Abstract
S1D (residues 636–789) is a neutralizing epitope region on the spike protein (S) of porcine epidemic diarrhea virus (PEDV). To accurately identify epitopes on S1D, the S1-phage library containing the gene encoding the S1D region of PEDV S protein was micropanned by six specific monoclonal antibodies (McAbs) against the S1D region. These micropanned epitope regions (MER) were focused on 696–779 amino acids of the S protein. To further map epitopes of the MER, seven overlapping mini-fragments covering MER nucleotides were separately synthesized and expressed in Escherichia coli BL21 with a GST tag. These mini-GST fusion proteins were scanned by ELISA and Western blotting with the six McAbs, and the result showed that S1D5 (residues 744–759) and S1D6 (residues 756–771) are two linear epitopes of the PEDV S protein. The antisera of the epitopes S1D5 and S1D6 could react with the native S protein of PEDV. Furthermore, Pepscan of the two linear epitopes demonstrated that SS2 (748YSNIGVCK755) and SS6 (764LQDGQVKI771) are two core epitopes on S1D5 and S1D6, respectively, located on the S protein of PEDV.
Keywords: PEDV, Spike protein, Epitope, Monoclonal antibody, Phage display, Pepscan
1. Introduction
Porcine epidemic diarrhea (PED) is a highly contagious, enteric disease of swine caused by porcine epidemic diarrhea virus (PEDV). The disease is characterized by severe diarrhea, vomiting, dehydration, and death, and has a mortality rate of up to 90% (Pensaert and Debouck, 1978, Timoney et al., 1998). Since PED was first reported in Belgium and the UK in 1978, the disease has frequently broken out in many swine-raising countries and has led to severe economic losses in Asia, notably in China, Japan and Korea (Chasey and Cartwright, 1978, Takahashi et al., 1983, Kweon et al., 1993, Fan and Li, 2005).
PEDV is an enveloped, positive-sense, single-stranded RNA virus, which belongs to the genus Coronavirus, in the family Coronaviridae, order Nidovirales. The genome of PEDV is approximately 30 kb in length. Subgenomic mRNAs of PEDV are transcribed from its genome RNA, and then are translated to produce viral proteins such as spike protein (S, 180–220 kDa), membrane protein (M, 27–32 kDa) and nucleocapsid protein (N, 55–58 kDa) (Egberink et al., 1988, Holmes, 2001, Brian and Baric, 2005). The S protein, a glycoprotein peplomer on the viral surface, plays an important role in induction of neutralizing antibodies, specific receptor binding and cell membrane fusion (Duarte and Laude, 1994, Li et al., 2007, Ma et al., 2005). These properties make the S protein a suitable candidate for genetic engineering of vaccines and diagnostic reagents.
Previously, we used antisera of PEDV to test the antigenicity of four overlapping fragments spanning the major immunodominant region (residues 1–789) of S protein, and identified one antigenic determinant (S1D: residues 636–789). The anti-S1D antibodies showed neutralizing activity for PEDV-infected Vero cells (Sun et al., 2007a). In order to provide some basis for the development of vaccines and diagnostic reagents, it is necessary to map the antigenic epitopes of the S1D region and accurately determine important residues in these epitopes. In this study, one micropanned epitope region of S1D (MER: residues 696–779) was identified by fd-tet phage display using six monoclonal antibodies (McAbs). Further analysis of the MER was carried out by ELISA, Western blotting and Pepscan using the six McAbs. Two novel B cell epitopes, SS2 (residues 748–755) and SS6 (residues 764–771), were identified in the S protein of PEDV.
2. Materials and methods
2.1. Viruses, plasmid, bacterial strains and expression vector
PEDV strain CV777 was kindly provided by Pensaert MB. Plasmid DNA (pMD18-TS1) containing the S gene (1–2367 bp) of PEDV strain CV777 had been constructed previously (Sun et al., 2007b). A gene-targeted fd-tet phage display library (S1-phage library) containing DNA fragments ranging from approximately 50 to 400 bp of the gene encoding the S1 region of PEDV S protein was established by our laboratory, and the neutralizing epitope region S1D located in S1 region of PEDV S protein (Sun et al., 2007a). Escherichia coli strains MC1061 and K91BluKan, and the phage display vector fUSE 1, were generous gifts from Prof. George P. Smith, University of Missouri, Columbia, USA.
