Highlights
-
•
No cross-neutralization was detected between PEDV and PdCV.
-
•
A two-way cross-reactivity was detected between PEDV and PdCV.
-
•
Conserved epitope(s) in viral N proteins may contribute to antigenic cross-reactivity.
-
•
Prevention of PEDV and PdCV will require the development of separate virus-specific vaccine products.
Abstract
Porcine epidemic diarrhea virus (PEDV) and porcine deltacoronavirus (PdCV) cause indistinguishable clinical signs and pathological changes in swine. Here we investigated the antigenic relationship between PEDV and PdCV. We provide the first evidence that conserved epitope(s) on the respective viral nucleocapsid proteins cross-react with each other although virus neutralization cross-reactivity was not observed. As a practical matter, prevention of these two very similar diseases of swine will require the development of separate virus-specific vaccine products.
1. Introduction
Porcine epidemic diarrhea virus (PEDV), a member of genus Alphacoronavirus in the family Coronaviridae, was first identified in the United Kingdom in 1971 (Pensaert and de Bouck, 1978). For the classic European strain (CV777) of PEDV, mortality rates, while significant, are not viewed as a severe threat to European pork production (Coussement et al., 1982, Pensaert and de Bouck, 1978). Since 2010, variant strains of PEDV that differ from the classic European strain appeared in China, South Korea, Japan, and many Asian countries, causing up to 100% mortality in suckling piglets (Chen et al., 2010, Song and Park, 2012). In May 2013, PEDV was identified as a new cause of neonatal diarrhea in Iowa and rapidly spread to more than 30 states, Canada and Mexico and caused significant economic losses in the swine industry (Huang et al., 2013, Stevenson et al., 2013). Sequence analyses suggest that US PEDV strains originated from China (Huang et al., 2013).
Members of the Deltacoronavirus are largely restricted to avian species (Chu et al., 2011, Dong et al., 2007). However, porcine deltacoronavirus (PdCV) specifically the Hong Kong (HK) strains HKU15-155 and HKU15-44 were first recovered from Asian piglets in 2009, suggesting that the tropism(s) of the Deltacoronavirus are expanding to mammals (Woo et al., 2012). In February 2014 a novel PdCV was discovered first in Ohio and Indiana, and then spread rapidly in other states and Canada (Wang et al., 2014). The clinical signs of PdCV infection (profuse watery diarrhea, emesis and resultant dehydration) are indistinguishable from PEDV with the exception of the mortality rate (40–60%) for PdCV is lower than PEDV (80–90%) (Jung et al., 2014, Ma et al., 2015).
Recently, we and others demonstrated that PdCV caused severe gastrointestinal disease in both gnotobiotic and conventional piglets (Chen et al., 2015, Jung et al., 2015, Ma et al., 2015). In the US and Canada, PdCV and PEDV are “reportable diseases”. In spite of the similarity of clinical signs and pathologic changes, the serologic relationships between PEDV and PdCV are not known in detail. Understanding this fundamental question will facilitate the development of diagnostic tools and vaccine product(s) that will be effective against both viral pathogens.
2. Results and discussion
To investigate the serologic relationships between PEDV and PdCV, we prepared high quality porcine-origin PEDV and PdCV-specific polyclonal antibodies. Cross reactivity was first explored using a virus-serum neutralization (VN) assay. For this, PEDV or PdCV-specific VN titers were determined using a plaque reduction neutralization assay. As shown in Table 1 , the VN titer of four PdCV sera ranged from 382 to 544 and no VN titer was detected in PEDV and control sera when PdCV was used as antigens. Thus, only PdCV immune serum neutralized infectivity of PdCV; neither control sera nor PEDV specific antisera neutralized PdCV. A similar homologous relationship was found when PdCV serum was tested for neutralizing activity against PEDV; cross-neutralizations were not observed. The PEDV-specific VN titer of four PEDV-infected piglets ranged from 230 to 256, and no PEDV-specific VN titer was detected in PdCV and control sera.
Table 1.
