance of IPI-2I cells. Monolayers of IPI-2I cells were treated with different concentrations (0, 1.25, 2.5, 5, 7.5 and 10 μg/ml) of trypsin and the results showed that the optimal concentration of trypsin added in IPI-2I cell culture medium was 2.5 μg/ml. We then investigated the susceptibility of IPI-2I cells to PDCoV infection under this trypsin concentration. As shown in Fig. 2 , typical CPE, characterized by shrunken, rounded and clustered cells, was observed at 12–30 hpi in PDCoV-infected IPI-2I cells (Fig. 2A). Infection was also confirmed by IFA staining with mAb against PDCoV S protein (Fig. 2B). By comparing the growth curves of PDCoV in IPI-2I and LLC-PK1 cells (MOI = 0.1), the viral titers in IPI-2I cells were lower than those in LLC-PK1 cells (Fig. 2C). It should be noted that the trypsin concentration used in LLC-PK1 cells was 7.5 μg/ml.
Fig. 2.
Infection and proliferation characteristics of PDCoV in IPI-2I cells. (A) IPI-2I cells were mock-infected or infected with PDCoV at a MOI of 1. Cytopathic effect was observed at 6, 12, 18, 24 and 30 hpi. (B) The infected IPI-2I cells were fixed at different time points post-infection and indirect immunofluorescence assay was performed with a monoclonal antibody against PDCoV S protein. (C) IPI-2I cells or LLC-PK1 cells were infected with PDCoV (MOI = 0.1). The trypsin concentrations used in IPI-2I cells and LLC-PK1 cells were 2.5 μg/ml and 7.5 μg/ml, respectively. Cells were collected at 6, 12, 18, 24 and 30 hpi to determine the viral titers by TCID50 assay.
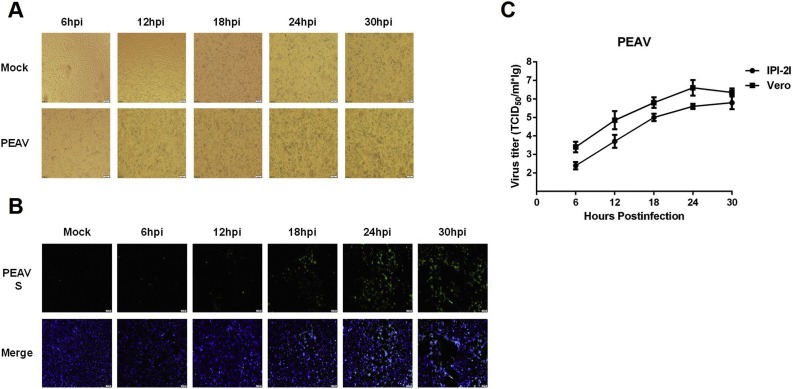
We further investigated the susceptibility of IPI-2I cells to PEAV. Typical CPE (Fig. 3 A) and PEAV S protein-specific immunofluorescence (Fig. 3B) could be observed. Similar to PDCoV infection, the growth curves showed that viral titers of PEAV (MOI = 0.1) in IPI-2I cells were lower than those in Vero cells (the trypsin concentration was 7.5 μg/ml) (Fig. 3C).
Fig. 3.
Infection and proliferation characteristics of PEAV in IPI-2I cells. (A) IPI-2I cells were mock-infected or infected with PEAV at a MOI of 1. Cytopathic effect was observed at 6, 12, 18, 24 and 30 hpi. (B) The infected IPI-2I cells were fixed at different time points post-infection and indirect immunofluorescence assay was performed with a monoclonal antibody against PEAV S protein. (C) IPI-2I cells or Vero cells were infected with PEAV (MOI = 0.1). The trypsin concentrations used in IPI-2I cells and Vero cells were 2.5 μg/ml and 7.5 μg/ml, respectively. Cell were collected at 6, 12, 18, 24 and 30 hpi to determine the viral titers by TCID50 assay.