2.2. Development of monoclonal antibodies (McAbs)
The PEDV particles were purified by sucrose density gradient centrifugation. Female 6-week-old BALB/c mice were immunized with 50 μg of purified PEDV emulsified in complete Freund's adjuvant (Sigma, St. Louis, MO, USA). At 3-week intervals two boosters of 50 μg of purified PEDV emulsified in incomplete Freund's adjuvant (Sigma, St. Louis, MO, USA) were administered, and mice were sacrificed 3 days after the last booster inoculation. Spleen cells from immunized mice were fused with SP2/0 myeloma cells using 50% (v/v) of PEG1450 (Sigma, St. Louis, MO, USA), and the fused cells were cultured in DMEM supplemented with 20% FCS, HAT medium. Positive hybridoma clones were selected by indirect ELISA using purified S1D protein with a glutathione S-transferase (GST) tag as coating antigen. The subtype of McAbs secreted by the final hybridoma clones was identified using the SBA Clonotyping™ System/HRP kit (SouthernBiotech, Birmingham, AL, USA). Ascites fluid was produced in primed BALB/c mice with incomplete Freund's adjuvant. Reactivity of the McAbs with the native S protein was tested by ELISA and Western blotting. Neutralizing activity of the McAbs was tested according to the methods described by Hofmann and Wyler (1989).
2.3. Micropanning of the S1-phage library with McAbs
Ascites fluid containing six strains of McAbs, named 2C4, 3G3, 5F8, 3G5, 6E6 and 3C3, was diluted to 1:200 in 0.1 M NaHCO3 (pH 9.6) and 100 μl of the dilution was added to each well of the ELISA plate (Nunc, Roskilde, Denmark). The ELISA plate was incubated at 4 °C for 12 h. The wells were then blocked with 5% skimmed milk at 37 °C for 1 h. After washing three times with TBST (50 mM Tris·Cl pH 7.5, 150 mM NaCl and 0.5% Tween 20), 100 μl S1-phages (106 TUs) were added to each well and then incubated at 37 °C for 1 h. After washing 10 times with TBST, 20 μl elution buffer (0.2 M glycine–HCl, pH 2.2) was added to the wells and incubated at room temperature for 10 min. Twenty microliters of elution buffer was added to blank wells to serve as a mock eluate. Eluted products were mixed with 4 μl neutralizing solution (1 M Tris·Cl pH 9.1); 2 μl of the mixture was used for TUs titration. Remnant elutes were used to infect 20 ml of starved K91BluKan cells, and then binding-phages in culture supernatant were purified by PEG8000/NaCl (2.8%/0.55 M). The binding-phages were used for micropanning as described above. After the third binding-phages were titrated, 10 random clones of each McAb were isolated. The DNA of infected cells from the above single colonies was sequenced with primer: fd-up 5′-TTTTGGAGATTTTCA ACGTG-3′, which anneals at 1562–1581 bp of the fUSE 1 vector, to identify phage containing the correct open reading frame and the correct orientation of the S1D gene.
2.4. Phage ELISA
Two phage clones, which displayed 696–770 amino acids and 712–779 amino acids, were designated as phage696 and phage712, respectively. The overlapping region displayed by phage696 and phage712 overlaid all other micropanned epitopes. Phage696 and phage712 were purified by PEG8000/NaCl, and their TUs were titrated for standardization. The ELISA plate wells were coated with 50 μl purified phage696 and phage712 containing 108 TUs/ml in coating buffer (50 mM Tris·Cl, 150 mM NaCl, pH 9.0) and incubated at 4 °C for 12 h. After washing the plate three times with TBST, 200 μl of blocking solution (5% skimmed milk in TBST) was added to each well and incubated at 37 °C for 1 h, and then 100 μl of McAbs from ascites fluid diluted 1/200 was added and incubated at 37 °C for 1 h. After washing three times with TBST, 100 μl of HRP-conjugated sheep anti-mouse IgG diluted 1/8000 in TBST was added, and then incubated at 37 °C for 1 h. After the addition of 100 μl substrate TMB solution and incubation at room temperature for 10 min, the reaction was stopped by 50 μl of 2 M H2SO4. The absorbance at 450 nm was measured. Wild phage and S1D recombinant protein were used as negative and positive controls, respectively, and the mixtures of the McAbs were used as their primary antibodies.
