Highlights
-
•
Inter-laboratory comparison of anti-PEDV antibody detection by different ELISAs.
-
•
Multiple panels of porcine sera have been tested for antibodies against PEDV.
-
•
Assay sensitivity and specificity has been estimated using Bayesian analysis.
-
•
The various assays detect infection of pigs at herd level by different PEDVs.
-
•
Not all the assays seem suitable for demonstrating freedom from disease.
Keywords: ELISA, Porcine coronavirus, PEDV, Ring trial, Serology
Abstract
Porcine epidemic diarrhea virus (PEDV) has caused extensive economic losses to pig producers in many countries. It was recently introduced, for the first time, into North America and outbreaks have occurred again in multiple countries within Europe as well. To assess the properties of various diagnostic assays for the detection of PEDV infection, multiple panels of porcine sera have been shared and tested for the presence of antibodies against PEDV in an inter-laboratory ring trial. Different laboratories have used a variety of “in house” ELISAs and also one commercial assay. The sensitivity and specificity of each assay has been estimated using a Bayesian analysis applied to the ring trial results obtained with the different assays in the absence of a gold standard. Although different characteristics were found, it can be concluded that each of the assays used can detect infection of pigs at a herd level by either the early European strains of PEDV or the recently circulating strains (INDEL and non-INDEL). However, not all the assays seem suitable for demonstrating freedom from disease in a country. The results from individual animals, especially when the infection has occurred within an experimental situation, show more variation.
1. Introduction
Porcine epidemic diarrhoea virus (PEDV) is a member of the Alphacoronavirus genus within the family Coronaviridae. Infection of swine by this virus causes disease characterized by diarrhoea and vomiting which can lead to severe dehydration and results in high (90–100%) mortality in newborn piglets (Stevenson et al., 2013). Older animals (≥14 days) normally recover from the infection and seroconvert against the virus. The disease was initially identified within the United Kingdom (UK) in 1971 and the detection of the aetiological agent, PEDV, was first achieved in Belgium (Pensaert and de Bouck, 1978); afterwards the virus spread within Europe and also to Asia (reviewed in Jung and Saif, 2015, Lee, 2015). In 2013, outbreaks of the disease occurred for the first time in the USA (Huang et al., 2013, Chen et al., 2014) and rapidly spread within both North and South America. About 7 million piglets died in a single year as a result of the outbreaks in the USA alone (Jung and Saif, 2015, Lee, 2015).
The genome of PEDV, like other coronaviruses, is a single-stranded positive sense RNA of about 28 kb. The virus produces a number of sub-genomic mRNAs which encode the various structural and non-structural proteins within infected cells. The virus particles include the spike (S) protein, the envelope (E) protein, the membrane (M) protein and the nucleocapsid (N) protein (Brian and Baric, 2005). The spike protein is exposed on the virus surface and gives the virus particles their characteristic morphology. A number of different variants of PEDV are known (see Huang et al., 2013, Wang et al., 2014, Lee, 2015). The early European viruses are represented by the CV777 strain (these are sometimes classified, on the basis of the S gene sequence, as being within genogroup 1a) while two different variants of PEDV (classified, on the same basis, within genogroups 1b and 2b) have been identified within the USA. The US PEDVs are also referred to as “INDEL” (e.g. OH 851, from the genogroup 1b) and “non-INDEL” (e.g. MN, 1A1 and 1A2 strains from 2013, in genogroup 2b); these differ by the presence of certain deletions and insertions within the S gene sequence (Huang et al., 2013, Wang et al., 2014, Lee, 2015).
Within infected pigs, antibodies are generated against the PEDV proteins and these can be detected by a variety of methods including ELISA, immunoblotting and immunostaining of infected cells.
The recent reappearance of PEDV (closely related to the INDEL OH 851 strain) infections within Europe, including in Germany, France, Italy, Portugal, The Netherlands and Slovenia (Hanke et al., 2015, Grasland et al., 2015, Boniotti et al., 2016, Mesquita et al., 2015, Toplak et al., 2016, EFSA, 2014), has led to the need for an assessment of existing diagnostic assays for the detection of PEDV infections. The PEDV can be identified by RT-PCR (e.g. Kim et al., 2007, Chen et al., 2014) in faecal or intestinal samples from acutely infected animals and a low level of viremia has also been observed in serum from acutely infected pigs (Jung et al., 2014, Jung et al., 2015, Lohse et al., 2016). However, the virus is only present in infected animals for a limited period (typically less than 1 month, see Lee, 2015) while the serological response can be expected to be much longer lasting (Crawford et al., 2015).
