Highlights
-
•
Two different disease manifestations in 2 fur seals associated with P. multocida.
-
•
Whole genome sequencing revealed the presence of within-host diversity of P. multocida, a novel finding for this important pathogen.
-
•
The isolates from several internal organs of the two seals belonged to the same genetic lineage.
-
•
Results suggest that transiting birds could have been responsible for introducing the outbreak strain.
Keywords: Whole genome sequencing, Bioinformatics, Pasteurella multocida pinnipeds, Pneumonia, Septicemia
Abstract
Pasteurella multocida is a heterogeneous bacterium, which has the capacity to cause disease in a wide range of host species and is also recognized as an important zoonotic pathogen. Two sequential deaths in captive fur seals occurred at Sea World, Australia during December 2017. A fibrinosuppurative bronchopneumonia in a Subantarctic fur seal (Arctocephalus tropicalis) resulted in death within 24 h of nonspecific signs of illness, whereas a septic peritonitis in a New Zealand fur seal (Arctocephalus forsteri) resulted in death within 12 h of clinical presentation. The cases happened within three days in two different pool locations, although both had previously been housed in the same area. A total of six Pasteurella multocida isolates were obtained from several internal organs at necropsy in both cases and were subjected to whole genome sequencing and phylogenomic analysis. In-silico typing of the isolates revealed that all belonged to Multi-Locus Sequence Type 7 and carried lipopolysaccharide outer core biosynthesis loci Type 3. Phylogenomic analysis of the isolates confirmed that the isolates were near identical at the core genome level, suggesting acquisition from a common source. The results also revealed the presence of within host and across animal diversity of P. multocida isolates for the first time even in a clearly connected outbreak.
1. Introduction
Pasteurella multocida is a Gram-negative bacterium that can cause disease in a wide range of animals, being associated with fowl cholera in avian species, haemorrhagic septicaemia in bovine species, atrophic rhinitis in swine, and snuffles in lagomorphs (Harper et al., 2006). As well, P. multocida is an opportunistic organism which lives as a commensal in the upper respiratory system of different animal species including mammals, birds and reptiles (Wilson and Ho, 2013). The organism is also an important zoonotic pathogen and responsible for the majority of human wound infections following animal bites (Kimura et al., 2004). It has been suggested that P. multocida can even infect a human host through secretion droplets via scratching, licking and kissing (Kimura et al., 2004).
Several members of the family Pasteurellaceae including Pasteurella canis and Pasteurella stomatis have been found to be part of the normal flora of healthy pinnipeds kept in captivity (Hansen et al., 2012). In marine mammals, pasteurellosis has been reported as an acute or peracute septicemia, sometimes with no obvious clinical signs other than sudden death (Dunn et al., 2001). P. multocida has been associated with acute hemorrhagic bronchopneumonia, hemorrhagic tracheitis, enteritis, bacteraemia and intestinal hemorrhage in cetaceans (Dunn et al., 2001). In pinnipeds, pasteurellosis has been reported as causing disease in different species including both phocids and otariids (Dunn et al., 2001). Indeed, it has been suggested that all pinniped species are susceptible to septicemia caused by P. multocida (Dunn et al., 2001).
P. multocida isolates are classified into five capsular serogroups (A, B, D, E and F) in the Carter scheme (Carter, 1955; Rimler and Rhoades, 1987) and into sixteen somatic (Heddleston) serovars (1–16) based on their lipopolysaccharide (LPS) antigens (Brogden et al., 1978; Heddleston et al., 1972). Recently, an LPS multiplex PCR (LPS-mPCR) has been developed to assign P. multocida isolates to one of the 8 LPS genotypes (L1 to L8) based on their LPS outer core biosynthesis locus (Harper et al., 2015). There has been no report on the Heddleston serovar of the P. multocida isolates associated with disease in marine mammals to the authors’ knowledge. However, Carter type A P. multocida has been reported previously in association with fatal septicaemia in a harbor seal in Canada (Vedros and Giard, 1982).
Here we report on the usage of whole genome sequencing to examine the strain relatedness of P. multocida isolates obtained from two sudden, sequential cases of death in captive fur seals at Sea World Australia. A fibrinosuppurative bronchopneumonia in a Subantarctic fur seal (Arctocephalus tropicalis) resulted in death within 24 h of nonspecific signs of illness, whereas a septic peritonitis in a New Zealand fur seal (Arctocephalus forsteri) resulted in death within 12 h of clinical presentation.
2. Materials and methods
2.1. Case history
Twelve pinnipeds of various species were housed in the relevant Sea World Australia pool system at the time of these cases. There were five different enclosures within the pool with a flow through water system with some recirculation.