2.5. Expression of the micropanned epitope region (MER)
Seven overlapping mini-fragments, which covered the micropanned epitope regions (2085–2336 bp), were separately synthesized according to the methods described by (Table 1 ) Hua et al. (2004). These mini-fragments were cloned into pGEX-6p-1 vector (Amersham Pharmacia Biotech, New Jersey, USA), and recombinant plasmids were transformed into E. coli BL21. The fusion proteins expressed in E. coli BL21 were purified according to the manufacturer's protocols (Pierce, Rockford, IL, USA). Using the same procedure, GST protein was also purified for the controlled trial.
Table 1.
Synthesized mini-fragments and deduced amino acid sequences
| Mini-fragment names | Sequences of synthesized mini-fragments (5′ → 3′) | Deduced amino acid sequences a |
|---|---|---|
| S1D1 | (+) gatccccatgttctttttcagagcaggctgcatatgttgatgatgatatagtgtaag | 696PCSFSEQAAYVDDDIV711 |
| (−) gggtacaagaaaaagtctcgtccgacgtatacaactactactatatcacattcagct | ||
| S1D2 | (+) gatccgatgatatagtgggtgttatttctagtttgtctaactccacttttaactaag | 708DDIVGVISSLSNSTFN723 |
| (−) gctactatatcacccacaataaagatcaaacagattgaggtgaaaattgattcagct | ||
| S1D3 | (+) gatcctccacttttaacaatacctgggagttgcctggtttcttctaccattcttaag | 720STFNNTRELPGFFYHS735 |
| (−) gaggtgaaaattgttatggaccctcaacggaccaaagaagatggtaagaattcagct | ||
| S1D4 | (+) gatccttctaccattctaatgatggctccaattgtacagagcctgtgttggtgtaag | 732FYHSNDGSNCTEPVLV747 |
| (−) gaagatggtaagattactaccgaggttaacatgtctcggacacaaccacattcagct | ||
| S1D5 | (+) gatcccctgtgttggtgtatagtaacataggtgtctgtaaatctggcagtatttaag | 744PVLVYSNIGVCKSGSI759 |
| (−) gggacacaaccacatatcattgtatccacagacatttagaccgtcataaattcagct | ||
| S1D6 | (+) gatcctctggcagtattggctatgtcccacttcaggatggccaagtcaagatttaag | 756SGSIGYVPLQDGQVKI771 |
| (−) gagaccgtcataaccgatacagggtgaagtcctaccggttcagttctaaattcagct | ||
| S1D7 | (+) gatcccttcaggatggccaagtcaagattgcacccatggttactgggaatatttaag | 764LQDGQVKIAPMVTGNI779 |
| (−) ggaagtcctaccggttcagttctaacgtgggtaccaatgacccttataaattcagct | ||
Note: (+) sign denotes sense strands and (−) sign denotes anti-sense strands. At 5′ and 3′ terminal of each sense strand there is a sequence of “gatcc” (underlined) and “g” (shading), respectively. And at 3′ and 5′ terminal of anti-sense strand there is a sequence of “g” (shading) and “cagct” (underlined), respectively. When the sense and anti-sense oligo-nucleotides annealed they would form a cohesive BamHI site at 5′ terminus and a cohesive SalI site at 3′ terminus. At the end of mini-fragment encoding gene there is an additional taa sequence to form a stop codon (bold).
Location of the polypeptides encoded by these mini-fragments based on the sequence of S protein of PEDV strain CV777 (GenBank accession no. AF353511).
2.6. ELISA of the recombinant proteins
The ELISA plate wells were coated with 1 μg/ml purified recombinant protein in 0.1 M carbonate buffer (pH 9.6) at 4 °C for 12 h and blocked with 5% skimmed milk at 37 °C for 1 h. After washing three times with PBST, 100 μl of McAbs from ascites fluid, diluted 1/200, were added to the wells and incubated at 37 °C for 1 h. The plates were washed three times and incubated with 100 μl of HRP-conjugated sheep anti-mouse IgG diluted 1/8000 in TBST at 37 °C for 1 h. Procedures of coloration, termination and absorbance measurement were as for the phage ELISA (Section 2.4).