Summary of virus-serum neutralizing antibody and serum IgG ELISA antibody.
| SerumA | Virus-serum neutralizing assayB |
Serum IgG ELISAC |
||
|---|---|---|---|---|
| PEDV | PdCV | PEDV | PdCV | |
| PEDV 1 | 256a | <20 | 5120a | 80a |
| PEDV 2 | 230a | <20 | 5120a | 40a |
| PEDV 3 | 256a | <20 | 5120a | 80a |
| PEDV 4 | 256a | <20 | 5120a | 80a |
| PdCV 1 | <20 | 382a | 40b | 10240b |
| PdCV 2 | <20 | 544a | 40b | 10240b |
| PdCV 3 | <20 | 544a | 80b | 10240b |
| PdCV 4 | <20 | 544a | 80b | 10240b |
| Control 1 | <20 | <20 | <20 | <20 |
| Control 2 | <20 | <20 | <20 | <20 |
| Control 3 | <20 | <20 | <20 | <20 |
| Control 4 | <20 | <20 | <20 | <20 |
Gn piglets were immunized intramuscularly twice (two weeks apart) with 0.3 mg of inactivated PEDV or PdCV antigen. For antigen preparation, 1.2 mg of purified PEDV and PdCV stocks were inactivated by 0.05% formalin, and mixed with 3 ml of Alhydrogel aluminum Hydroxide Gel Adjuvant in 1 ml of PBS. The control piglets were immunized with 1 ml of adjuvant alone. Sera were collected from PEDV antigen, PdCV antigen, or mocked-inoculated Gn piglets at week 4 post-immunization and were heat inactivated at 56 °C for 30 min.
Virus-serum neutralizing antibody titer was determined by a plaque reduction neutralization assay using PEDV or PdCV. Data are average of three independent VN assay. Value within a column followed by the different lowercase letters (a and b) are significantly different (P < 0.05).
Serum IgG titer was determined by ELISA using purified PEDV or PdCV as coated antigens.
Serum cross-reactivity was also measured by standard ELISA. Ninety-six-well plates were coated with 50 μl of highly purified PEDV or PdCV (2.5 μg/ml). Individual serum samples were tested for PEDV or PdCV-specific IgG on viral antigen-coated plates. Interestingly, PEDV antigen had an obvious cross-reactivity with all tested PdCV sera (Table 1). Three out of four PdCV sera had an ELISA titer of 80, and one PdCV sera had a titer of 40, when PEDV was used as the coating antigen. Similarly, PdCV antigen had a detectable cross-reactivity with all PEDV-immune sera (Table 1). Two out of four PEDV sera had an ELISA titer of 80, and two PEDV sera had a titer of 40, when PdCV was used as the coating antigen. In all cases, sera from control piglets were negative for ELISA titer (<20). Thus, a two-way cross-reactivity between PEDV and PdCV was detected by ELISA.
The above results clearly demonstrated that two-way cross-reactivity of PEDV and PdCV was detectable by ELISA but not VN assay. To address the possible reasons contributed to this difference, we decided to perform additional serological assays. Serologic cross-reactivity was next determined by immunofluorescence assay (IFA) in virus-infected cells. Confluent ST cells were infected with PEDV VBS2 strain or PdCV Michigan/8977/2014 strain at a multiplicity of infection (MOI) of 1.0. At 24 h post-infection, the cells were fixed and reacted with PEDV or PdCV polyclonal antibody. As shown in Fig. 1 A, strong fluorescent signals were detected in PEDV-infected ST cells whereas no signal was detected in PdCV-infected or mock-infected ST cells when stained with PEDV polyclonal antibody. Similarly, strong fluorescent signals were detected in PdCV-infected ST cells whereas no signal was detected in PEDV-infected or mock-infected ST cells when stained with PdCV polyclonal antibody. Thus, no cross-reactivity was detected by IFA.
Fig. 1.
