Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is difficult to control due to a high mutation rate and the emergence of virulent strains. The objective of this study was to analyze the immunological and pathological responses after infection with the European subtype 3 strain Lena in comparison to subtype 1 strains Belgium A and Lelystad-Ter Huurne (LV). Sixteen pigs were inoculated per strain, and sixteen pigs with PBS. At days 7 and 21 post-inoculation (p.i.), four pigs per group were immunized with an Aujeszky disease vaccine (ADV) to study the immune competence after PRRSV infection. Infection with the Lena strain resulted in fever and clinical signs. This was not observed in the Belgium A or LV-infected pigs. Infection with the Lena strain resulted in high virus titers in serum, low numbers of IFN-γ secreting cells, a change in leukocyte populations and a delayed antibody response to immunization with ADV. Levels of IL-1β, IFN-α, IL-10, IL-12, TNF-α and IFN-γ mRNA of the Lena-infected pigs were increased during the first week of infection. For pigs infected with the Belgium A or LV strain, the effects of infection on these parameters were less pronounced, although for the Belgium A-infected pigs, the level of the analyzed cytokines, except for TNF-α, and leukocyte populations were comparable to the Lena-infected pigs. These results suggest that while the outcome of infection for the three strains was comparable, with mostly clearance of viremia at day 33 p.i, differences in immune responses were observed, perhaps contributing to their virulence.
Keywords: Genetic subtypes, Immune response, Pathogenesis, Porcine reproductive and respiratory syndrome virus
1. Introduction
Porcine reproductive and respiratory syndrome virus (PRRSV) is a single stranded RNA virus of the Arteriviridae family (Meulenberg et al., 1994). This virus is widespread and causes disease characterized by abortions and stillbirth, increased pre-weaning mortality and respiratory disorders in growing pigs. Although PRRSV alone is considered a primary pathogen, co-infections with bacterial and viral pathogens like porcine circovirus type 2 (PCV2), swine influenza, porcine respiratory coronavirus, Mycoplasma hyopneumoniae or Streptococcus suis commonly occur and exacerbate clinical symptoms in growing pigs (Dorr et al., 2007). As a consequence, PRRSV is one of the most significant causes of economic losses in the swine industry worldwide (Neumann et al., 2005).
PRRSV shows a high degree of genetic variation. Two genotypes are recognized, represented by two prototypes: Lelystad virus (European type or genotype I PRRSV) and VR-2332 (American type or genotype II PRRSV) (Wensvoort et al., 1991, Collins et al., 1992). The European type strains can be further divided into at least three subtypes: Pan-European subtype 1 and Eastern European subtypes 2 and 3 (Stadejek et al., 2008). Between the European subtypes, differences have been described in virulence of strains. One Eastern European subtype 3 strain, Lena, was described to be more virulent than a subtype 1 strain, as determined by clinical manifestations in infected pigs under experimental conditions (Karniychuk et al., 2010). The mechanisms by which a PRRSV strain exerts its virulence are unknown, but it has been suggested to be related to in vivo replication capacity, tissue distribution, or immunomodulatory properties (Haynes et al., 1997, Johnson et al., 2004, Loving et al., 2008).
PRRSV infects in vivo specific subsets of differentiated macrophages in lungs, lymphoid tissues and placenta (Van Breedam et al., 2010). Through interactions with these cells, the virus may influence the host immune response. In comparison to other viral infections such as porcine influenza virus or porcine coronavirus, an altered innate immune response was observed with lower levels of IFN type I after infection with some PRRSV strains (Van Reeth et al., 1999). The adaptive immune response is usually weak and delayed. Although an abundant virus-specific antibody response is induced, this exhibits minimal virus neutralization activity (Kimman et al., 2009). The cell-mediated immune response, as measured by PRRSV-specific IFN-γ secreting cells, is late compared to other pathogens (Meier et al., 2003). As a consequence, a previous infection with PRRSV may not protect against a homologous re-infection.