2.7. Western blotting
The purified proteins and native S protein of PEDV were separated by SDS-15% polyacrylamide gel. After electrophoresis the separated proteins were transferred to nitrocellulose membrane (NC). The NC membranes were blocked with 5% skimmed milk in PBS at 37 °C for 1 h, and then incubated with 100 μl of McAbs from ascites fluid diluted 1/200 at 37 °C for 1 h. After washing three times with PBS, the NC membrane was incubated with 100 μl of HRP-conjugated sheep anti-mouse IgG diluted 1/2000 in PBS at 37 °C for 1 h. The NC membrane was washed three times with PBST and then coloration was carried out in 3,3′-diaminobenzidine (DAB) substrate solution.
2.8. Analysis of immunogenicity of the epitopes S1D5 and S1D6
The GST fusion proteins of the epitopes S1D5 and S1D6 were purified according to the manufacturer's protocols. Female 8-week-old BALB/c mice were immunized with 50 μg of purified fusion proteins emulsified in complete Freund's adjuvant (Sigma, St. Louis, MO, USA). At 3-week intervals two boosters of 50 μg of purified fusion proteins emulsified in incomplete Freund's adjuvant (Sigma, St. Louis, MO, USA) were administered. Antiserum of GST protein was also prepared for the controlled trial using the same procedures. Three mice were immunized with each protein. The antibody levels were examined by ELISA using the S1D-GST fusion protein as coating antigen (data not shown). To further verify whether antibodies against the epitopes S1D5 and S1D6 could reacted with native S protein of PEDV, an ELISA was carried out using the purified PEDV as coating antigen. In ELISA, antisera of the epitopes S1D5 and S1D6 were diluted 100 times with PBS for detection, other procedures were as for ELISA of the recombinant proteins (Section 2.6). Neutralizing activity of antibodies against the epitopes S1D5 and S1D6 was tested according to the methods described by Hofmann and Wyler (1989).
2.9. Pepscan of the epitopes S1D5 and S1D6
In order to identify core amino acids on the epitopes S1D5 and S1D6, seven polypeptides were synthesized by CL Bio-Scientific Co., Ltd. (XiAn, China). Location and sequences of the synthetic peptides are shown in Table 2 . Six polypeptides (SS1–SS6) with four overlapping amino acids covered the epitope region of S1D5 and S1D6; the SS7 polypeptide comprised all the peptides (SS1–SS6). The Pepscan procedure was as follows. The ELISA plate wells were coated with the synthetic peptides in 0.1 M carbonate buffer (pH 9.6) at 10 μg/ml at 4 °C for 12 h. Other operations were the same as the phage ELISA (Section 2.4).
Table 2.
Sequence of the synthesized peptides corresponding to the epitopes S1D5 and S1D6
| Names | Amino acids sequences and location on S proteina |
|---|---|
| SS1 | 744PVLVYSNI751 |
| SS2 | 748YSNIGVCK755 |
| SS3 | 752GVCKSGSI759 |
| SS4 | 756SGSIGYVP763 |
| SS5 | 760GYVPLQDG767 |
| SS6 | 764LQDGQVKI771 |
| SS7 | 744PVLVYSNIGVCKSGSIGYVPLQDGQVKI771 |
Location of the synthesized peptides based on the sequence of S protein of PEDV strain CV777 (GenBank accession no. AF353511).
3. Results
3.1. Screening of S1-phage library with McAbs
Six McAbs against the S1D region were prepared by lymphocyte hybridoma technique. All six McAbs could reacted with the native spike protein of PEDV and had no neutralizing activity for PEDV-infected Vero cells (data not shown). The S1-phage library was micropanned using the six McAbs against the S1D region. Sixty phage clones, comprising 10 clones selected randomly from the binding-phages of each McAb, were sequenced by the dideoxy method. The result showed that polypeptides displayed by 55 phage clones (with the exception of five meaningless sequences) were focused on amino acids 696–779 (MER) of the S protein. Further analysis of the MER demonstrated that 30 phage clones displayed amino acids 696–770 (SME I) and 25 phage clones displayed amino acids 712–779 (SME II) (Fig. 1A). The reactivity of the micropanned phage clones with the McAbs was tested by ELISA. The result indicated that phage696 and phage712, which displayed SME I and SME II, respectively, were both able to react with the six McAbs (Fig. 1B and C). These results clearly demonstrated that the crucial epitopes on the S1D region were located within amino acids 696–779 of the PEDV S protein.