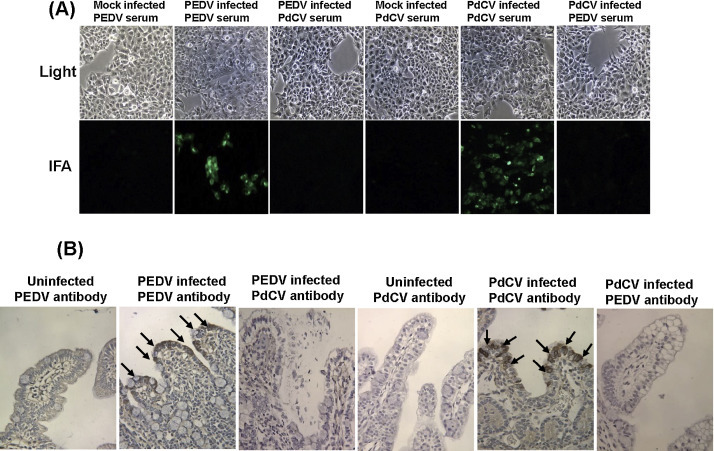
Serological cross-reactivity of PEDV and PdCV virus-infected cells and virus-infected intestinal tissues. (A) Immunofluorescence assay (IFA) in PEDV or PdCV-infected ST cells. Confluent ST cells were infected with PEDV VBS2 strain or PdCV Michigan/8977/2014 strain at a multiplicity of infection (MOI) of 1.0. After 1 h absorption, the inoculum was removed, the cells were washed once with DMEM, and fresh DMEM (supplemented with 5 μg/ml trypsin 1:250) was added, and the infected cells were incubated at 37 °C. When cytopathic effects (CPE) were observed, the cells were fixed with 4.0% (v/v) paraformaldehyde-0.2% (v/v) glutaraldehyde in 0.1 M potassium phosphate buffer (PPB), pH 7.4, for 15 min, 22 °C, followed by washing 3 times with PBS. After permeabilization with 0.1% Triton X-100 in PBS for 15 min, the cells were washed with PBS, blocked with PBS containing 2% bovine serum albumin for 1 h, 22 °C. Then cells were incubated with PEDV or PdCV polyclonal antibody. After 3 washes with PBS, the cells were further incubated with FITC-labeled rabbit anti-pig secondary antibody overnight at 4 °C. After 3 washes with PBS, samples were examined under Olympus fluorescent microscope system at The Ohio State University. Photos at upper panels were taken under a light microscope (100×). Photos at lower panels were taken under an immunofluorescent microscope (100×). (B). Immunohistochemistry (IHC) analysis of small intestine sections from Gn piglets. PEDV VBS2 infected duodenal tissue at 72 h PI, PdCV infected Michigan/8977/2014 duodenal tissue at 72 h PI, or uninfected controls were stained with anti-PEDV serum or anti-PdCV serum. Black arrows indicate positive antigens. Hematoxylin, 300×. Five-micron sections of paraffin-embedded tissues were placed onto positively charged slides. After deparaffinization, sections were incubated with target retrieval solution (Dako, Carpinteria, CA) for antigen retrieval. After blocking, a primary anti-PdCV or PEDV serum antibody was incubated for 30 min, 22 °C followed by incubation with a biotinylated horse anti-pig IgG secondary antibody (Vector Laboratories, Burlingame, CA). Slides were further incubated with ABC Elite complex to probe biotin (Vector Laboratories) and then developed using a 3,3′-diaminobenzidine (DAB) chromogen Kit (Dako); hematoxylin was used as a counterstain.
Cross-reactivity was also determined in virus-infected intestinal tissues. Twelve Gn piglets were divided into three groups (n = 4) and housed in three separate isolators. Gn piglets in group 1 and 2 were orally inoculated with 104 PFU of PEDV VBS2 and PdCV Michigan/8977/2014 strain respectively. The remaining four piglets in group 3 were orally inoculated with DMEM as controls. As expected, PEDV and PdCV-inoculated Gn piglets developed severe watery diarrhea, vomiting, and dehydration at 24 h post-inoculation. These two viruses caused indistinguishable clinical signs in gnotobiotic piglets. However, PEDV caused more severe diseases than PdCV in gnotobiotic piglets. 72 h post-inoculation, piglets inoculated with PEDV reached 20% body weight losses. Thus, all piglets were terminated at this time point. Replicate sections of duodenum, jejunum, ileum and colon were subjected to immunohistochemical (IHC) staining. In tissue sections, anti-PdCV antibody reacted only with virus-infected intestinal epithelial cells (duodenum, jejunum and ileum) in tissue sections collected from PdCV-infected Gn piglets and not with either PEDV-infected or uninfected control tissue sections (Fig. 1B, Table 2 ). In a similar fashion, the PEDV-specific antibody reacted only with PEDV-infected tissue sections. No cross-reactivity was detected by IHC (Fig. 1B, Table 2).
Table 2.