Case 1 involved a male Subantarctic fur seal that had been found stranded on a beach near Napier, New Zealand in November 2006. The animal was deemed non-releasable and a decision was made to keep it in the collection at Marineland, Napier. In 2014, the animal was transported to Sea World Australia, Gold Coast, Queensland, weighing 87 kg at the time of transportation. Prior to transport, several tests including biochemistry, complete blood count, PCR for the detection of Mycobacterium tuberculosis complex and serology for phocine morbillivirus were performed (the latter two giving negative results). The seal’s right eye was enucleated under general anaesthesia in 2015. The seal incurred minor bite wounds to the ventral abdomen inflicted by conspecifics and treated with oral amoxicillin/clavulanic acid in 2017. This seal also had mild recurring neurologic episodes with apparent seizures over the prior years.
Case 2 involved a male New Zealand fur seal that was found at Brunswick Heads, New South Wales, Australia in August 2011. The seal was transported to Sea World Australia for treatment as it was underweight and dehydrated. It was deemed non-releasable after treatment that included the administration of dexamethasone and subcutaneous fluids to stabilize the animal’s condition and prophylactic amoxicillin/clavulanic acid therapy for minor wound management. Two negative intradermal skin tests for Mycobacterium tuberculosis complex were obtained during the quarantine period. The seal was estimated to be born in 2010 and weighed 14.18 kg at admission.
2.2. Gross pathology
A blood sample was collected from the Subantarctic fur seal prior to death for biochemical and hematological analysis. Both animals’ bodies were subjected to necropsy examination at Sea World, Australia. Tissue samples from adrenal glands, thyroid glands, femoral and tibial bone marrow, brain, kidney, liver, lung, lymph node (axillary lymph nodes in case one, and mesenteric lymph nodes in case two), pancreas, spleen, testicle and epididymis were obtained for histopathological evaluation. Fluid from the pleural cavity (case 1) and peritoneal cavity (case 2) as well as urine samples, fecal swabs, sections of liver, lung, spleen, heart and kidney were obtained for bacteriological culture.
2.3. Histopathology
All tissues were fixed in 10% neutral buffered formalin and embedded in paraffin. Sections were stained with haematoxylin and eosin for histological evaluation.
2.4. Bacteriology, parasitology and virology
Fresh tissue samples were transferred on ice to the laboratory where sterile swabs were inoculated onto Columbia agar with 5% horse blood (HBA) and MacConkey agar No. 2 (bioMerieux Australia Pty Ltd, Murarrie, QMA) and incubated aerobically at 37 °C for 48 h. Potentially significant organisms were single colony picked. Bacterial identification was based on colony morphology, Gram-stain, the capacity of organisms to produce cytochrome C oxidase, catalase and MALDI-TOF mass spectrometer (bioMerieux).
Fecal swab samples were inoculated onto xylose lysine deoxycholate (XLD) agar and into selenite F broth (bioMerieux Australia Pty Ltd, Murarrie, Queensland, Australia), and incubated aerobically at 35 °C for 24 h. The incubated selenite F broth was then streaked onto XLD agar and incubated aerobically at 35 °C for 24 h. The plates were then monitored for the possible presence of Salmonella. Faeces from Case 2 was tested using a canine multiplex PCR for detection of Salmonella species, Campylobacter jejuni, Giardia species, Cryptosporidium canis, Coronavirus and Parvovirus.
Multiple water samples and swabs of the pool system as well as swabs of shade structures and haul-out platforms were collected as well as food fish, all as the possible sources of the infection. The samples were processed as described above and screened for P. multocida.
2.5. Whole genome sequencing
The six P. multocida isolates obtained from the two seals were subjected to whole genome sequencing (WGS). DNA was extracted using the DNeasy UltraClean Microbial Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions from overnight cultures grown on 5% sheep blood agar. The DNA samples were then sequenced on the Illumina NextSeq 500 platform (150 bp paired ends) by the Australian Centre for Ecogenomics at the University of Queensland (St Lucia QLD 4072, Australia). Raw Illumina sequencing data was quality filtered to remove Illumina adaptor sequences, low quality reads and reads shorter than 75 bp using Nesoni v0.132 (Victorian-Bioinformatics-Consortium, 2013). The qualities of the reads were assessed using FastQC v0.11.5 (Andrews, 2010). Quality-trimmed paired end reads were then assembled using SPAdes v3.10.1 (Bankevich et al., 2012). In-silico multi-locus sequence typing (MLST) profiling was performed using MLST v2.8 (Jolley and Maiden, 2010; Seemann) and the P. multocida RIRDC-MLST scheme (Subaaharan et al., 2010). A local database consisting of the gene sequences representing the 16 Heddleston serovar LPS outer core biosynthesis loci was prepared and Blastn v2.2.31+ was used to perform in-silico LPS typing. The relevant contig containing the outer core biosynthesis locus was extracted, Geneious 10.0.6 was used for annotation of the region and ClustalW v2.1 for alignments and comparision to the corresponding loci in P. multocida type strain PM70. As well, the quality trimmed raw Illumina reads were mapped to PM70 complete genome and the LPS gene coordinates were monitored for changes.