Fig. 1.
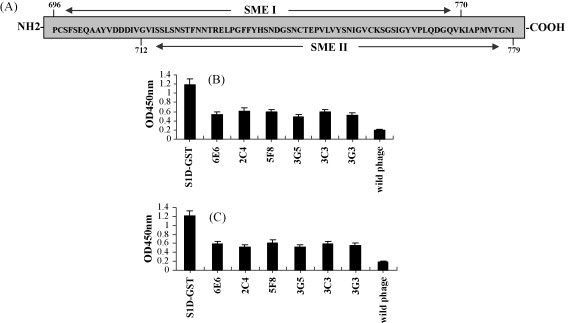
The result of micropanning of the S1-phage library and the reactivity of phage696 and phage712 with McAbs. Schematic diagram of the micropanned epitope region (MER) located in the PEDV S protein. (A) Gray histogram represents the whole MER, which includes two overlapping regions, SME I and SME II. (B) The result of the phage696 ELISA. (C) The result of the phage712 ELISA.
3.2. Mapping of the MER with McAbs by Western blotting and ELISA
To further identify epitopes on MER, seven overlapping mini-fragments (Table 1) were expressed in E. coli BL21 with a GST tag, and then these recombinant proteins were purified (Fig. 2A). The reactivity of the purified proteins with the six McAbs was detected by ELISA and Western blotting, respectively. Using Western blotting, the S1D5-GST protein showed a strong reaction with McAbs 2C4, 3C3 and 5F8 (Fig. 2B), and the S1D6-GST protein showed a strong reaction with McAbs 3G3, 6E6 and 3G5 (Fig. 2C). The results of the ELISA were in accordance with those of Western blotting (Fig. 2D). These results indicated that S1D5 (744PVLVYSNIGVCKSGSI759) and S1D6 (756SGSIGYVPLQDGQVKI771) are two linear epitopes of the PEDV S protein.
Fig. 2.
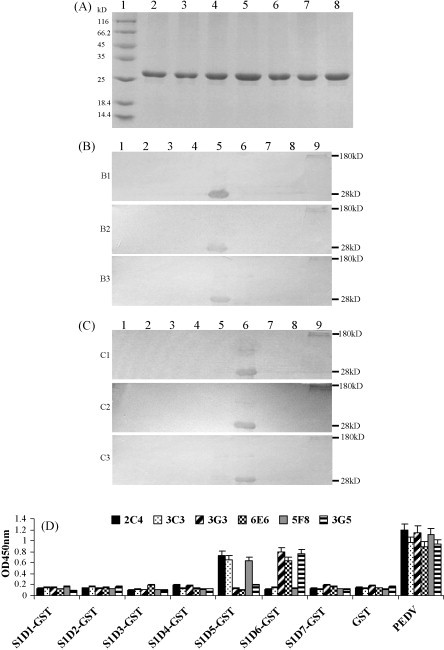
Purification of the recombinant proteins and detection of their antigenicity for McAbs by Western blotting and ELISA. (A) Electrophoresis pattern of the purified recombinant protein on SDS-15% polyacrylamide gel. Lanes: (1) protein marker; (2–8) purified proteins of S1D1-GST, S1D2-GST, S1D3-GST, S1D4-GST, S1D5-GST, S1D6-GST and S1D7-GST, respectively. (B) Western blotting analysis of the recombinant proteins with McAbs 2C4 (B1), 3C3 (B2) and 5F8 (B3). Lanes: (1–9) S1D1-GST, S1D2-GST, S1D3-GST, S1D4-GST, S1D5-GST, S1D6-GST, S1D7-GST, GST and PEDV, respectively. (C) Western blotting analysis of the recombinant proteins with McAbs 3G3 (C1), 6E6 (C2) and 3G5 (C3). Lanes: (1–9) S1D1-GST, S1D2-GST, S1D3-GST, S1D4-GST, S1D5-GST, S1D6-GST, S1D7-GST, GST and PEDV, respectively. (D) Detection of reactivity of the recombinant proteins with six McAbs by ELISA.