Summary of immunohistochemical (IHC) staining of intestinal tissues.
| Pig number | Anti-PEDV serum |
Anti-PdCV serum |
||||||
|---|---|---|---|---|---|---|---|---|
| Duodenum | Jejunum | Ileum | Colon | Duodenum | Jejunum | Ileum | Colon | |
| PEDV I | 4 | 4 | 4 | 2 | 0 | 0 | 0 | 0 |
| PEDV II | 4 | 4 | 4 | 1 | 0 | 0 | 0 | 0 |
| PEDV III | 4 | 4 | 4 | 2 | 0 | 0 | 0 | 0 |
| PEDV IV | 4 | 4 | 4 | 2 | 0 | 0 | 0 | 0 |
| PdCV I | 0 | 0 | 0 | 0 | 3 | 4 | 4 | 0 |
| PdCV II | 0 | 0 | 0 | 0 | 3 | 4 | 4 | 0 |
| PdCV III | 0 | 0 | 0 | 0 | 3 | 4 | 4 | 0 |
| PdCV IV | 0 | 0 | 0 | 0 | 3 | 4 | 4 | 0 |
| Control I | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Control II | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Control III | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Control IV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Note: IHC staining magnitude was scored for each piglet. 0: no antigen; 1: mild, less than 20% of epithelial cells were positive for antigen; 2: mild to moderate, 20–50% of epithelial cells were positive; 3: moderate, 50–80% of epithelial cells were positive; 4: severe, more than 80% epithelial cells were positive. No antigens were detected for PEDV or PdCV-infected intestinal tissues using control serum.
The cross reactivity detected by serum ELISA promoted us to perform further analyses. Next, we sought to determine whether anti-PEDV or anti-PdCV antibodies can cross-react with purified PEDV and PdCV virion proteins. For this, 2 μg of purified PEDV and PdCV was analyzed by 15% SDS-PAGE and subjected to Western blot using commercially available monoclonal PEDV and PdCV nucleocapsid (N)-specific antibody (Medgene Labs, Brookings, SD). PEDV N (49.0 kDa) and PdCV N proteins (37.0 kDa) were detected by PEDV and PdCV N monoclonal antibody, respectively; and no cross-reactivity was observed (Fig. 2 A). A similar Western blot was performed using either anti-PEDV or anti-PdCV polyclonal antibody raised in Gn piglets. As shown in Fig. 2B, PEDV antibody reacted strongly with PEDV proteins. Interestingly, a protein band with molecular weight corresponding to PdCV nucleocapsid (N) protein was detected by anti-PEDV antibody. Similarly, anti-PdCV antibody had a strong reaction with PdCV proteins but also reacted with PEDV N protein. Thus, a two-way of cross reactivity in N proteins between PEDV and PdCV was detected, a finding consistent with the cross-reactivity detected by ELISA.
Fig. 2.
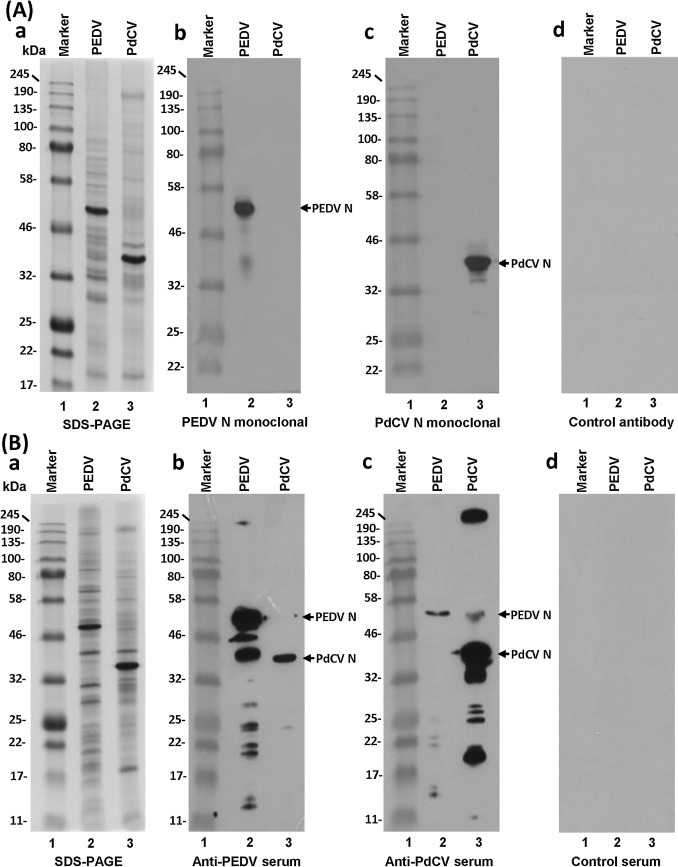
Western blot analysis of PEDV and PdCV virion proteins. (A) Western blot analysis using viral N monoclonal antibody. Panel a: SDS-PAGE analysis. 2 μg of purified PEDV and PdCV was analyzed by 15% SDS-PAGE, and stained with Coomassie blue. Panel b: Western blot analysis using PEDV N monoclonal antibody; Panel c: Western blot analysis using PdCV N monoclonal antibody; Panel d: Western blot analysis using control monoclonal antibody. The proteins were transferred onto a Hybond ECL nitrocellulose membrane (Amersham, Piscataway, NJ) in a Trans-Blot semidry electrophoretic transfer cell (Bio-Rad, Hercules, CA). The blots were blocked in 5% skim milk in PBST (phosphate-buffered saline supplemented with 0.02% Tween) and incubated with each antibody at a dilution of 1:100 in blocking buffer. Afterwards, the blot was incubated with horseradish peroxidase (HRP)-conjugated anti-pig IgG secondary antibody (1:2,500 dilution). After washing in PBST for (three times, 15 min each), the blots were developed with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Pittsburgh, PA) and exposed to Kodak BioMax MR film (Kodak, Rochester, NY). (B) Western blot analysis using anti-PEDV and PdCV serum antibody. Panel a: SDS-PAGE analysis. 