The induction of an insufficient adaptive immune response is also a feature of current modified live PRRSV vaccines. None of the European genotype vaccines on the market, containing subtype 1 strains, can claim to provide full protection (Murtaugh and Genzow, 2011). Furthermore, the genetic diversity of PRRSV is thought to influence the efficacy of vaccines (Labarque et al., 2004). In experimental studies, PRRSV vaccines were often not efficacious against infection with heterologous strains (Diaz et al., 2006, Zuckermann et al., 2007, Darwich et al., 2010). To aid the development of more efficacious vaccines, an improved knowledge of the immune response against PRRSV is necessary.
Until now, most studies investigating the immune response of pigs after infection with field strains of PRRSV used American genotype strains (Meier et al., 2003, Johnson et al., 2004, Klinge et al., 2009, Wang et al., 2011). Only a limited number of studies have used European type strains for infection (Diaz et al., 2005, Gómez-Laguna et al., 2009), and no information is available about immunological responses to the more virulent European subtype 3 strains. The aim of the present study was to characterize the immune response after infection with two European subtype 1 strains, Belgium A and the prototype Lelystad-Ter Huurne (LV) virus, and a virulent subtype 3 strain, Lena. This subtype 3 strain had caused severe clinical signs and 40% mortality in a previous study (Karniychuk et al., 2010). We demonstrated between the strains significant differences in fever induction, viral load, IFN-γ secreting cells, cytokine levels, blood cell populations and the humoral response to an unrelated secondary antigen.
2. Materials and methods
2.1. Viruses
PRRSV strain Lena is a recent Eastern European subtype 3 PRRSV isolate and was used at the third passage on porcine alveolar macrophages (PAM). Lena was isolated from a Belarusian farm with reproductive and respiratory failure (Karniychuk et al., 2010). Strains Belgium A and LV are European subtype 1 PRRSV isolates. PRRSV strain Belgium A has also been described as strain 07V063 (Vanhee et al., 2010). This strain was isolated from a stillborn piglet, derived from a Belgian farm during an outbreak of PRRSV associated reproductive disorders (Karniychuk et al., 2010). This strain was used at the third passage on PAM. PRRSV strain LV-Ter Huurne was isolated during the 1991 epizootic from a clinical case of PRRS in the Netherlands (Wensvoort et al., 1991). This strain was used at the seventh passage on PAM.
2.2. Animals and housing
Sixty-four 5 week old male pigs, cross-breeds between the Great Yorkshire, Large White type and Dutch Landrace, were obtained from a PRRSV-free farm in the Netherlands with a high health status. All pigs were confirmed negative for PCV2 by PCR one week prior to transport, since PRRSV and PCV2 co-infection can increase the severity of disease and cause post-weaning multisystemic wasting syndrome (Grau-Roma et al., 2011). To harmonize the genetic background of pigs across experimental groups, four male pigs of each sixteen sows were selected, and evenly distributed between four groups. The four groups were housed in different rooms of an isolation unit. Between rooms, clothing, footwear and gloves were changed and materials needed for sampling and rectal temperature monitoring were provided separately for each room. Standard feed for finishing pigs was provided once a day, and the pigs had unlimited access to water.
2.3. Experimental protocol
After one week of acclimatization, the pigs were inoculated intranasally with 1.5 ml containing 105 50% tissue culture infectious dose (TCID50) of either PRRSV European subtype 3 strain Lena (group Lena), PRRSV European subtype 1 strain Belgium A (group Belgium A), PRRSV European subtype 1 strain LV-Ter Huurne (group LV) or an equal volume of PBS (group control). To study the humoral immune competence of pigs after PRRSV infection, four pigs per group were immunized intramuscularly in the neck, behind the ear, with 2 ml of ADV-vaccine on days 7 and 21 post inoculation (p.i.). The inactivated ADV vaccine, Suvaxyn I-Aujeszky, was kindly supplied by Pfizer Animal Health. This vaccine contained an oil-in-water adjuvant. At days 3 and 7 p.i. four pigs per group were euthanized. All remaining pigs were euthanized at the termination of the experiment on day 35 p.i. The experiment was approved by the Ethics Committee for Animal Experiments of the Central Veterinary Institute of Wageningen UR.