3.3. Immunogenicity of the epitopes S1D5 and S1D6
To detect immunogenicity of the epitopes S1D5 and S1D6, BALB/c mice were immunized using the GST fusion protein S1D5 and S1D6. After immunization three times, antibodies against the epitopes S1D5 and S1D6 could react with native S protein of PEDV in ELISA (Fig. 3 ). But the antibodies induced by the epitopes S1D5 and S1D6 had no neutralizing activity for PEDV-infected Vero cells (data not shown).
Fig. 3.
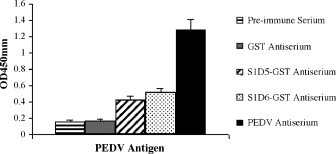
Reactivity of antisera against the epitopes S1D5 and S1D6 with PEDV by ELISA.
3.4. Minimization of the epitopes S1D5 and S1D6 with McAbs by Pepscan
In order to identify core amino acids on the epitopes S1D5 and S1D6, Pepscan of seven synthesized peptides (Table 2) was carried out with the six McAbs. In ELISA, the SS2 polypeptide was recognized by McAbs 2C4, 5F8 and 3C3; the SS6 polypeptide was recognized by McAbs 6E6, 3G5 and 3G3; and the SS7 polypeptide showed a strong reaction with all the McAbs. These data demonstrate that SS2 (748YSNIGVCK755) and SS6 (764LQDGQVKI771) are core amino acids of the epitopes S1D5 and S1D6, respectively (Fig. 4 ).
Fig. 4.
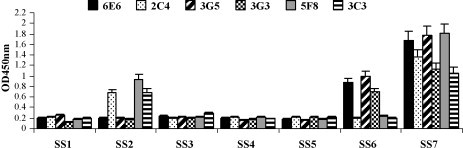
Pepscan of the epitopes S1D5 and S1D6 with McAbs.
4. Discussion
PEDV is an important pathogen causing viral diarrhea in the swine industry (Carvajal et al., 1995, Chae et al., 2000). Although much research has been carried out on the general characteristics of PEDV, few reports have been published on the functions of the PEDV structural proteins, especially S protein. Like those of other coronaviruses, the PEDV S protein plays an important role in inducing neutralizing antibodies (Duarte and Laude, 1994, Kang et al., 2006). Therefore, the screening and identification of antigenic epitopes on the PEDV S protein is essential for developing epitope-based vaccines and diagnostic reagents.
In this study, phage display and Pepscan methods were adapted for mapping epitopes of the neutralizing epitope region (S1D, residues 636–789) of the PEDV S protein. First, one epitope region (MER) was identified on the S1D region of PEDV S protein by phage display. Epitopes could not be determined accurately because the inserts expressed on the surface of the recombinant phages were overlength (approximately 200–300 bp). Second, more accurate identification of the epitopes was performed based on the method described by Hua, and two linear epitopes, S1D5 and S1D6, were determined in the MER (Hua et al., 2004). Antibodies against the epitopes S1D5 and S1D6 could react with native S protein of PEDV in ELISA, but the antibodies induced by the epitopes S1D5 and S1D6 had no neutralizing activity for PEDV-infected Vero cells. The epitopes S1D5 and S1D6 are 16 amino acids in length and have four overlapping amino acids. Generally, linear epitopes are short stretches of the primary structure of proteins and consist of six to nine or more continuous amino acid residues. The Pepscan method was used to identify core amino acids on epitopes S1D5 and S1D6, the result showed that the core amino acids were located at amino acids 748–755 (SS2) and amino acids 764–771 (SS6), respectively. It was found that epitopes SS2 and SS6 were adjacent to epitope S1P3 (residues 697–742) in the primary structure of S protein. The epitope S1P3 was identified previously by phage display using native antisera against PEDV (Sun et al., 2007b). This result suggests that epitopes SS2, SS6 and S1P3 represent a considerable immunodominant region of PEDV S protein.