2 μg of purified PEDV and PdCV was analyzed by 15% SDS-PAGE, and stained with Coomassie blue. Panel b: Western blot analysis using anti-PEDV serum; (c) Western blot analysis using anti-PdCV serum; (d) Western blot analysis using control pig serum.
The results of ELISA and Western blot suggest that N protein is responsible for the two-way cross-reactivity. Thus, we performed a sequence alignment of N proteins of selected PEDV and PdCV strains. It was found that four regions (47-GYW-49, 67-FYYTGTGPRGNLKY-82, 194-PKG-197, and 329-EWD-332) are conserved between PEDV and PdCV (Supplementary Fig. 1). These regions likely contain the N protein epitopes that account for the cross-reaction observed between these two viruses in both ELISA and Western blot assays.
Porcine coronaviruses are significant enteric and respiratory pathogens of swine. In 1946, porcine transmissible gastroenteritis virus (TGEV), an Alphacoronavirus (α-CoV), was identified as the cause of a devastating enteric disease of pigs in the United States (Doyle and Hutchings, 1946). A second US porcine α-CoV, porcine respiratory coronavirus (PRCV) was officially identified in 1984 (Wesley et al., 1990). PRCV is a deletion mutant of TGEV that alters viral tropism from intestinal to respiratory epithelia. In 1971, a new α-CoV, PEDV was first identified in the United Kingdom (Pensaert and de Bouck, 1978). In 2014, a porcine Deltacoronavirus, PdCV was characterized in the US (Ma et al., 2015). TGEV, PEDV, and PdCV replicate in small intestinal enterocytes causing life-threatening acute enteric disease in suckling piglets. Currently, the antigenic relationship among these porcine enteric coronaviruses is still poorly understood. Recently, it was reported that no cross-neutralization was observed between TGEV and PEDV despite the fact that both of them belong to Alphacoronavirus (Lin et al., 2015). Interestingly, one-way cross-reactions were observed between TGEV Miller strain hyperimmune pig antisera and PEDV strains using a cell culture immunofluorescent (CCIF) assay (Lin et al., 2015). Specifically, PEDV antigens can be detected using hyperimmune TGEV Miller antiserum. Sequence analysis suggested at least one epitope on the N-terminal region of PEDV/TGEV N protein that contributed to this cross-reactivity (Lin et al., 2015).
Our study is the first study to systemically assess the antigenic relationship between PEDV and PdCV, both of which are currently prevalent in the US. We found that no virus neutralization cross-reactivity was observed between PEDV and PdCV. Interestingly, a two-way of cross-reactivity was detected between PEDV and PdCV despite the fact that they belong to two different coronavirus genera. Our study contributes to our understanding of the antigenic relationship among porcine coronaviruses and will facilitate the development of more specific serological diagnostic assays and effective vaccines to control PEDV and PdCV outbreaks.