3.2. IPI-2I cells can be infected with PEDV, but with low efficiency
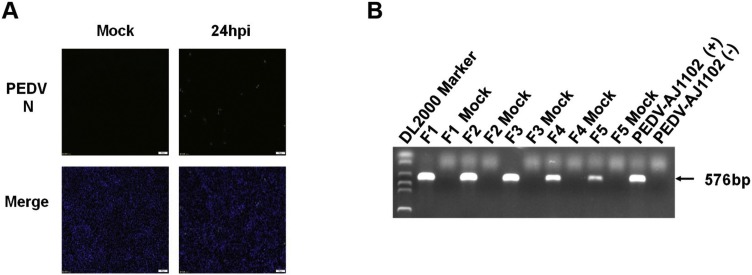
To investigate whether IPI-2I cells are susceptible to PEDV, IPI-2I cells were infected with PEDV at an MOI of 5 to examine CPE. CPE was undetectable by 2–4 days after PEDV infection, which was different from the three CoVs tested above. IFA with mAb against PEDV N protein showed that only a small number of PEDV-positive cells could be detected (Fig. 4 A). We also analyzed the genomic RNA of PEDV passaged in IPI-2I using PEDV N gene-specific primers for RT-PCR, and the results showed that the amount of viral RNAs gradually decreased with continuous passages (Fig. 4B). Taken together, we concluded that, although IPI-2I cells can be infected with PEDV, the infection efficiency is very low.
Fig. 4.
Susceptibility of IPI-2I cells to PEDV infection. (A) IPI-2I cells were mock-infected or infected with PEDV at a MOI of 5. At 24 hpi, indirect immunofluorescence assay was performed with a monoclonal antibody against PEDV N protein. (B) PEDV was passaged for five generations in IPI-2I cells. Viral genomic RNA was detected for each passage by RT-PCR.
3.3. IPI-2I-dervied homogeneous IPI-FX cells are highly susceptible to PEDV infection
In a previous study, Zhang et al. tested the susceptibility of IPEC-J2 cells to PEDV infection and also found that only a few cells were infected by PEDV (Zhang et al., 2018), similar to our observations in PEDV-infected IPI-2I cells. They speculated that it was due to the non-homogeneity of IPEC-J2 cells. Thus, they sub-cloned IPEC-J2 cells by limited serial dilutions and obtained a homogeneous cell population (IPEC-DQ) that could support efficient and productive infection of PEDV (Zhang et al., 2018). Thus, we hypothesized that IPI-2I cells were likely to have heterogeneous characteristics similar to those of IPEC-J2 cells. We subcloned IPI-2I cells by limited serial dilutions as illustrated in Fig. 5 A. A total of 24 sub-cloned homogeneous cell populations exhibited higher susceptibility to PEDV infection, as demonstrated by PEDV N protein-specific IFA (data not shown). A homogeneous cell population possessing higher infection efficiency, designated IPI-FX, was chosen for further experiments. As shown in Fig. 5B, typical CPE could be observed in IPI-FX cells infected with PEDV, and over 80% of cells could be infected, as demonstrated by IFA with mAb against PEDV N protein (Fig. 5B). The viral titers reached to 106.0TCID50/ml at 30 hpi (Fig. 5C).
Fig. 5.
Sub-cloning screening of IPI-2I cells and susceptibility of IPI-FX cells to PEDV infection. (A) The procedure for sub-cloning screening of IPI-2I cells. (B) IPI-FX cells were mock-infected or infected with PEDV at a MOI of 5. At 24 hpi, cytopathic effect was observed and infection was confirmed by indirect immunofluorescence assay with a monoclonal antibody against PEDV N protein. The titers of PEDV in IPI-FX cells (MOI = 0.1) at different time points (6, 12, 18, 24 and 30 hpi) were determined by TCID50 assay.
3.4. IPI-FX cells are also highly susceptible to TGEV, PDCoV, and PEAV
As IPI-FX cells could be efficiently infected by PEDV, we investigated whether the remaining three swine enteric CoVs (TGEV, PDCoV, PEAV) could also infect IPI-FX cells. To this end, IPI-FX cells were infected with TGEV, PDCoV or PEAV, respectively. As shown in Fig. 6 , typical CPE and specific immunofluorescence could be observed in IPI-FX cells at 24 h post-infection with TGEV (Fig. 6A), PDCoV (Fig. 6B), and PEAV (Fig. 6C). The results of the growth curve indicated that all three CoVs could efficiently proliferate in IPI-FX cells (Fig. 6A–C).