In order to investigate the relationship between the seal isolates and the other P. multocida strains, a total of twenty-five complete genomes were obtained from Genbank using ncbi-genome-download (https://github.com/kblin/ncbi-genome-download). Parsnp v1.2 (Treangen et al., 2014) was used for core genome alignment and SNP calling in the draft genome assemblies with P. dagmatis strain NCTC11617 as an outgroup as previously described (LeCount et al., 2018). A phylogenetic tree was then constructed with RAxML x8.2.9 using the core genome SNP alignment after removal of the recombination sites by Gubbins v2.1.0 (Stamatakis, 2014). A general-time reversible nucleotide substitution model with a GAMMA correction for site variation was used for tree construction (bootstrap 1000 with Lewis ascertainment correction). The generated phylogenetic trees were visualised using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) and Evolview (He et al., 2016). PHYLOViZ v2.0 was used to build the minimum spanning tree for the seal isolates (Nascimento et al., 2017).
3. Results
3.1. Clinical findings (Case 1)
On 5th of December 2017, a Subantarctic fur seal was found unresponsive at the bottom of the pool. Animal care workers had previously reported anorexia, lethargy and signs of a potential seizure over the preceding 24–48 h. Once stimulated to move to the surface from the bottom of the pool, the seal was observed floating right side up and holding onto the side of the pool with its right fore and hind flippers. When submerged under water, the seal made minimal effort to return to the surface, and in doing so would return to hold on to the side of the pool. Cefovecin, diazepam, flunixin meglumine and subcutaneous fluids were administered poolside. Following a one hour period of observation, concern that the seal was beginning to aspirate water resulted in the decision to move the animal to the Veterinary Quarantine Centre (VQC) on-site at Sea World, Australia. Once dry-docked, amikacin, dexamethasone, thiamine, intravenous fluids and oxygen therapy was instituted. The seal remained depressed and prostrate with marked respiratory difficulty evidenced by open-mouth breathing and significant thoracic excursions, which worsened through the night. On December 6th, following a 14 h therapeutic period, the patient remained moribund in a critical condition. Due to a poor prognosis, a decision was made to euthanize the seal with an intramuscular overdose of tiletamine-zolazepam (Zoletil®).
3.2. Clinical findings (Case 2)
On 8th of December 2017, a New Zealand fur seal was seen with diarrhoea and regurgitation following feeding. Animal care workers noted anorexia and lethargy persisting for 6 to 12 h prior to the onset of these symptoms. The seal was retrieved from the pool and found to be bright, alert and responsive; a brief dry dock at the VQC facilitated administration of cefovecin, and the seal was returned to a dry enclosure at 1 pm for day and overnight watch. At 3:30 pm, the seal was found dead in the enclosure.
3.3. Hematology and biochemistry
Full details of the biochemistry and hematology results for the Subantractic fur seal (Case 1) are presented in Supplementary Table 1. An in-house blood smear at the VQC determined erythrocytes to be normochromic and normocytic, and platelets morphologically normal with only one band neutrophil observed in the whole slide. In-house cytology of pleural fluid confirmed a significant presence of polymorphonuclear cells, predominantly lytic neutrophils and a few macrophages, in addition to a Gram-negative bacterial population.
More detailed results from a reference laboratory corrected the leukocyte parameters for the presence of nucleated erythrocytes. Neutrophils were found to contain Döhle bodies, and the cell line exhibited a marked left shift with toxic change. These findings confirmed in-house laboratory observations of a peripheral panleukopenia. A low protein level with concurrent decreases in albumin and globulin levels and an inverted calcium-phosphorus ratio were the most striking observations on the biochemical analysis.
For Case 2 (the New Zealand fur seal), reference laboratory testing following necropsy revealed that the turbid peritoneal fluid contained erythrocytes (45.6 × 1012/L), leukocytes (135 × 1012/L) and protein (83.8 g/L). Both cytospin and direct smears of the abdominal fluid presented with high cellularity, containing predominantly degenerate neutrophils (>90%), with lower numbers of large mononuclear cells and lymphocytes amidst moderate numbers of erythrocytes in a lightly stippled background with numerous lysed cells and cell debris. A significant bacterial population consisting of bipolar staining short rods were found both free and phagocytised by leukocytes. Neither erythrophagia nor haemosiderin were identified.