Extensive serological screening of swine (2500 samples/yr) within Denmark during the period 2000 to 2006 did not detect any sign of PEDV infection. More recently, following the disease outbreaks in the USA, additional Danish sera (2400 samples in 2014 and 3960 samples in 2015) were also tested and, again, all gave negative results. It is important to ensure that national diagnostic laboratories are able to detect PEDV infection efficiently when outbreaks of disease occur. For PEDV-free countries, like Denmark, Sweden and UK, or when planning to export animals from PEDV negative herds it is also important to be able to declare freedom from disease. Therefore, assays with both high sensitivity and high specificity are needed. Different laboratories use a variety of “in-house” assays; in addition, commercial tests for the detection of antibodies to PEDV are available. However, the properties of these different tests have not been analysed, in parallel, previously. An assessment of a range of tests, performed in different reference laboratories from Denmark (DK), Italy (IT), France (FR), The Netherlands (NL), Sweden (SE) and the UK using shared panels of porcine sera, collected from animals in the field and from experimental infection studies, has now been undertaken and the results of these analyses are presented.
2. Material and methods
2.1. Description of porcine serum panels
Panel 1 included 54 sera collected in different countries including known positive sera (diluted or neat) from pigs experimentally infected with the Br1/87 or CV777 early European strains of PEDV, negative control sera plus field sera from farms with PED clinical disease (in the US and Canada,) and also field sera from farms without clinical disease (in DK, FR and SE).
Panel 2 included 8 sera collected in DK from experimentally infected pigs at either 14 or 28 days post inoculation (dpi) with either the early European (Br1/87 strain) or a recent “non-INDEL” US strain of PEDV (described in detail by Lohse et al., 2016).
Panel 3 included 20 sera from finisher pigs from a single herd in Italy that had experienced clinical signs of PED a few weeks prior to sampling; they were collected in 2015. The presence of an INDEL strain of PEDV (closely related to OH 851) on this farm was confirmed by RT-qPCR and sequencing (data not shown).
Panel 4 included two sets of 40 sera collected from farms in Italy, during 2015. One set of 40 sera was collected from 6 different farms that had each experienced clinical signs of PED and from which PEDV (very closely related to the INDEL OH 851 strain) had been identified. The second set of 40 samples was collected from 5 other Italian farms, localized in PEDV-free areas, that had no history of enteric signs, and which had tested negative by a PEDV specific RT-PCR and using a PEDV-Ab ELISA (see below) (note: due to limitations in availability of sera, this panel was only tested in two different laboratories using three separate assays).
2.2. “In house” blocking ELISA (DK); ELISA 1
The presence of anti-PEDV antibodies in sera was determined (as in Lohse et al., 2016) using an “in-house” blocking ELISA (analogous to that used for PRRSV (Sørensen et al., 1997)) using antigen prepared from PEDV (Br1/87, closely related to CV777)-infected Vero cells. Briefly, Vero cells were infected with the Br1/87 strain of PEDV and after 24–48 h, when CPE was apparent, the cells (and medium) were frozen. After thawing, cell debris was removed by centrifugation at 5500 × g for 10 min at 5 °C. The virus antigen was harvested from the supernatant by further centrifugation (30000 × g for 4 h) and resuspended in PBS (1/100th of initial volume). The antigen was coated (typically at 1:1000 dilution but titrated for each batch) onto 96-well ELISA plates, washed and then stored frozen until use. Sera (diluted 1:10) were added to the wells and incubated overnight at 20 °C, prior to further incubation for 1 h with a biotin-conjugated pig anti-PEDV polyclonal antibody (diluted 1:100 in 10% normal pig serum) prepared essentially as described previously (Sørensen et al., 1997). Following washing, the bound biotinylated-antibody was detected using avidin-conjugated horseradish peroxidase (eBioscience, diluted as recommended by manufacturer) plus 3,3′,5,5′- tetramethylbenzidine substrate and the OD was measured at 450/630 nm. The cut-off value for a positive reaction is set at 40% blocking, values below 35% are considered negative while intermediate values are considered inconclusive.
2.3. “In-house” blocking ELISA (NL and UK); ELISA 2
Sera were tested using the ELISA essentially as described by van Nieuwstadt and Zetstra (1991). For this assay, ELISA plates coated with cell culture grown virus antigen (CV777) were incubated with serum and then unblocked virus is detected using two different monoclonal antibodies. Samples were tested using two-fold dilutions and the presented results were obtained using 1:2 or 1:4. Blocking values >50% are positive, values <40% are negative and values of 40–50% are considered inconclusive.
2.4. “In-house” blocking ELISA (IT); ELISA 3
Sera were tested using an in-house blocking ELISA based on a double antibody sandwich that has been described previously (Sozzi et al., 2010). In brief, the ELISA microplates were coated with the 1F12 capture monoclonal antibody (MAb). Serum samples diluted 1:2 or 1:4 were mixed with equal volumes of whole PEDV (CV777), inactivated with ß-propiolactone, and pre-incubated in an auxiliary microplate for 1 h at 37 °C. Then, 50 μl of the pre-incubated mixtures were transferred into the 1F12 MAb-coated plate and the conjugated horseradish peroxidase MAb 4C3 was added. Following a further 1 h incubation at 37 °C the plate was washed. The colorimetric reaction was performed and optical densities (OD) were measured at 492 nm. Results were calculated by determining the absorbance value reduction, expressed as percentage inhibition (PI) having the control wells as reference. The antibody-blocking reaction was considered positive if the PI was ≥60%.