2.4. Clinical signs and body temperature
Rectal temperature and clinical signs were recorded daily. Fever was defined as body temperature higher than 40.5 °C for one day or 40 °C for two consecutive days. For quantitative assessment of the severity of disease, a list of eleven PRRSV-relevant criteria was used (Table 1 ). For each criterion, a score was recorded as either normal (score 0), slightly altered (score 1), distinct clinical sign (score 2), or severe PRRSV sign (score 3). These scores were added up to obtain a total score per pig per day.
Table 1.
Clinical score list (adapted from Mittelholzer et al., 2000). All the 11 categories were scored for each individual animal and summed resulting in a maximum score of 28.
| # | Parameter | Criteria | Score |
|---|---|---|---|
| No abnormalities. | 0 | ||
| 1 | Liveliness | Slightly reduced (stands up hesitantly, but without help) | 1 |
| Tired, gets up only when forced to, lies down again | 2 | ||
| Dormant, will not stand up | 3 | ||
| 2 | Body shape | Emaciated, backbone and ribs visible | 1 |
| 3 | Coughing | Mild (few incidences 1–5 observation duration) | 1 |
| Severe (more frequent incidences, >5 observation duration) | 2 | ||
| 4 | Sneezing | Mild (few incidences 1–5 observation duration) | 1 |
| Severe (more frequent incidences, >5 observation duration) | 2 | ||
| 5 | Breathing (judge before approaching pig) | Increased frequency, snoring, barely visible chest movement | 1 |
| Increased frequency, snoring, distinct chest and abdominal movement | 2 | ||
| Increased frequency, problems breathing, panting, breathing through open mouth | 3 | ||
| 6 | Skin changes body (not the ear) | Hair coat standing up | 1 |
| Discolored skin: red, pale, gray or yellow | 2 | ||
| Blue–purple discoloration of the skin | 3 | ||
| 7 | Skin changes ears | Discolored skin of the ears: red, pale, gray or yellow | 1 |
| Blue–purple discoloration of the ears | 2 | ||
| Ear necrosis | 3 | ||
| 8 | Eyes/conjunctiva | Reddened, clear secretion | 1 |
| Inflamed eyes, turbid secretion or swelling of the eyelids | 2 | ||
| Highly inflamed eyes, purulent secretion, accentuated blood vessels | 3 | ||
| 9 | Nasal discharge | Clear nasal discharge | 1 |
| Discolored nasal discharge | 2 | ||
| 10 | Appetite | Eat slowly when fed | 1 |
| Does not eat when fed, but tastes food | 2 | ||
| Does not eat at all, shows no interest for food | 3 | ||
| 11 | Defecation | Reduced amount of feces, dry | 1 |
| Only small amount of dry, fibrin-covered feces or diarrhea | 2 | ||
| No feces, mucus in rectum, or watery or bloody diarrhea | 3 | ||
2.5. Sampling collection and pre-treatment
Serum samples were collected at days 0, 3, 5, 7, 10, 14, 21, 26 and 33 p.i. to determine virus titers and antibody levels. These samples were stored directly at −70 °C until testing. Whole blood samples of the non-ADV immunized pigs that remained in the experiment until day 35 p.i. were collected in Paxgene-RNA tubes (Becton, Dickinson and Company) at days 0, 3, 7, 14, and 33 p.i. In Paxgene® RNA tubes (PreAnalytiX), cells are lysed and the RNA is stabilized. Samples were incubated for 2 h at room temperature and then stored at − 70 °C until RNA isolation. Heparin stabilized blood samples were collected at days 0, 3, 7, 10, 14, 21, 26 and 33 p.i. Peripheral blood mononuclear cells (PBMC) were isolated from these blood samples and used for IFN-γ ELISPOT assay and flow cytometry. Isolation of PBMC was performed by density gradient centrifugation using 50 ml Leucosep® tubes (Greiner Bio-One). In brief, the heparinized whole-blood samples were diluted with equal volumes of PBS, and 30 ml of the diluted blood was added to a Leucosep® tube. The cell separation tubes were centrifuged at room temperature for 20 min at 1380 × g without braking. The cell suspension was collected, and remaining red blood cells were lysed with ACK lysing buffer containing 0.15 M NH4Cl, 10 mM KHCO3 and 0.1 mM Na2EDTA. The cells were then washed twice in PBS (centrifugation for 15 min at 640 × g) and resuspended in 1 ml PBS for counting with the Z2 Coulter Counter (Beckman Coulter).