3.4. Necropsy and histopathology
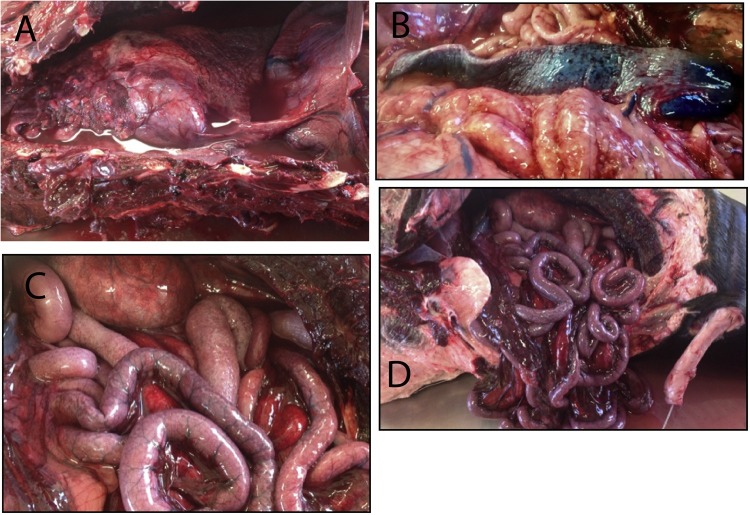
The Subantarctic fur seal was an eleven-year old male weighing 118 kg at necropsy. The axillary lymph nodes were bilaterally enlarged. Subcutaneous emphysema and hemorrhage were found caudo-dorsally proximal to the hind flippers, and subcutaneous edema was prominent ventrally. A 2 by 20 mm pink-red lesion characterized by thinning mucosa was found at the ventral aspect of the oesophagus in the mid cervical region. The trachea and bronchi contained a frothy pink-white fluid. The lung parenchyma was consolidated and contained multifocal to coalescing asymmetric pink-red foci ranging from 5 to 20 mm in size; these foci contained small bubbles (Fig. 1 A) with cut sections of the lung sinking in 10% buffered formalin. Petechial hemorrhages were visualized on the surface of the epicardium and pericardial sac, with a mild endocardiosis at the mitral valve characterized by irregular nodules ranging from 3 to 5 mm in size. The spleen was contracted and blue-purple in colour, with petechial hemorrhages found on the visceral surface (Fig. 1B). The descending colon and rectum contained a yellow-white fluid diarrhea. Morphological diagnoses included a peripheral panleukopenia (severe, acute), panhypoproteinemia (moderate, acute) and pleural effusion (severe, acute to subacute; effusive, exudative, hemorrhagic). Histopathological evaluation of several lung sections revealed a severe, acute diffuse fibrinosuppurative bronchopneumonia with pleuritis and numerous small Gram-negative bacilli. In the heart, there was mild diffuse epicardial mesothelial cell hyperplasia and hypertrophy. In the liver, there was centrilobular congestion and mild hepatocellular fatty change.
Fig. 1.
Necropsy images of the Subantarcticractic fur seal (panels A and B) and New Zealand fur seal (panels C and D). The lung parenchyma was consolidated and emphysematous, presenting with fibrinous adhesions and a contracted, mottled pink-red-white appearance. The lung lobes were surrounded by an effusive pyoserosanguinous pleural fluid (panel A). The blue-purple spleen of the Subantractic fur seal, contracted with dark red petechial hemorrhages on its surface (panel B). The serosal surface of the stomach, small intestine and large intestine of the New Zealand fur seal were mottled brown-pink-purple-red-tan with a coalescing divot pattern recognizable at the antimesenteric surface (panel C). Severe diffuse congestion and inflammation was observed in the alimentary tract of the New Zealand fur seal, the urethra was catheterized for urine collection (panel D).
The New Zealand fur seal was a seven year old male which weighed 85 kg at necropsy. Upon opening the abdominal cavity, 2–3 L of brown-red exudative fluid containing white flocculent material was observed. The serosal surface of the stomach was mottled brown-pink-red with prominent vasculature, and the mesentery and omentum were severely congested (Fig. 1C). Both the small and large intestine were congested and had a pink-purple-red-tan coalescing divot pattern recognizable at the antimesenteric surface (Fig. 1C and D). The mesenteric lymph nodes were enlarged and vessels of the alimentary tracts were dilated. The spleen was contracted and blue-purple in color and petechial hemorrhages were found on the visceral surface. Bile imbibition was present along the serosal surface of the liver underlying the rib cage. No significant gross lesions were detected in the tongue, tonsils, salivary glands, thyroid glands, pharynx, larynx, stomach, testicles, epididymis and urinary bladder. Histopathological changes were limited to minimal inflammation in abdominal adipose tissue accompanied by numerous bacteria (mainly small Gram-negative bacilli), as well as congestion of many tissues.
3.5. Microbiology
P. multocida was cultured as either a pure growth or the predominant organism in a mixed growth after overnight incubation from lung, thoracic fluid, heart, kidney, liver and spleen of the Subantarctic fur seal (Case 1), and from abdominal fluid, left lung, mesenteric lymph node, heart, kidney, liver and spleen of the New Zealand fur seal (Case 2). The heaviest growths of P. multocida were from lung and thoracic fluid of Case 1 and abdominal fluid of Case 2. All the environmental samples were negative for P. multocida using routine diagnostics procedures.
Faecal analysis revealed the presence of a few white blood cells suggesting a possible infectious process was underway. However, neither pathogenic bacteria nor viruses were detected by routine culture or PCR of the faeces.