2.5. Biovet PEDV ELISA (as used in DK, FR, SE & IT); ELISA 4
Serum samples were tested in this indirect ELISA as described by the manufacturer (Biovet, Quebec, Canada). This assay detects antibodies that bind to the PEDV nucleoprotein. Results are presented as a ratio of: the OD for the sample (S)/OD for the positive control (P). S/P ratios >0.4 are considered positive.
2.5.1. Statistical methods
A Bayesian approach was used to provide an estimation of the sensitivity (se) and specificity (sp) for three serological tests under consideration at one time. The method, described by Branscum et al. (2005), was applied to estimate the characteristics of three conditionally dependent tests in a single population and without a gold standard. The model parameters were therefore the three sensitivities, three specificities, covariances between test results in seropositive and seronegative sera and one true prevalence. So our data consisted of the cross-classified test results for the n tested sera from the population: e.g. being the number of sera that were found positive with the three tests, the number of sera that were found positive with test 1 and 2 and negative with test 3. The 8 combinations of the cross-classified results between the three tests were defined as such.
Beta distributions Be(a,b) were used as priors for the parameters of interest (sensitivities, specificities, proportion of seropositive sera). We used non informative priors for sensitivity (uniform distributions) and mildly informative priors for specificities, considering that the expected specificities of the serological test would be >0.4 with 95% certainty and with a mode equal to 0.9 according to relative specificities that we estimated between the different tests. For true prevalence, we estimated that the prior distribution should represent a prevalence >0.4 with 95% certainty and mode equal to 0.6 according to the previous information we had on sera (experimental or field sera from a farm without any report of clinical disease). The same methodology, as described by Branscum et al. (2005), was used to define priors for the covariances (uniform prior distribution over the ranges of covariances).
The models were run using the WinBUGS freeware program (Spiegelhalter et al., 1996) commanded by the R package R2Winbugs. Parameter estimates were based on analytical summaries of 10,000 iterations of the Gibbs sampler with a burn-in phase of 1000 iterations. Three parallel chains were run with different starting values randomly chosen from uniform distributions (0,1). Confirmation of the lack of convergence was required before the posterior distributions produced by the Gibbs sampler could be used. The R-CODA package (Best et al., 1995) by R software (R Development Core Team, 2008) was used to assess convergence of the resulting Monte Carlo Markov Chain (MCMC) objects. Successive trace plots were examined to detect slow mixing, both the Heidelberger test (Heidelberger and Welch, 1983) and the Raftery and Lewis tests (Raftery and Lewis, 1992) for the convergence of single chains were applied. The Gelman-Rubin (Brooks and Gelman, 1998) diagnosis was carried out to assess convergence of the 3 parallel chains and autocorrelations were also checked.
All the results obtained, for the serological assays being compared, except for those using pre-diluted sera (in Table 1A, Table 1B ) were included in these analyses.
Table 1A.
Detection of anti-PEDV antibodies in sera from the field and from experimental studies.
 |
Negative results are highlighted in bold.
1The blocking (%) values are indicated together with the conclusion (Con.). A positive (Pos) reaction is defined as >40% block, while a negative (Neg) is defined as <35% block and results of 35–40% blocking are defined as inconclusive (Inc).
2Samples were tested using 2-fold dilutions, results are presented from 1:2 or 1:4 dilutions and the conclusion is shown. Blocking values >50% are Pos, values <40% are Neg and values of 40–50% are considered Inc.
3Assayed at dilutions of 1:4 and 1:8, the respective blocking (%) values are shown together with the conclusion (Con.) A positive (Pos) reaction is defined as ≥60% block, while a negative reaction (Neg) is defined as <60% block.
4In this test the ratio of the Sample(S)/Positive (P) control values were calculated. S/P values >0.4 are Pos while those below are Neg.
Table 1B.
Detection of anti-PEDV antibodies in sera from the field from herds without clinical signs of disease.
 |
Negative results are highlighted in bold.
1The blocking (%) values are indicated together with the conclusion (Con.). A positive (Pos) reaction is defined as >40% block, while a negative (Neg) is defined as <35% block and results of 35–40% blocking are defined as inconclusive (Inc).
2Samples were tested using 2-fold dilutions, results are presented from 1:2 or 1:4 dilutions and the conclusion is shown. Blocking values >50% are Pos, values <40% are Neg and values of 40–50% are considered Inc.
3Assayed at dilutions of 1:4 and 1:8, the respective blocking (%) values are shown together with the conclusion (Con.) A positive (Pos) reaction is defined as ≥60% block, while a negative reaction (Neg) is defined as <60% block.
4In this test the ratio of the Sample(S)/Positive (P) control values were calculated. S/P values >0.4 are Pos while those below are Neg.