3. Conclusion
We report for the first time that a two-way cross-reactivity was detected between PEDV and PdCV by serum ELISA and Western blot assays, but only when performed with polyclonal immune serum. These data provide the first evidence that conserved epitope(s) in N proteins may contribute to this cross-reactivity. Antigenic cross-reactivity was not detected in either virus-infected cells or intestinal tissues using IFA or IHC nor in cross-neutralization assays. Sensitivity differences in these latter assays may account for this finding. Alternatively, both ELISA and blots employ an ionic detergent (SDS) that may uncover hidden epitopes on viral N proteins by virtue of protein denaturation. As a practical matter, given that cross-reactivity occurs only in a viral protein that is not involved in neutralization of viral infectivity, development of vaccine product(s) for use against these two very similar diseases of young swine will require the development of separate virus-specific vaccine products.
4. Materials and methods
4.1. Cell culture adapted PEDV strain
Plaque-purified PEDV VBS2 strain (isolated from an Ohio farm) were grown in Vero CCL-81 cells in DMEM supplemented with 0.018% (w/v) Tryptose Phosphate Broth (TPB) (Sigma), 0.02% yeast extract (Sigma), 5 μg/ml Trypsin 1:250 (Sigma, catalog number T0646), 10 UI/ml penicillin–streptomycin, 0.05 mg/ml gentamicin, and 0.05 mg/ml Kanamycin. At 48 h post-infection, supernatant was harvested, and virus titer was determined by plaque assay in Vero CCL-81 cells.
4.2. Cell culture adapted PdCV strain
Cell culture adapted PdCV Michigan/8977/2014 strain (PdCV MI) was purchased from National Veterinary Services Laboratories at USDA (Ames, IA). The PdCV MI strain was grown in swine testicular (ST) cells in DMEM containing 0.2 μg/mL TPCK-trypsin (Invitrogen). At 72 h post-infection, supernatant was harvested, and virus titer was determined by plaque assay in ST cells.
4.3. Purification of PEDV and PdCV
Stocks of PEDV VBS2 and PdCV Michigan/8977/2014 were purified by sucrose gradient ultracentrifugation. Briefly, 15 flasks of Vero CCL-81 and ST cells were infected by PEDV VBS2 and PdCV Michigan/8977/2014 at an MOI of 0.1. When an extensive CPE was observed, the media from multiple flasks were combined, and residual cell debris was removed by centrifugation (3000 × g, 30 min) at 4 °C. Virus was then concentrated by ultracentrifugation in a Ty50.2 rotor (Beckman Coulter, Fullerton, CA) at 30,000 × g for 2 h at 4 °C. The pellet was resuspended in 500 μl of NTE buffer (100 mM NaCl, 10 mM Tris, 1 mM EDTA, pH 7.4) and further purified through a 10% (wt/vol) sucrose NTE cushion by centrifugation for 2 h at 50,000 × g at 4 °C in an SW50.1 rotor (Beckman). The final virus-containing pellet was resuspended in 500 μl of NTE buffer overnight. Infectious PEDV or PdCV titer was quantitated by plaque assay. Viral protein content was measured by Bradford reagent (Sigma Chemical Co.). The virus stocks were confirmed to be free of other porcine enteric viruses by virus-specific RT-PCR or PCR described previously (Ma et al., 2015).
4.4. Preparation of aluminum formulated PEDV and PdCV inactivated antigens
For formalin inactivation, purified PEDV and PdCV stocks were incubated with 0.05% formalin (Fisher Scientific) at 37 °C for 18 h. The formalin was then neutralized by 0.05 M Sodium Metabisulfite 24 h at 4 °C. Loss of viral infectivity was confirmed by titration of inactivated virus preparations in cell culture. For vaccine preparation, 1.2 mg of formalin inactivated PEDV or PdCV was formulated with 3 ml of Alhydrogel aluminum Hydroxide Gel Adjuvant (InvivoGen) in 1 mL of PBS. The products were stored at 4 °C and inoculated to pigs on the same day.