In the case of PEDV, the S protein could not be cleaved into S1 and S2 subunits, due to a lack of proteolytic cleavage sites (Duarte and Laude, 1994, Park et al., 2006, Yeo et al., 2003). However, the S1 domain (residues 1–789) and the S2 domain (residues 790–1383) have been artificially defined by the conserved nonamer and the GxCx motifs at the position of S protein cleavage sites in coronavirus group II members (Follis et al., 2006). Based on bio-informatics analysis of the S protein structure of other coronaviruses, the S1 domain is thought to form the spherical head of the S protein, and is easily accessible to neutralizing antibodies. The S2 domain forms the stem portion and trans-membrane structure of the S protein, and it seldom induces antibodies because of the inaccessibility of this region in intact virions (Holmes, 2001, Wu et al., 2004, De Haan et al., 2006). The epitopes SS2 and SS6 are presumed to be located in the spherical head of the S protein of PEDV.
Recently, Chang reported one neutralizing epitope region (residues 499–638) on the S protein of PEDV (Chang et al., 2002). In addition, Cruz recently identified a mocking epitope (1368GPRLQPY1374) that has antigenic similarities with PEDV neutralizing epitopes (Cruz et al., 2006). The epitopes SS2 and SS6 reported here are located in different regions of these epitopes, as identified formerly, and they represent two novel B cell antigenic epitopes of PEDV S protein. The results of this study will provide some basic information for further characterization of the antigenic structure of PEDV S protein, and identified epitopes have a potential use for developing diagnostic reagents and effective vaccines for PEDV.
Acknowledgements
We thank Prof. George P. Smith for provision of the phage display kit. This work was supported by a grant from the National Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute (HVRI), The Chinese Academy of Agricultural Sciences (CAAS).
References
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. In: Enjuanes L., editor. Coronavirus Replication and Reverse Genetics. Springer-Verlag; Berlin: 2005. pp. 2–22. [Google Scholar]
- Carvajal A., Lanza I., Diego R., Rubio P., Girrnenes P. Seroprevalence of porcine epidemic diarrhea virus infection among different types of breeding swine farms in Spain. Prev. Vet. Med. 1995;23:33–40. [Google Scholar]
- Chae C., Kim O., Choi C., Min K., Cho W.S., Kim J., Tai J.H. Prevalence of porcine epidemic diarrhea virus and transmissible gastroenteritis virus infection in Korean pigs. Vet. Rec. 2000;147:606–608. doi: 10.1136/vr.147.21.606. [DOI] [PubMed] [Google Scholar]
- Chang S.H., Bae J.L., Kang T.J., Kim J., Chung G.H., Lim C.W., Laude H., Yang M.S., Jang Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells. 2002;14:295–299. [PubMed] [Google Scholar]
- Chasey D., Cartwright S.F. Virus-like particles associated with porcine epidemic diarrhea. Res. Vet. Sci. 1978;25:255–256. doi: 10.1016/S0034-5288(18)32994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz D.J., Kim C.J., Shin H.J. Phage-displayed peptides having antigenic similarities with porcine epidemic diarrhea virus (PEDV) neutralizing epitopes. Virology. 2006;354:28–34. doi: 10.1016/j.virol.2006.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haan C.A., Te Lintelo E., Raaben M., Wurdinger T., Bosch B.J., Rottier P.J. Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range. J. Virol. 2006;80:10909–10918. doi: 10.1128/JVI.00950-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte M., Laude H. Sequence of the spike protein of the porcine epidemic diarrhea virus. J. Gen. Virol. 1994;75:1195–1200. doi: 10.1099/0022-1317-75-5-1195. [DOI] [PubMed] [Google Scholar]
- Egberink H.F., Ederveen J., Callebaut P., Horzinek M.C. Characterization of the structural proteins of porcine epizootic diarrhea virus, strain CV777. Am. J. Vet. Res. 1988;49:1320–1324. [PubMed] [Google Scholar]