3. Results
3.1. Inter-laboratory comparison of PEDV serology tests using ELISA (panel 1)
As an initial assessment of the properties of the serology-based diagnostic tests used in the different laboratories, a combined panel of 54 sera was distributed between the participating laboratories and tested by ELISA for the presence of anti-PEDV antibodies (see Table 1A, 1B). The different assays that were used comprised a mixture of “in-house” tests (ELISAs 1-3) and a commercial test (ELISA 4). The samples included known positive samples (from experimentally infected pigs), field samples collected from countries never known to have had cases of PEDV infection (SE and DK) and also field samples from herds in the US and Canada known to be infected with PEDV. It should be noted that the sera from SE had been selected following pre-screening using the ELISA 4 and found to be negative in this assay (Table 1B). In addition, some samples from FR were field samples that were specifically selected because they were collected at the beginning of 2014 from clinically normal herds but (with one exception) had given unexpected positive results in the ELISA 4 (see below). These selected sera were collected prior to the re-appearance of PEDV in FR (Grasland et al., 2015) and are not considered representative of pig sera from FR in general. The results of all the tests are presented in Table 1A, 1B.
The ELISA 1 identified anti-PEDV antibodies in the sera of 7 experimentally infected pigs (inoculated with an early European strain of PEDV, either Br1/87 or CV777) and still obtained a positive result when such sera were diluted 50, 200 or even 400 times. In addition, known positive field sera from the US and Canada also tested positive in this assay. In contrast, negative control sera, from the field or experimental studies, collected in DK (5 samples) and SE (17 samples), which have never experienced cases of PED, all proved negative. Furthermore, the 10 selected field sera, collected from clinically normal pigs from France, also all tested negative in this assay.
The same samples were also tested using ELISA 2. When testing sera from animals infected with early European strains of PEDV, two different laboratories, correctly identified 4/4 sera from pigs experimentally infected with the Br1/87 strain of PEDV (see Table 1A, Table 1B) but did not efficiently detect the pre-diluted (50–400×) samples of such sera. Furthermore, both of these laboratories obtained a positive result with either 6 or 7 of the 7 sera from CV777-infected pigs (from FR). Negative results were obtained on all 22 known negative sera in one laboratory but 2 of these negative sera (DK13 and DK17) were scored positive in another. Some 7 sera from pigs infected with US or Canadian strains of PEDV were also tested with ELISA 2 and 4 or 5 of them scored positive in the two laboratories. There was a greater disparity using the selected FR field sera, there was a single positive reaction in one lab however, 7 were scored positive in the other using this assay system.
A subset of these panel 1 samples, including sera from the Br1/87-infected pigs, was also tested using ELISA 3, another “in house” blocking assay, as described in Materials and Methods, (see Table 1A, 1B). This assay successfully identified each of the undiluted pig sera that were from animals experimentally infected with PEDV as being positive. However, when the sera were diluted (50–400×), prior to assay, then negative results were obtained in each case. Sera from animals infected with both the early European strains of PEDV and the recent INDEL US strain (from DK experimental studies and field sera from the US and Canada) were scored as positive, except for one serum (GVB/04) from a pig experimentally infected with the CV777 strain, but the 6 others were correctly identified. Of the 10 selected French field sera, 5 were scored positive while 5 were negative, including GVB/16, which was scored as negative in most of the other tests including the ELISA 4 (see below).
A commercial assay (ELISA 4) was used in 3 separate laboratories to test the whole panel of sera (Table 1A, 1B). This system successfully identified (in 18 of 21 tests) the sera from 7 pigs experimentally infected with the CV777 strain but, unexpectedly, was less efficient (only 4 of 12 tests of the undiluted sera were positive) at detecting the infection in 4 pigs by the Br1/87 strain that is very closely related to CV777 (these two strains had the same origin but have been passaged separately over time). The ELISA 4 did not detect antibodies in any of the pre-diluted sera from experimentally infected pigs. Sera from 7 pigs infected with the recent US strain of PEDV were successfully detected as positive (in 19 out 21 tests using this assay). However, using 5 sera from uninfected pigs in DK (with no history of PEDV infection), in the 15 tests using ELISA 4, some 5 were scored positive, 1 test was inconclusive and only 9 were scored negative. The 10 selected French field sera, collected from herds without clinical signs of PED, included 9 samples that had unexpectedly scored positive in this test during initial testing in one of the laboratories. A very similar pattern of results (8 or 9 positive) was obtained for these 10 sera when tested in this assay in each of the 3 laboratories (Table 1B, ELISA 4(a, b, c)) but these data contrast with the negative results obtained using ELISA 1, see Table 1B. All the negative sera (17 in total) from SE tested negative in the ELISA 4 in each of the 3 laboratories but they had been pre-selected for distribution in this panel based on the results from this assay.
3.2. Detection of experimental infections by old European and recent US strains of PEDV using ELISA (panel 2)
A further round of testing was performed on a second panel of 8 sera that were collected at either 14 or 28 dpi from animals that had been experimentally infected with either the European Br1/87 strain of PEDV (pigs 4 and 5) or with a recent US strain (a non-INDEL strain from Iowa) of PEDV (pigs 12 and 15). These experimental infections have been described previously (Lohse et al., 2016). The results from the testing of these sera, using the different assays, are shown in Table 2 . The ELISA 1 scored 4 out of 4 sera from Br1/87-infected pigs as seropositive (at both 14 and 28 dpi) but only 2 out of 4 sera from the US PEDV-infected pigs tested positive with 2 being inconclusive. These results were derived from 9 to 11 repeat tests of these sera and the mean blocking values were used for the overall interpretation (Table 2). To give an indication of the spectrum of results obtained in these tests, the number of individual tests scored as positive, inconclusive and negative are indicated (Table 2). The profile of these results closely reflects the overall interpretation. The positive sera generally gave uniformly positive results whereas the sera scored as inconclusive provided results that were scored in each of the 3 categories.