4.5. Preparation of anti-PEDV and anti-PdCV immune sera in gnotobiotic (Gn) piglets
Anti-PEDV and anti-PdCV immune sera were prepared in Gnotobiotic (Gn) piglets (Ma et al., 2015). The animal protocol used in this study was approved by the Institutional Laboratory Animal Care and Use Committee of The Ohio State University. Gn piglets were derived into a sterile environment via Cesarean section from an specific-pathogen-free gravid sow as previously described (Eaton et al., 1998) and housed in germfree isolation units. Piglets were fed a milk-replacement diet (PermalatR) and maintained as described elsewhere (Eaton et al., 1998). Twelve Gn piglets were divided into three groups (n = 4) and housed in three separate isolators. Gn piglets in group 1 and 2 were immunized twice (two weeks apart) intramuscularly with 0.3 mg of inactivated PEDV and PdCV antigens respectively. The remaining four piglets in group 3 were immunized with adjuvant alone and served as controls. Four weeks after immunization, all piglets were terminated and serum were collected from each piglet, heat-inactivated, and stored at −80 °C for subsequent serologic assays.
4.6. Virus neutralization (VN) assay
Serum samples were collected from each piglet and tested for the presence of PEDV or PdCV-specific VN antibodies. Briefly, two-fold dilutions of the serum samples were incubated with 100 PFU/well of PEDV or PdCV at 37 °C for 1 h. The mixtures were then transferred to confluent Vero CCL-81 or ST cells in 6-well plates for plaque assay. After fixation and staining, the plaques were counted and 50% plaque reduction titers were calculated as the PEDV or PdCV-specific VN antibody titers. The assay was repeated for 3 times for each sample.
4.7. Detection of PEDV and PdCV-specific antibody by ELISA
Ninety-six-well plates were coated with 50 μl of highly purified PEDV or PdCV (2.5 μg/ml) in 50 mM NaCO3 buffer (pH 9.6) at 4 °C overnight. Briefly, serum samples were 2-fold-serially diluted and added to PEDV or PdCV antigen-coated wells. After incubation at room temperature for 1 h, the plates were washed five times with PBS-Tween (0.05%), followed by incubation with 50 μl of goat anti-swine IgG horseradish peroxidase-conjugated secondary antibody (Sigma) in 4% normal goat serum in PBST at a dilution of 1:80,000 for 1 h. Plates were washed and developed with 75 μl of 3,3′,5,5′-tetramethylbenzidine (TMB), and the optical density (OD) at 450 nm was determined using an enzyme-linked immunosorbent assay (ELISA) plate reader. End point titer values were determined as the reciprocal of the highest dilution that had an absorbance value greater than background level (DMEM control).
4.8. Immunofluorescence assay (IFA)
Confluent Vero CCL-81 and ST cells were infected with PEDV VBS2 and PdCV Michigan/8977/2014 at a multiplicity of infection (MOI) of 1.0, respectively. Before inoculation, the cells were washed twice using DMEM with 5 μg/ml trypsin 1:250 (Gibco). After 1 h of absorption, the inoculum was removed, the cells were washed one time with DMEM, and fresh DMEM (supplemented with 5 μg/ml trypsin 1:250) was added, and the infected cells were incubated at 37 °C. When cytopathic effect (CPE) were observed, the cells were washed twice with PBS and fixed with 4.0% (v/v) paraformaldehyde-0.2% (v/v) glutaraldehyde in 0.1 M potassium phosphate buffer (PPB), pH 7.4, for 15 min, 22 °C, followed by washing 3 times with PBS. After permeabilization with 0.1% Triton X-100 in PBS for 15 min, the cells were washed with PBS, blocked with PBS containing 2% bovine serum albumin for 1 h at RT. Then cells were incubated with anti-PEDV sera, anti-PdCV sera, FITC-labeled mouse anti-PEDV N protein monoclonal antibody, FITC-labeled mouse anti-PdCV N protein monoclonal antibody (Medgene, SD) at dilution of 1:200 overnight at 4 °C. The cells detected by immune sera were further detected by FITC-rabbit anti-swine IgG secondary antibody (Abcam) at a dilution of 1:100 for 3 h. After 3 washes with PBS, samples were examined under Olympus fluorescent microscope system at The Ohio State University.
4.9. Challenge of gnotobiotic piglets by PEDV and PdCV
Twelve one-week-old Gn piglets were divided into three groups (n = 4) and housed in three separate isolators. Gn piglets in group 1 and 2 were orally inoculated with 104 PFU of PEDV VBS2 and PdCV Michigan/8977/2014 strain respectively. The remaining four piglets in group 3 were orally inoculated with DMEM as controls. After virus challenge, the piglets were observed and evaluated daily for body weight and temperature changes, and clinical signs of PEDV or PdCV infection. At 72 h post-inoculation, all piglets were terminated. Duodenum, jejunum, ileum, and colon were collected and fixed in 10% (v/v) phosphate-buffered formalin for immunohistochemical staining.