3.6. Genomic analysis
Two of the isolates from Case 1 (isolates PM2371 and PM2372, obtained from the lung and thoracic fluid, respectively) as well as four isolates from Case 2 (isolates PM2373 and PM2375 obtained from the lung and mesenteric lymph node, respectively, and isolates PM2374 and PM2376, both obtained from the abdominal fluid) were subjected to further analysis via WGS. The in-silico MLST typing grouped all six isolates into MLST sequence Type 7. Use of the in-house LPS database of the eight previously described LPS outer core biosynthesis loci types suggested that all six isolates were carrying an identical LPS outer core biosynthesis locus, closely related to that of LPS Type 3. Detailed analysis showed that the LPS loci of the seal isolates was 6191 bp in length and had 184 nucleotide differences with that from PM70. The natC gene in the seal isolates appeared to carry a non-synonymous change at position 290 which results in introduction of a stop codon. In addition to that, the isolates harbored an insertion at position 777 of the gatG gene resulting in a CDC frame shift.
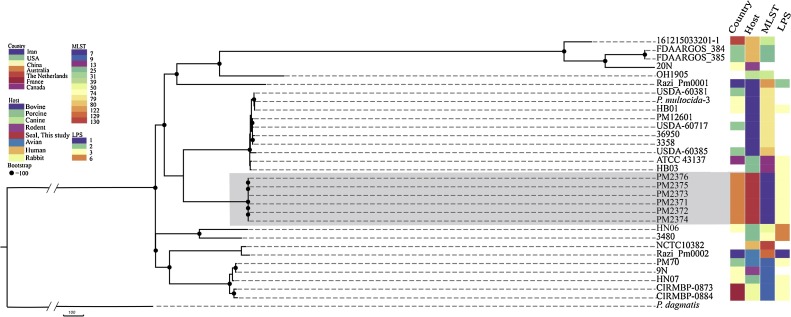
Maximum likelihood analysis of core genome SNPs clustered all the P. multocida isolates obtained in the current study together as a well-supported subclade, that was distinct from the other twenty five isolates (Fig. 2 ). The seal isolates differed by roughly 550 SNPs from the next nearest clade (Fig. 2). This second clade which shared the same ancestor with seal isolates consisted of 8 bovine and 2 porcine isolates, with three of the bovine isolates (sequences GCA_002859525, GCA_002859285, and GCA_002859465) randomly picked from a study on naturally occurring respiratory disease in a herd that consisted of multisourced feedlot cattle, thus none of the GenBank isolates were from outbreak situations (DeDonder et al., 2016) (Supplementary Table 2).
Fig. 2.
Maximum likelihood tree of the core genome SNP of the seal isolates compared to those of the complete genomes obtained from GenBank. A total of 16,525 core genome SNPs were identified, using the complete genomes of P. dagmatis as outgroup. The seal isolates made a separate cluster (highlighted with the grey box).
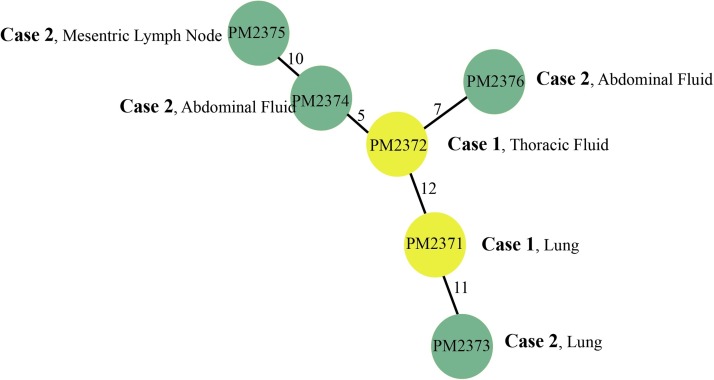
In order to investigate the isolate relatedness within the outbreak, draft genome assemblies of the seal isolates were then aligned to the complete genome of P. multocida PM70 for SNP calling with a total number of 45 SNPs identified between the six seal isolates. The minimum spanning tree demonstrated the case one and case two isolates of P. multocida share a very recent ancestor (Fig. 3 ).
Fig. 3.
Minimum spanning tree of the seal isolates. Yellow circles represent case one isolates and green circles represent case two isolates. Numbers on the branches represent number of core genome SNP differences between the isolates.