Table 2.
Detection of anti-PEDV antibodies in sera from experimentally infected pigs.
1: Samples are from pigs infected with PEDV Br1/87 or PEDV US (non-Indel strain) as described by Lohse et al. (2016).
2: DPI = days post infection.
3: The assay was run on 9–11 occasions with the indicated sera. The mean blocking% results are shown and the number of tests providing Pos, Neg or Inc results are indicated.
Negative results are highlighted in black. Inconclusive results are highlighted in grey.
The ELISA 3 scored 3 of the 4 sera from Br1/87 infected pigs as positive, the only negative result was for pig 4 at 14 dpi and this same animal was scored positive at 28 dpi. In contrast, 1 animal (pig 12) infected with an INDEL US strain of PEDV scored negative at both 14 and 28 dpi whereas another (pig 15) was positive at 14 dpi but was scored negative at 28 dpi.
The ELISA 4 was used in two different laboratories and detected the infection by the US strain of PEDV more efficiently (8/8 tests scored positive, i.e. 4/4 in both laboratories) than the Br1/87 infections (only 3 out of 8 tests scored positive). Pig 4 tested negative in this assay at both 14 and 28 dpi in both laboratories.
The same negative result for pig 4 was also obtained using ELISA 2 (see Table 2) but this test did produce a positive result for one of the Br1/87 infected pigs at 28 dpi. The ELISA 2 also produced a negative result for the sera from the pigs experimentally infected with US PEDV. However, 4 of the 7 field sera from PEDV-infected herds from Canada and the US did score positive in this assay (Table 1A).
3.3. Analysis of Italian field sera from a single farm with a recent outbreak of PEDV (panel 3)
A panel of 20 field sera from Italy was collected in 2015 and distributed to the different laboratories. These samples originated from a single farm that had experienced a PEDV infection a couple of months before the sampling. Sequence analysis of the S gene sequence from the virus involved in the disease outbreak indicated that it was closely related to the OH 851 strain of PEDV (data not shown) and to the PEDV strain detected recently in Germany (Hanke et al., 2015). The results from the analysis of these assays are shown in Table 3 . It could be expected that these sera are predominantly positive.
Table 3.
Analysis of field sera collected from a PEDV-infected farm in Italy during 2015.
 |
1: The blocking (%) values are indicated together with the conclusion (Con.). A Pos reaction is defined as >40% block, while a negative (Neg) is defined as <35% block and results of 35–40% blocking are defined as inconclusive (Inc).
2: Samples were tested at dilutions of 1:2 and 1:4 and the conclusion is shown. Blocking values >50% are Pos, values <40% are Neg and values of 40–50% are considered Inc.
3: Assayed at dilutions of 1:4 and 1:8, the respective blocking (%) values are shown together with the conclusion (Con.) A positive (Pos) reaction is defined as ≥60% block, while a negative (Neg) is defined as <60% block.
4: In this test the ratio of the Sample(S)/Positive (P) control values were calculated. Values >0.4 are Pos.
Negative results are highlighted in black. Inconclusive results are highlighted in grey.
In the ELISA 1, 19 of these 20 samples tested positive with one sample (sample ID 20) scored as “inconclusive”. In the ELISA 2, 14 of these samples were tested as positive while in the ELISA 3, 17 of these 20 sera tested positive with 3 being negative (including sample ID 20). The 17 positive samples included all of those scored as positive in the ELISA 2. One sample (ID 17) scored positive in ELISA 3 but was considered inconclusive in ELISA 2. The single inconclusive sample identified in ELISA 1 was scored as negative in ELISAs 2, 3 and 4.
The panel 3 sera were tested in the ELISA 4 in two laboratories. It was found that 11 of these 20 sera scored positive in this assay in one laboratory with 9 being negative. Similar results were obtained in the other laboratory; but samples with ID 9 and ID 23 were scored negative in this assay in one but positive in the other
3.4. Analysis of Italian field sera from multiple PEDV-infected and uninfected farms (panel 4)
Forty serum samples were collected in Italy during 2015 from 6 different farms at 2–9 weeks following detection of an outbreak of PEDV (OH 851-like strain). It could be expected that these sera would also be predominantly positive. These samples were tested in ELISAs 1 and 3 and also in the commercial ELISA 4 (Table 4 ). Of the 40 samples, 39 were scored as positive in the ELISA 3 and the same 39 samples were also scored positive in the ELISA 1 with the same single negative sample (sample ID 37). In the ELISA 4, 33 sera were scored as positive with the remaining 7 scored negative; the positive samples included sample ID 37 that was identified as negative in the other 2 assays. Interestingly, 4 of the 7 samples collected from farm 4, sample IDs 23-26, all scored negative in this test, whereas 3 other sera from the same farm were positive and all 7 sera from this farm were scored as positive by the other assays (note, it was demonstrated that the PEDV on this farm was an OH 851-like strain of the virus).