4.10. Immunohistochemical staining (IHC)
Five-micron sections of paraffin-embedded tissues were placed onto positively charged slides. After deparaffinization, sections were incubated with target retrieval solution (Dako, Carpinteria, CA) for antigen retrieval. After blocking, a primary anti-PdCV serum antibody from PdCV-infected convalescent sows was incubated for 30 min, 22 °C followed by incubation with a biotinylated horse anti-pig IgG secondary antibody (Vector Laboratories, Burlingame, CA). Slides were further incubated with ABC Elite complex to probe biotin (Vector Laboratories) and then developed using a 3,3'-diaminobenzidine (DAB) chromogen Kit (Dako); hematoxylin used as a counterstain. Tissue sections from PdCV-infected and uninfected samples were used as positive and negative controls respectively. Tissue sections from PdCV-infected and uninfected samples were incubated with hyperimmune serum against PEDV in IHC assay. Reciprocally, IHC assays were also performed using intestinal tissue sections from PEDV-infected samples using anti-PdCV serum antibody.
4.11. Statistical analysis
Statistical analysis of one-way ANOVA was performed by using Minitab statistical analysis software (Minitab, Inc., State College, PA), a P-value of <0.05 was considered statistically significant.
Acknowledgments
We thank Susan Ringler and Chelssie Breece for their excellent technical assistance. Funding for this work was supported by the Department of Veterinary Biosciences, College of Veterinary Medicine, The Ohio State University, and Guangzhou Science & Technology Program (No. 201508020039) and Natural Science Foundation of China (No. 31500127). We thank members in Li’s laboratory for helping the Gn pig study and critical reading for this manuscript. The animal protocol used in this study was approved by the Institutional Laboratory Animal Care and Use Committee of The Ohio State University.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vetmic.2016.02.004.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Chen J., Wang C., Shi H., Qiu H., Liu S., Chen X., Zhang Z., Feng L. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010;155:1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R., Welch M., Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Leung C.Y., Gilbert M., Joyner P.H., Ng E.M., Tse T.M., Guan Y., Peiris J.S., Poon L.L. Avian coronavirus in wild aquatic birds. J. Virol. 2011;85:12815–12820. doi: 10.1128/JVI.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussement W., Ducatelle R., Debouck P., Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets: I. Histological and histochemical study. Vet. Pathol. 1982;19:46–56. doi: 10.1177/030098588201900108. [DOI] [PubMed] [Google Scholar]
- Dong B.Q., Liu W., Fan X.H., Vijaykrishna D., Tang X.C., Gao F., Li L.F., Li G.J., Zhang J.X., Yang L.Q., Poon L.L., Zhang S.Y., Peiris J.S., Smith G.J., Chen H., Guan Y. Detection of a novel and highly divergent coronavirus from asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 2007;81:6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle L.P., Hutchings L.M. A transmissible gastroenteritis in pigs. J. Am. Vet. Med. Assoc. 1946;108:257–259. [PubMed] [Google Scholar]
- Eaton K.A., Ringler S.S., Krakowka S. Vaccination of gnotobiotic piglets against Helicobacter pylori. J. Infect. Dis. 1998;178:1399–1405. doi: 10.1086/314463. [DOI] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4:e00737–00713. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Eyerly B., Lu Z., Chepngeno J., Saif L.J. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 2015;21:650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Wang Q., Scheuer K.A., Lu Z., Zhang Y., Saif L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014;20:662–665. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Gao X., Oka T., Vlasova A.N., Esseili M.A., Wang Q.H., Saif L.J. Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J. Virol. 2015;89:3332–3342. doi: 10.1128/JVI.03196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6:e00064. doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, ohio, USA, 2014. Emerg. Infect. Dis. 2014:20. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley R.D., Woods R.D., Hill H.T., Biwer J.D. Evidence for a porcine respiratory coronavirus, antigenically similar to transmissible gastroenteritis virus, in the United States. J. Vet. Diagn. Invest. 1990;2:312–317. doi: 10.1177/104063879000200411. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