4. Discussion
While several members of the family Pasteurellaceae have been identified as part of the normal flora of healthy pinnipeds, P. multocida has been isolated from cases of septicaemia, mortality and pneumonia in pinnipeds as well as cetaceans (Dunn et al., 2001; Hansen et al., 2012; Nielsen et al., 2013). Fatal septicaemic pasteurellosis has been reported in cetaceans and pinnipeds, where the most common origin of generalized pasteurellosis in captive and wild pinnipeds is bite wounds inflicted by conspecifics (Hansen et al., 2012). Pasteurellosis in marine mammals is often presented as an acute or peracute septicaemia resulting in fatality with or without clinical symptoms (Dunn et al., 2001). The clinical signs, if present, can include nonspecific signs such as altered behavior, anorexia, lethargy and listless swimming a few hours preceding death (Dunn et al., 2001). Typical necropsy findings include pneumonia, enteritis, myocarditis with epicardial/pericardial petechiation, hemorrhage, hepatitis, nephritis, splenitis, necrotic peritonitis, cervical inflammation and necrosis of adipose overlying the cervical esophagus (Dunn et al., 2001). However, the per-acute nature of pasteurellosis might result in few gross lesions.
In the seal in Case 1, in-house tests and necropsy findings such as lymphadenopathy and cardiac changes suggested a severe inflammatory response and an acute sepsis due to an invasive bacterial infection creating a pyogenic focus in the thorax via an oesophageal portal of entry. Severe secretory diarrhoea was consistent with the biochemical findings of panhypoproteinemia characterized by concurrent decreases in albumin and globulin, suggesting blood loss and/or protein-losing enteropathy.
A severe effusive, exudative, hemorrhagic and septic peritonitis was observed upon necropsy in the fur seal in Case 2, with bacteria being present in extracellular and intracellular fluids. Given the apparently good health of this seal through its years of residence at Sea World, and the peracute onset of clinical signs resulting in sudden death, a severe bacteraemic sepsis as a sequelae to peritonitis was suspected. Isolation of P. multocida mostly as pure culture, from several internal sites and organs, clearly identified this Gram-negative organism as the cause of septicaemia and death in the two seals.
In-silico MLST typing revealed all isolates from both cases belonged to ST 7. Historically, ST 7 consists of six isolates, all obtained from Australian animals, five from chickens suffering fowl cholera and one isolate from a pig suffering from septicaemia due to P. multocida (https://pubmlst.org/pmultocida/). To the authors’ knowledge, no seal isolates have been examined by MLST or any other genotyping method.
The clinical presentation of septic pasteurellosis is suspected to appear most probably after the levels of endotoxin reaches a certain concentration (Wilkie et al., 2012). The key antigenic component of the endoxtoxin of P. multocida is the LPS, which also forms the basis of the Heddleston serotyping scheme (Heddleston et al., 1972). Our in-silico typing showed that all the seal isolates carried LPS outer core biosynthesis loci type 3, which is the most prevalent LPS type carried by the P. multocida isolates associated with fowl cholera in Australia (Harper et al., 2013). However, our data suggested that the LPS of the seal isolates have a unique gatG sequence which has not been previously reported. Consequently, the results obtained from MLST and LPS typing of the seal isolates are possibly suggesting that the birds transiting through the pools are responsible for introducing the outbreak isolate to the seals. Several species of seabirds frequent the pools at Sea World Australia, which supports this hypothesis. However, more studies including sampling the guano of the visiting birds would be needed to get more information about the source of the infection.
P. multocida is relatively resistant to environmental stresses being known to survive in turkey blood dried on glass for eight days and in soil held at 20 °C, pH7 and moisture content of 50% for up to 100 days (Glisson et al., 2013). Hence, despite routine disinfection protocols, risk factors including exposure via wild seabird guano which is often deposited on the shade sails above the pools will remain. Guano can be a potential source of P. multocida as the organism has been previously isolated from the cloaca of apparently healthy web-footed birds in a Danish study of commercial poultry production systems (Muhairwa et al., 2000). In addition to that, an association between prevalence of pasteurellosis in seals in an aquarium in the USA with waterfowl migration has been suggested earlier (Dunn et al., 2001).
Two out of twelve animals were affected in December 2017 and the peracute nature of disease, resulting mortality linked to the same bacterial species led to an initial belief that the two cases represented an outbreak of pasteurellosis. Our use of WGS and the comparison of the draft genome assembly of the six seal isolates to those obtained from the public database provided the needed resolution to confirm the isolates belonged to the same genetic lineage, thus confirming that an outbreak of pasteurellosis had indeed occurred. This is beyond what routine MLST would provide. As WGS is only now starting to be applied to outbreaks of pasteurellosis, there are limited similar studies. However, in a study on two on-going outbreaks in turkeys, each in a different US state, whole genome sequencing clustered isolates from each outbreak into distinct clusters (LeCount et al., 2018).
In addition to that, the whole genomic data allowed insights into the level of diversity of P. multocida from within an animal in an outbreak as well as across animals in an outbreak. The minimum spanning tree demonstrated the case one and case two isolates of P. multocida share a very recent ancestor and the possibility of transmission through fomites from a common source. It also showed genetic diversity even within the same seal with the Case 1 thoracic fluid and lung isolates differing by 12 SNPs. This finding suggests that this seal has been originally infected with at least two different strains. As single colonies were initially picked from the primary plates for both tissues, it is not possible to determine if both of the genetic types seen in Case 1 were present in both sampled tissues. However, in Case 2, the two available isolates from the abdominal fluid (two separate fluid collections were cultured) yielded isolates that differed by 5 or 7 SNPs from the Case 1 thoracic fluid isolate. Hence, this analysis demonstrated that the Case 2 isolates are closely related with the two isolates obtained from Case 1, suggesting a common source.