Table 4.
Detection of anti-PEDV antibodies in sera collected from PEDV-infected farms.
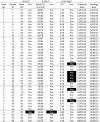 |
Negative results are highlighted in bold.
1In the ELISA 1, the blocking (%) values are indicated together with the conclusion (Con.). A Pos reaction is defined as >40% block, while a negative (Neg) is defined as <35% block and results of 35–40% blocking are defined as inconclusive (Inc).
2In ELISA 3, values ≥60% are scored positive (Pos). Values <60% are scored negative (Neg).
3In the ELISA 4, values >0.4 are scored Pos.
Another collection of 40 sera, obtained from 5 different farms without clinical signs of PED, were also tested by the same two laboratories (Table 5 ). All 40 sera scored as negative in the “in house” ELISA 1 and ELISA 3 and of these 38 also scored negative in the ELISA 4 (Table 5). However, 2 of the sera tested positive in this assay.
Table 5.
Detection of anti-PEDV antibodies in sera collected from 5 farms without evidence of PEDV infection.
 |
Negative results are highlighted in bold.
1In the ELISA 1, the blocking (%) values are indicated together with the conclusion (Con.). A Pos reaction is defined as >40% block, while a negative (Neg) is defined as <35% block and results of 35–40% blocking are defined as inconclusive (Inc).
2In ELISA 3, values ≥60% are scored positive (Pos). Values <60% are scored negative (Neg).
3In the ELISA 4, values >0.4 are scored Pos.
3.5. Statistical analysis of sensitivity and specificity
The diagnosis of convergence for each model indicated good convergence and that the number of iterations was sufficient. Trace plots indicated good mixing with a low level of autocorrelations (data not shown). In the comparison of the ELISAs 1, 2 and 4 using the results from Tables 1–3 (but excluding the results from the pre-diluted sera as analysed in Table 1), the results (Table 6A ) showed that the ELISA 1 provided high sensitivity (median 95%) and has a median specificity of 86%. The ELISA 2 is less sensitive (75%) but has a high specificity (97%). In contrast, the commercial assay, ELISA 4, has sub-optimal characteristics both in terms of sensitivity (83%) and specificity (68%).
Table 6A.
Bayesian analysis of sensitivity and sensitivity of the indicated ELISAs. This analysis used non-informative priors for sensitivities using data from Table 2, Table 3 (except for pre-diluted samples used in Table 1A, 1B).
 |
In a separate analysis, the ELISAs 1, 3 and 4 were compared for their performance on the samples tested; this included a larger selection of serum samples (data from Tables 1–5). This analysis (Table 6B ) showed that both ELISA 1 and ELISA 3 have good characteristics with high sensitivity (95 or 97%) and specificity (94–96%). As in the more restricted analysis (Table 6A), the ELISA 4 was less satisfactory with a calculated sensitivity of 79% and specificity of 80%.
Table 6B.
Bayesian analysis of sensitivity and sensitivity of the indicated ELISAs. This analysis used non-informative priors for sensitivities using data from Table 2, Table 3, Table 4, Table 5 (except for pre-diluted samples used in Table 1A, 1B).
 |
For both sets of results (as in Table 6A, B), a sensitivity analysis was carried out to evaluate the influence of priors on the final estimates by comparing different options including informative priors for all parameters, mildly informative priors for sensitivities, non-informative priors for sensitivities and non-informative priors for all parameters except prevalence estimate. Median estimates were not very different between the various models and were not strongly modified when non informative priors were used (data not shown). The option presented (using non-informative priors for sensitivities) was a good compromise in terms of precision of the estimates and the informative characteristics of the priors.
4. Discussion
In this study, multiple panels of porcine sera, collected from the field and from experimental studies within different countries from Europe and North America, have been analysed for the presence of anti-PEDV antibodies using 3 different “in house” ELISAs and a widely used commercial ELISA. Overall, it is apparent that, on a “herd-basis”, each of the assays is able to successfully detect infection by both the old European strains and also by the two, more recent, strains (non-INDEL and INDEL) that have infected herds within the USA in 2013/2014. The INDEL PEDV strain has also occurred in a number of European countries in 2014 and 2015. However, there can be significant differences in the detection of antibodies in individual animals by the different serological assays, and so it is clearly necessary to test several animals from a herd to ensure that successful detection of an infection of the herd is achieved. This should not be a problem since it can be considered best practice to test a representative sample and it is known that the virus spreads rapidly through an infected herd.