Fig. 6.
IPI-FX cells can be effectively infected by TGEV, PDCoV, and PEAV. IPI-FX cells were mock-infected or infected with TGEV (A), PDCoV (B), PEAV (C) at a MOI of 1 for 24 h. Cytopathic effect was observed and indirect immunofluorescence assays were performed to confirm infection with specific monoclonal antibodies. The growth kinetics curves of TGEV (A), PDCoV (B), PEAV (C) in IPI-FX cells were determined by collecting virus-infected cells (MOI = 0.1) at different time points (6, 12, 18, 24 and 30 hpi), followed by TCID50 assay.
4. Discussion
Enteric CoVs are the main pathogens causing viral diarrhea in pigs. The primary target cells for swine enteric CoVs in vivo are villous epithelial cells of the intestinal tract. Thus, cell lines derived from primary porcine intestinal epithelial cells are the ideal cell models to study interactions between these swine enteric CoVs and the host in vitro. In this study, we characterized the susceptibility of porcine IPI-2I intestinal epithelial cells to infection with four different swine enteric CoVs. We found that IPI-2I cells are highly susceptible to TGEV, PDCoV and PEAV. Although the non-homogeneous IPI-2I cells were not susceptible to PEDV, higher susceptibility could be achieved in IPI-FX, a homogeneous cell population obtained by sub-clone screening of the parental cell line.
Although IPI-2I cells are highly susceptible to PDCoV and PEAV, the viral titers in the infected IPI-2I cells were lower than those in the commonly used susceptible cells, LLC-PK1 and Vero cells, respectively. We speculated that the lower trypsin tolerance of IPI-2I may account for the lower viral titers of PDCoV and PEAV. The maximum concentration of trypsin tolerated by IPI-2I cells was 2.5 μg/ml, while LLC-PK1 and Vero cells are highly resistant to trypsin, and 5–10 μg/ml of trypsin concentrations are usually used for PDCoV and PEAV infection (Dong et al., 2016; Hu et al., 2015; Pan et al., 2017). Previous studies have suggested that trypsin is crucial for cell membrane fusion and entry of swine enteric CoVs, and can mediate the membrane fusion activation of virions by lysis of glycoprotein spikes (Dong et al., 2016; Wicht et al., 2014). The higher viral titers of TGEV in infected IPI-2I cells compared with ST cells appears to support our speculations. There is no need for trypsin addition for TGEV proliferation, and the higher viral titers of TGEV in IPI-2I cells were achieved in the absence of exogenous trypsin. We also determined the viral titers of PDCoV and PEAV in LLC-PK1 and Vero cells, respectively, under the trypsin concentration of 2.5 μg/ml, and found that similar viral titers could be achieved compared to those in IPI-2I cells (data not shown). The trypsin tolerance concentrations of IPI-FX cells were also measured and found that the same trypsin concentrations were tolerated by IPI-FX and IPI-2I cells. Work to obtain a homogeneous cell population with higher trypsin tolerance from IPI-FX cells is currently ongoing in our laboratory.
In this study, we found that PEDV can infect IPI-2I cells; however, only a few cells appeared to be PEDV-positive, which may be due to the heterogeneity of IPI-2I cells, because the homogeneous IPI-FX cells obtained by sub-cloning from IPI-2I cells were highly susceptible to PEDV infection. Likewise, the non-homogeneous IPEC-J2 cells exhibit lower susceptible to PEDV, while the homogeneous IPEC-DQ which was sub-cloned from IPEC-J2 cells by limited serial dilutions, can be readily infected by PEDV (Zhang et al., 2018). Recently, Jung et al. tested the susceptibility of IPEC-J2 to infection with PDCoV and found that viral RNA could be detected in the cell culture supernatant after two continuous passages in the IPEC-J2 cells supplemented with 10 μg/ml of trypsin; however, no CPE was observed (Jung et al., 2018). By modifying the procedure of inoculation, that is, omitting the washing of cells after virus adsorption, the 4th passaged PDCoV in IPEC-J2 cells showed CPE characterized by enlarged, rounded and densely granular cells (Jung et al., 2018). In our present study, evident CPE could be observed in IPI-2I cells infected with the 1st passage of PDCoV, even when supplemented with only 2.5 μg/ml of trypsin in the cell culture medium. It appears that IPI-2I cells are more susceptible than IPEC-J2 cells to PDCoV infection. Based on the results that homogeneous IPEC-DQ from IPEC-J2 as well as IPI-FX from IPI-2I cells are highly susceptible to PEDV, it is possible that homogeneous IPEC cell lines susceptible to PDCoV, and even to all four swine enteric CoVs, can be obtained by the sub-cloning method. This work has been conducted in our laboratory.