Given the peracute nature of pasteurellosis in these two cases, it is very unlikely to assume one strain infected case 1 and then underwent microevolution within seal one before transferring to seal two. Rather, our results suggest that the seals most probably obtained an infecting population from a common external source such as seabird guano or fomites. Mixed colonization by several bacterial lineages is relatively common among human bacterial pathogens such as Staphylococcus aureus (Didelot et al., 2016) and Psuedomonas aeruginosa in cystic fibrosis patients (Sherrard et al., 2017). Mixed colonization by different genetic types within the one host has not previously reported in P. multocida.
Future investigations into pasteurellosis outbreaks would help in providing more insight if multiple single colonies were selected from multiple tissues in multiple animals. Sampling the oropharyngeal of healthy seals is also needed to see whether seals can be asymptomatic carriers of such pathogenic P. multocida strains. As well, where relevant resources and capacities are available, future studies should use a metagenomic approach in which the full genetic diversity of P. multocida in tissues and relevant environmental samples could be determined.
Studies in rabbits have demonstrated that direct contact with nasal secretions is an important route of transmission of pasteurellosis (DiGiacomo et al., 1987). Given the two seals were kept in different but adjacent pools since 17th of November 2017, it is unlikely that there were any direct contact with nasal secretion from Case 1 to Case 2. However, the fact that water moved freely between the pools would have made it possible for nasal secretions from the seal in Case 1 one to reach the adjacent pools. Excess rainfall in the region of Sea World Australia in November and December 2017 qualifies as another risk factor, allowing stagnant freshwater to accumulate and salinity in the pinniped pools to decrease, potentially facilitating overgrowth of bacterial organisms.
5. Conclusion
This study describes two different disease manifestations in two fur seals associated with P. multocida. Our use of whole genome sequencing has allowed a confirmation that the two cases were directly connected with all six available isolates being nearly identical at the core genome level. As well, the whole genome data analysis has revealed the presence of genetic diversity within animals and across animals even in a clearly connected outbreak with only days between the two animal sampling times.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Acknowledgments
The authors thank Dr. John Mackie from QML Pathology Vetnostics, Murarrie, QLD 4172, Australia for the histopathology report and the trainers and veterinary assistants at Sea World Australia for their assistance during the treatment of the two animals.
The authors also thank Dr. Marina Harper from Monash University, Clayton, VIC 3800, Australia for providing us the sequences for the LPS from the P. multocida type strains P1997, P2100, P903 and P2237.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.vetmic.2019.03.017.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data. [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden K.A., Rhoades K.R., Heddleston K.L. A new serotype of Pasteurella multocida associated with fowl cholera. Avian Dis. 1978;22:185–190. [PubMed] [Google Scholar]
- Carter G.R. Studies on Pasteurella multocida. I. A hemagglutination test for the identification of serological types. Am. J. Vet. Res. 1955;16:481–484. [PubMed] [Google Scholar]
- DeDonder K.D., Apley M.D., Li M., Gehring R., Harhay D.M., Lubbers B.V., White B.J., Capik S.F., KuKanich B., Riviere J.E., Tessman R.K. Pharmacokinetics and pharmacodynamics of gamithromycin in pulmonary epithelial lining fluid in naturally occurring bovine respiratory disease in multisource commingled feedlot cattle. J. Vet. Pharmacol. Ther. 2016;39:157–166. doi: 10.1111/jvp.12267. [DOI] [PubMed] [Google Scholar]
- Didelot X., Walker A.S., Peto T.E., Crook D.W., Wilson D.J. Within-host evolution of bacterial pathogens. Nat. Rev. Microbiol. 2016;14:150–162. doi: 10.1038/nrmicro.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiacomo R.F., Jones C.D., Wathes C.M. Transmission of Pasteurella multocida in rabbits. Lab. Anim. Sci. 1987;37:621–623. [PubMed] [Google Scholar]
- Dunn J.L., Buck J.D., Robeck T.R. CRC Handbook of Marine Mammal Medicine. CRC Press; 2001. Bacterial diseases of cetaceans and pinnipeds. [Google Scholar]
- Glisson J.R., Hofacre C.L., Christensen J.P. Fowl cholera. In: Swayne D.E., Glisson J.R., McDougald L.R., Nolan L.K., Suarez D.L., Nair V.L., editors. Diseases of Poultry. Wiley-Blackwell; Ames: 2013. pp. 807–823. [Google Scholar]