In general, it seems that infection in the field generates a higher level of anti-PEDV antibodies than is achieved within experimental studies. This is well demonstrated by the panel of 20 sera collected from a herd that had recently been infected PEDV (see Table 3). The majority of sera were scored as positive in the assays used here, especially with the ELISAs 1 and 3. However, in contrast, using sera from experimentally infected pigs, the proportion of positive tests was much lower with some assays (see Table 2). It was also apparent that the level of antibody blocking observed with the ELISAs 1, 2 and 3 was higher with the field sera than with the experimental sera. This may reflect the route, level and frequency of the virus exposure that occurs in the field compared to that used experimentally.
In practice, it might be requested to use serological assays to demonstrate freedom of a country or herd from PED. For this purpose, it is particularly important that the assays demonstrate high specificity; otherwise “false-positive” reactions can require a lot of work to prove freedom from PED. It appears that the commercial ELISA 4 produces some false positive results. In Table 5, it is shown that 2 out of 40 sera from uninfected herds scored positive. These sera scored negative in both the ELISA 1 and ELISA 3 “in house” assays. Similarly, a group of 10 sera from France were selected for testing in the ring trial on the basis of unexpected positive reactions in the ELISA 4 (Table 1A, 1B) since they were all collected from herds with no clinical signs of PED. These samples all tested negative in the ELISA 1. These were collected at the beginning of 2014 prior to the introduction of the OH 851- related (INDEL) PEDV into France that was first detected in December 2014. Some of these sera also tested positive in other assays (ELISAs 2 and 3) and thus it may be that the pigs have been infected with another agent that cross-reacts, to some degree, in these assays. It should be noted that the ELISA 4 detects antibodies to the PEDV nucleoprotein only whereas the other assays are based on the use of whole virus preparations.
The ELISA 1 has been used on over 20,000 field sera from Denmark and there have been no positive reactions suggesting a high level of specificity for this assay; the statistical analysis presented in Table 6A, B confirms this. It seems unlikely that the non-specific reactions in certain other assays could be due to porcine respiratory coronavirus (PRCV) infection as this is widespread in Europe, including in DK, and thus would be expected to give positive results in a larger proportion of sera than has been observed here.
To declare a herd or region free of PED, a high level of sensitivity is required. The ELISA 1 appears to have the highest analytical sensitivity since it was the only one capable of detecting a positive reaction using pre-diluted serum samples (Table 1A, 1B). However, it was found that the antibody response to the experimental infection with the US PEDV did not induce a very strong antibody response, as determined by ELISA 1, and indeed the overall result was inconclusive in 1 of the 2 infected animals tested. For the sera from the experimentally infected pigs, it was possible, using this ELISA, to follow the generation of the immune response throughout the course of 28 days post-infection and it is clear that seroconversion did occur in each of the US PEDV inoculated pigs (see Lohse et al., 2016). The ELISA 2 and ELISA 3 also had difficulty in detecting the immune response in the animals experimentally infected with the US PEDV (see Table 2). The ELISA 4 was better in this respect (see Table 2). In contrast to these results, using field sera, there was a very high degree of agreement between the ELISA 1 and ELISA 3 (see Tables 3–5) and 39 of 40 sera from the OH 851-related PEDV-infected herd in Italy all scored positive in both of these tests (Table 4) while 40 sera from uninfected herds, also in Italy, were all negative in both assays.
5. Conclusions
There was a high degree of consistency in the performance of ELISA 4 when used in different laboratories (see Table 1A, 1B) but its overall performance in terms of sensitivity and specificity is sub-optimal (see Table 6A, B). ELISA 2 performed with good specificity (Table 6A) although with sub-optimal sensitivity. It was found that this assay can detect PEDV infection at a herd level. Both the ELISA 1 and ELISA 3 performed very well and exhibited both high sensitivity and specificity.
Acknowledgements
We gratefully acknowledge financial support from the CoVet Lab (a partnership of national veterinary public health institutes from Denmark, France, The Netherlands, Sweden and the United Kingdom). In addition we are grateful for excellent technical assistance from Jane Borch, Pernille Manley Hansen, Lionel Bigault and Daniela Bresciani.
Contributor Information
Bertel Strandbygaard, Email: bstr@vet.dtu.dk.
Antonio Lavazza, Email: antonio.lavazza@izsler.it.
Davide Lelli, Email: davide.lelli@izsler.it.
Yannick Blanchard, Email: Yannick.BLANCHARD@anses.fr.
Béatrice Grasland, Email: Beatrice.GRASLAND@anses.fr.
Sophie Le Poder, Email: slepoder@vet-alfort.fr.
Nicolas Rose, Email: Nicolas.ROSE@anses.fr.
Falko Steinbach, Email: Falko.Steinbach@ahvla.gsi.gov.uk.
Wim H.M. van der Poel, Email: wim.vanderpoel@wur.nl.
Frederik Widén, Email: frederik.widen@sva.se.
Graham J. Belsham, Email: grbe@vet.dtu.dk.
Anette Bøtner, Email: aneb@vet.dtu.dk.