Clinically, co-infections in pigs by two or even three kinds of swine enteric CoVs often occur. For example, PEDV and TGEV co-infections are frequently detected in piglets with severe diarrhea, and recently a chimeric swine enteric CoV containing the S-gene and 3a sequences from PEDV within a backbone of TGEV has been reported in Italy (Belsham et al., 2016). Similar chimeric viruses were also reported in Germany and Slovakia (Akimkin et al., 2016; Mandelik et al., 2018). It was proven experimentally that co-infections with PEDV and TGEV in cultured cells enhanced the damage to tight junctions and the remodeling of microfilaments compared with single infections with the same viruses (Belsham et al., 2016). Co-infections with PEDV and PDCoV in pigs have also be reported in several studies previously (Dong et al., 2015; Jang et al., 2017; Song et al., 2015). Before the outbreak of PEAV in farms in China in 2017, PEDV broke out in the same farms, indicating possible concurrent infections with PEDV and PEAV (Gong et al., 2017; Zhou et al., 2018). Together, co-infections with different swine enteric CoVs are an enormous threat to pig production. Thus, to dissect the mechanisms involved in co-infection with swine enteric CoVs in vivo or in vitro is receiving increasing attention. The IPI-FX cells established here are highly susceptible to all four swine enteric CoVs, and are derived from the ileum of porcine intestinal tracts, the common natural target of these enteric CoVs. Therefore, the IPI-FX cell line is an ideal cell model to study co-infections of swine enteric CoVs.
5. Conclusion
Our present study revealed that IPI-2I cells, a porcine ileum intestinal epithelial cell line, can be efficiently infected by TGEV, PDCoV and PEAV, demonstrating typical CPE. Although PEDV can infect IPI-2I cells, the infection efficiency is very low, which may be due to the heterogeneity of IPI-2I cells. A homogeneous cell line, IPI-FX, was obtained from IPI-2I cells by sub-cloning with limited serial dilutions. The established IPI-FX cells can be readily infected by PEDV, as well as PDCoV, TGEV and PEAV. Therefore, IPI-FX cells can be used as an excellent cell model for co-infection studies of different swine enteric CoVs.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31730095, 31672569), the National Key R&D Plan of China (2018YFD0500100), the Key Technology R&D Programme of China (2015BAD12B02), and the Major S&T Project of Hubei Province (2017ABA138).