- Fan J.H., Li Y.J. Cloning and sequence analysis of the M gene of porcine epidemic diarrhea virus LJB/03. Virus Genes. 2005;30:69–73. doi: 10.1007/s11262-004-4583-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follis K.E., York J., Nunberg J.H. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell–cell fusion but does not affect virion entry. Virology. 2006;350:358–369. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. Coronaviruses. In: Knipe D.M., Howley P.M., Griffin D.E., editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 1187–1197. [Google Scholar]
- Hofmann M., Wyler R. Quantitation, biological and physicochemical properties of cell culture-adapted porcine epidemic diarrhea coronavirus (PEDV) Vet. Microbiol. 1989;20:131–142. doi: 10.1016/0378-1135(89)90036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua R.H., Zhou Y.J., Wang Y.F., Hua Y.Z., Tong G.Z. Identification of two antigenic epitopes on SARS-CoV spike protein. Biochem. Biophys. Res. Commun. 2004;319:929–935. doi: 10.1016/j.bbrc.2004.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T.J., Han S.C., Yang M.S., Jang Y.S. Expression of synthetic neutralizing epitope of porcine epidemic diarrhea virus fused with synthetic B subunit of Escherichia coli heat-labile enterotoxin in tobacco plants. Protein Expr. Purif. 2006;46:16–22. doi: 10.1016/j.pep.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Kweon C.H., Kwon B.J., Jung T.S., Kee Y.J., Hur D.H., Hwang E.K., Rhee J.C., An S.H. Isolation of porcine epidemic diarrhea virus (PEDV) in Korea. Korean J. Vet. Res. 1993;33:249–254. [Google Scholar]
- Li B.X., Ge J.W., Li Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology. 2007;365:166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G.P., Feng Y.J., Gao F., Wang J.Z., Liu C., Li Y.J. Biochemical and biophysical characterization of the transmissible gastroenteritis coronavirus fusion core. Biochem. Biophys. Res. Commun. 2005;337:1301–1307. doi: 10.1016/j.bbrc.2005.09.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Song D.S., Ha G.W., Park B.K. Cloning and further sequence analysis of the spike gene of attenuated porcine epidemic diarrhea virus DR13. Virus Genes. 2006;35:55–64. doi: 10.1007/s11262-006-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., Debouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.B., Feng L., Shi H.Y., Chen J.F., Liu S.W., Chen H.Y., Wang Y.F. Spike protein region (aa 636–789) of porcine epidemic diarrhea virus is essential for induction of neutralizing antibodies. Acta Virol. 2007;51:327–338. [PubMed] [Google Scholar]
- Sun D.B., Lang H.W., Shi H.Y., Chen J.F., Cui X.C., Wang C.B., Tong Y.E., Feng L. Screening and identification of B cell epitopes of porcine epidemic diarrhea virus spike glycoprotein by phage display. Prog. Biochem. Biophys. 2007;34:971–977. [Google Scholar]
- Takahashi K., Okada K., Ohshima K. An outbreak of swine diarrhea of a new-type associated with coronavirus-like particles in Japan. Jpn. J. Vet. Sci. 1983;45:829–832. doi: 10.1292/jvms1939.45.829. [DOI] [PubMed] [Google Scholar]
- Timoney J.F., Gillespie J.H., Scott F.W., Barlough J.E. Comstock Publishing Associates; Ithaca: 1998. Hagan and Bruner's Microbiology and Infectious Diseases of Domestic Animals. pp. 897–898. [Google Scholar]
- Wu X.D., Shang B., Yang R.F., Yu H., Ma Z.H., Shen X., Ji Y.Y., Lin Y., Wu Y.D., Lin G.M., Tian L., Gan X.Q., Yang S., Jiang W.H., Dai E.H., Wang X.Y., Jiang H.L., Xie Y.H., Zhu X.L., Pei G., Li L., Wu J.R., Sun B. The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells. Cell Res. 2004;14:400–406. doi: 10.1038/sj.cr.7290240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo S.G., Hernandez M., Krell P.J., Nagy E.E. Cloning and sequence analysis of the spike gene of porcine epidemic diarrhea virus Chinju99. Virus Genes. 2003;26:239–246. doi: 10.1023/A:1024443112717. [DOI] [PMC free article] [PubMed] [Google Scholar]