References
- Best N.G., Cowles M.K., Vines S.K. MRC Biostatistics Unit; Cambridge, UK: 1995. CODA Manual. [Google Scholar]
- Boniotti M.B., Papetti A., Lavazza A., Alborali G., Sozzi E., Chiapponi C., Faccini S., Bonilauri P., Cordioli P., Marthaler D. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg. Infect. Dis. 2016;22:83–87. doi: 10.3201/eid2201.150544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branscum A.J., Gardner I.A., Johnson W.O. Estimation of diagnostic-test sensitivity and specificity through Bayesian modelling. Prev. Vet. Med. 2005;68:145–163. doi: 10.1016/j.prevetmed.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks S.P., Gelman A. Alternative methods for monitoring convergence of iterative simulations. J. Comput. Graphical Stat. 1998;7:434–455. [Google Scholar]
- Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E., Gauger P.C., Schwartz K.J., Madson D., Yoon K.J., Stevenson G.W., Burrough E.R., Harmon K.M., Main R.G., Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014;52:234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K., Lager K., Miller L., Opriessnig T., Gerber P., Hesse R. Evaluation of porcine epidemic diarrhea virus transmission and the immune response in growing pigs. Vet. Res. 2015;46:49. doi: 10.1186/s13567-015-0180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA AHAW Panel (EFSA Panel on Animal Health and Welfare), 2014. Scientific Opinion on porcine epidemic diarrhoea and emerging pig deltacoronavirus. EFSA Journal 12 (10), 3877. 10.2903/j.efsa.2014.3877. [DOI] [PMC free article] [PubMed]
- Grasland B., Bigault L., Bernard C., Quenault H., Toulouse O., Fablet C., Rose N., Touzain F., Blanchard Y. Complete genome sequence of a porcine epidemic diarrhea S gene indel strain isolated in France in December 2014. Genome Announc. 2015;3(3):e00535–e00615. doi: 10.1128/genomeA.00535-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke D., Jenckel M., Petrov A., Ritzmann M., Stadler J., Akimkin V., Blome S., Pohlmann A., Schirrmeier H., Beer M., Höper D. Comparison of porcine epidemic diarrhea viruses from Germany and the United States, 2014. Emerg. Infect. Dis. 2015;21:493–496. doi: 10.3201/eid2103.141165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger P., Welch P.D. Simulation run length control in the presence of an initial transient. Oper. Res. 1983;31:1109–1144. [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. 2013;4:e00737–e00813. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Saif L.J. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Wang Q., Scheuer K.A., Lu Z., Zhang Y., Saif L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014;20:662–665. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Annamalai T., Lu Z., Saif L.J. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs 26-day-old weaned pigs. Vet. Microbiol. 2015;178:31–40. doi: 10.1016/j.vetmic.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Kim I.J., Pyo H.M., Tark D.S., Song J.Y., Hyun B.H. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J. Virol. Methods. 2007;146:172–177. doi: 10.1016/j.jviromet.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse L., Krog J.S., Strandbygaard B., Rasmussen T.B., Kjær J., Belsham G.J., Bøtner A. Experimental infection of young pigs with an early European strain of porcine epidemic diarrhoea virus and a recent US strain. Transbound. Emerg. Dis. 2016 doi: 10.1111/tbed.12509. (epub May 2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita J.R., Hakze-van der Honing R., Almeida A., Lourenço M., van der Poel W.H., Nascimento M.S. Outbreak of porcine epidemic diarrhea virus in Portugal, 2015. Transbound. Emerg. Dis. 2015;62:586–588. doi: 10.1111/tbed.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2008. R: A language and environment for statistical computing. ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- Raftery A.L., Lewis S. Comment: one long run with diagnostics: implementation strategies for Markov chain Monte Carlo. Stat. Sci. 1992;7:493–497. [Google Scholar]
- Sørensen K.J., Bøtner A., Madsen E.S., Strandbygaard B., Nielsen J. Evaluation of a blocking Elisa for screening of antibodies against porcine reproductive and respiratory syndrome (PRRS) virus. Vet. Microbiol. 1997;56:1–8. doi: 10.1016/s0378-1135(96)01345-4. [DOI] [PubMed] [Google Scholar]
- Sozzi E., Luppi A., Lelli D., Martin A.M., Canelli E., Brocchi E., Lavazza A., Cordioli P. Comparison of enzyme-linked immunosorbent assay and RT-PCR for the detection of porcine epidemic diarrhoea virus. Res. Vet. Sci. 2010;88:166–168. doi: 10.1016/j.rvsc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelhalter D.J., Thomas A., Best N.G., Gilks W.R. MRC Biostatistics Unit; Cambridge: 1996. BUGS: Bayesian Inference Using Gibbs Sampling, Version 0.5 (version ii) [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Toplak I., Ipavec M., Kuhar U., Kušar D., Papić B., Koren S., Toplak N. Complete genome sequence of the porcine epidemic diarrhea virus strain SLO/JH-11/2015. Genome Announc. 2016;4(2):e01725–e01815. doi: 10.1128/genomeA.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nieuwstadt A.P., Zetstra T. Use of two enzyme-linked immunosorbent assays to monitor antibody responses in swine with experimentally induced infection with porcine epidemic diarrhea virus. Am. J. Vet. Res. 1991;52:1044–1050. [PubMed] [Google Scholar]