References
- Akimkin V., Beer M., Blome S., Hanke D., Hoper D., Jenckel M., Pohlmann A. New chimeric porcine coronavirus in swine feces, Germany, 2012. Emerg. Infect. Dis. 2016;22:1314–1315. doi: 10.3201/eid2207.160179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An K., Fang L., Luo R., Wang D., Xie L., Yang J., Chen H., Xiao S. Quantitative proteomic analysis reveals that transmissible gastroenteritis virus activates the JAK-STAT1 signaling pathway. J. Proteome Res. 2014;13:5376–5390. doi: 10.1021/pr500173p. [DOI] [PubMed] [Google Scholar]
- Belsham G.J., Rasmussen T.B., Normann P., Vaclavek P., Strandbygaard B., Botner A. Characterization of a novel chimeric swine enteric coronavirus from diseased pigs in Central Eastern Europe in 2016. Transbound. Emerg. Dis. 2016;63:595–601. doi: 10.1111/tbed.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J., Zeng S., Xiao S., Chen H., Fang L. Complete genome sequence of porcine epidemic diarrhea virus strain AJ1102 isolated from a suckling piglet with acute diarrhea in China. J. Virol. 2012;86:10910–10911. doi: 10.1128/JVI.01919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnahan A.J., Brown D.R. Porcine IPEC-J2 intestinal epithelial cells in microbiological investigations. Vet. Microbiol. 2012;156:229–237. doi: 10.1016/j.vetmic.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., An K., Xie L., Wu W., Zhang R., Wang D., Fang Y., Chen H., Xiao S., Fang L. Transmissible gastroenteritis virus infection induces NF-kappaB activation through RLR-mediated signaling. Virology. 2017;507:170–178. doi: 10.1016/j.virol.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine deltacoronavirus in mainland China. Emerg. Infect. Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Yang H., Liu H., Du T., Fang P., Wang D., Chen H., Xiao S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet. Microbiol. 2016;196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle L.P., Hutchings L.M. A transmissible gastroenteritis in pigs. J. Am. Vet. Med. Assoc. 1946;108:257–259. [PubMed] [Google Scholar]
- Gerdts V., Zakhartchouk A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet. Microbiol. 2017;206:45–51. doi: 10.1016/j.vetmic.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L., Li J., Zhou Q., Xu Z., Chen L., Zhang Y., Xue C., Wen Z., Cao Y. A new Bat-HKU2-like coronavirus in swine, China, 2017. Emerg. Infect. Dis. 2017;23:1607–1609. doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26:2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. 2013;4:e00737–00713. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G., Lee K.K., Kim S.H., Lee C. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014-2016. Transbound. Emerg. Dis. 2017;64:1364–1370. doi: 10.1111/tbed.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Miyazaki A., Hu H., Saif L.J. Susceptibility of porcine IPEC-J2 intestinal epithelial cells to infection with porcine deltacoronavirus (PDCoV) and serum cytokine responses of gnotobiotic pigs to acute infection with IPEC-J2 cell culture-passaged PDCoV. Vet. Microbiol. 2018;221:49–58. doi: 10.1016/j.vetmic.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeffer B., Bottreau E., Velge P., Pardon P. Epithelioid and fibroblastic cell lines derived from the ileum of an adult histocompatible miniature boar (d/d haplotype) and immortalized by SV40 plasmid. Eur. J. Cell Biol. 1993;62:152–162. [PubMed] [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis. 2012;18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wicht O., van Kuppeveld F.J., He Q., Rottier P.J., Bosch B.J. A single point mutation creating a furin cleavage site in the spike protein renders porcine epidemic diarrhea coronavirus trypsin independent for cell entry and fusion. J. Virol. 2015;89:8077–8081. doi: 10.1128/JVI.00356-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelik R., Sarvas M., Jackova A., Salamunova S., Novotny J., Vilcek S. First outbreak with chimeric swine enteric coronavirus (SeCoV) on pig farms in Slovakia - lessons to learn. Acta Vet. Hung. 2018;66:488–492. doi: 10.1556/004.2018.043. [DOI] [PubMed] [Google Scholar]
- Pan Y., Tian X., Qin P., Wang B., Zhao P., Yang Y.L., Wang L., Wang D., Song Y., Zhang X., Huang Y.W. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating of fifty percent endpoints. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- Song D., Zhou X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D., Wu Q., He H., Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound. Emerg. Dis. 2015;62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014;20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicht O., Li W., Willems L., Meuleman T.J., Wubbolts R.W., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 2014;88:7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ke H., Blikslager A., Fujita T., Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 2018;92:e01677–17. doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Gao J., Zhu L., Yang Q. Transmissible gastroenteritis virus and porcine epidemic diarrhoea virus infection induces dramatic changes in the tight junctions and microfilaments of polarized IPEC-J2 cells. Virus Res. 2014;192:34–45. doi: 10.1016/j.virusres.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Liu S., Wang X., Luo Z., Shi Y., Wang D., Peng G., Chen H., Fang L., Xiao S. Contribution of porcine aminopeptidase N to porcine deltacoronavirus infection. Emerg. Microbes Infect. 2018;7:65. doi: 10.1038/s41426-018-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]