- Hansen M.J., Bertelsen M.F., Christensen H., Bisgaard M., Bojesen A.M. Occurrence of Pasteurellaceae bacteria in the oral cavity of selected marine mammal species. J. Zoo Wildl. Med. 2012;43:828–835. doi: 10.1638/2011-0264R1.1. [DOI] [PubMed] [Google Scholar]
- Harper M., Boyce J.D., Adler B. Pasteurella multocida pathogenesis: 125 years after Pasteur. FEMS Microbiol. Lett. 2006;265:1–10. doi: 10.1111/j.1574-6968.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- Harper M., St Michael F., John M., Vinogradov E., Steen J.A., van Dorsten L., Steen J.A., Turni C., Blackall P.J., Adler B., Cox A.D., Boyce J.D. Pasteurella multocida Heddleston serovar 3 and 4 strains share a common lipopolysaccharide biosynthesis locus but display both inter- and intrastrain lipopolysaccharide heterogeneity. J. Bacteriol. 2013;195:4854–4864. doi: 10.1128/JB.00779-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper M., John M., Turni C., Edmunds M., St Michael F., Adler B., Blackall P.J., Cox A.D., Boyce J.D. Development of a rapid multiplex PCR assay to genotype Pasteurella multocida strains by use of the lipopolysaccharide outer core biosynthesis locus. J. Clin. Microbiol. 2015;53:477–485. doi: 10.1128/JCM.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Zhang H., Gao S., Lercher M.J., Chen W.-H., Hu S. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016;44:W236–W241. doi: 10.1093/nar/gkw370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston K.L., Gallagher J.E., Rebers P.A. Fowl cholera: gel diffusion precipitin test for serotyping Pasteruella multocida from avian species. Avian Dis. 1972;16:925–936. [PubMed] [Google Scholar]
- Jolley K.A., Maiden M.C.J. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura R., Hayashi Y., Takeuchi T., Shimizu M., Iwata M., Tanahashi J., Ito M. Pasteurella multocida septicemia caused by close contact with a domestic cat: case report and literature review. J. Infect. Chemother. 2004;10:250–252. doi: 10.1007/s10156-004-0331-5. [DOI] [PubMed] [Google Scholar]
- LeCount K.J., Schlater L.K., Stuber T., Robbe Austerman S., Frana T.S., Griffith R.W., Erdman M.M. Comparison of whole genome sequencing to restriction endonuclease analysis and gel diffusion precipitin-based serotyping of Pasteurella multocida. J. Vet. Diagn. Invest. 2018;30:42–55. doi: 10.1177/1040638717732371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhairwa A.P., Christensen J.P., Bisgaard M. Investigations on the carrier rate of Pasteurella multocida in healthy commercial poultry flocks and flocks affected by fowl cholera. Avian Pathol. 2000;29:133–142. doi: 10.1080/03079450094162. [DOI] [PubMed] [Google Scholar]
- Nascimento M., Sousa A., Ramirez M., Francisco A.P., Carriço J.A., Vaz C. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017;33:128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- Nielsen K.A., Owen H.C., Mills P.C., Flint M., Gibson J.S. Bacteria isolated from dugongs (Dugong dugon) submitted for postmortem examination in Queensland, Australia, 2000-2011. J. Zoo Wildl. Med. 2013;44:35–41. doi: 10.1638/1042-7260-44.1.35. [DOI] [PubMed] [Google Scholar]
- Rimler R.B., Rhoades K.R. Serogroup F, a new capsule serogroup of Pasteurella multocida. J. Clin. Microbiol. 1987;25:615–618. doi: 10.1128/jcm.25.4.615-618.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, T. mlst Github (https://github.com/tseemann/mlst).
- Sherrard L.J., Tai A.S., Wee B.A., Ramsay K.A., Kidd T.J., Ben Zakour N.L., Whiley D.M., Beatson S.A., Bell S.C. Within-host whole genome analysis of an antibiotic resistant Pseudomonas aeruginosa strain sub-type in cystic fibrosis. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subaaharan S., Blackall L.L., Blackall P.J. Development of a multi-locus sequence typing scheme for avian isolates of Pasteurella multocida. Vet. Microbiol. 2010;141:354–361. doi: 10.1016/j.vetmic.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedros N.A., Giard R. International Association for Aquatic Animal Medicine Conference. 1982. A potential for Pasturella multocida infections in marine mammals. [Google Scholar]
- Victorian-Bioinformatics-Consortium . 2013. Nesoni. [Google Scholar]
- Wilkie I.W., Harper M., Boyce J.D., Adler B. Pasteurella multocida: diseases and pathogenesis. In: Aktories K., Orth J.H.C., Adler B., editors. Pasteurella multocida: Molecular Biology, Toxins and Infection. Springer; Berlin Heidelberg, Berlin, Heidelberg: 2012. pp. 1–22. [Google Scholar]
- Wilson B.A., Ho M. Pasteurella multocida: from zoonosis to cellular microbiology. Clin. Microbiol. Rev. 2013;26:631–655. doi: 10.1128/CMR.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.