2.6. RNA isolation from Paxgene blood samples
Samples collected in PAXgene Blood RNA tubes were thawed for 2 h at room temperature. Tubes were centrifuged at 2800 × g for 10 min at room temperature and the supernatant discarded. The pellet was washed twice in RNase-free water and the pellet was dissolved in 1 ml TRIzol® reagent (Invitrogen) Further extraction of RNA was performed according to instructions of the manufacturer. The yield and purity of the RNA was assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific).
2.7. Virus isolation and titration
PRRSV isolation from serum was performed on PAM cells, obtained from 3 to 5 week old piglets from a PRRSV-free herd in the Netherlands. The cells were previously tested negative for PCV2 by PCR. The PAM cells were cultured in 24-well plates (Greiner) at a concentration of 5 × 105 cells/well in 1 ml RPMI 1640 medium supplemented with 10% FBS, 100 IU/ml penicillin and 100 μg/ml streptomycin (cRPMI; all from Gibco, Invitrogen). To each well, a volume of 250 μl serum was inoculated and plates were incubated at 37 °C in a 5% CO2 humidified atmosphere. After 3 days, the monolayers were washed in 0.15 M NaCl solution, dried and frozen. The monolayers were stained by immuno peroxidase monolayer assay (IPMA) as described by Van der Linden et al. (2003), using a 1:500 dilution of the monoclonal antibody SDOW17-A (Rural Technologies) against the nucleocapsid protein of PRRSV. Virus positive serum samples taken at days 3, 7, 10, 21 and 33 p.i. from the four non-immunized pigs that remained in the experiment until day 35 p.i. were titrated. Serum dilutions were made in four-fold after making five decimal dilutions. Virus titers were calculated as TCID50 using the Spearman-Kärber method (Finney, 1978).
2.8. Detection of PRRSV by quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Viral RNA concentrations in serum were analyzed by qRT-PCR using a standard curve. RNA was isolated from 200 μl serum using the High Pure viral RNA kit (Roche Applied Science) according the manufacturer's instructions. Standard curves were constructed for each virus strain by extracting RNA from five decimal dilutions of medium with known concentrations of infectious virus. After the RNA isolation, the nucleic acids were stored at −70 °C until analysis in the qRT-PCR.
A one-tube qRT-PCR was performed with the MX3005 (Stratagene) instrument using the Quantitect Probe RT-PCR kit from Qiagen. The reaction mixture (25 μl) contained 0.25 μl of kit-supplied enzyme, 12.5 μl of Quantitect Mix, 15 μM of each primer (Fw: 5′-GAT GAC RTC CGG CAY C-3′, Rev: 5′-CAG TTC CTG CGC CTT GAT-3′) and 10 μM of probe (5′-Fam-TGC AAT CGA TCC AGA CGG CTT-Tamra-3′) (Frossard et al., 2012). The RT-PCR was done for 30 min at 50 °C and 15 min at 95 °C. A two-step cycling protocol was used as follows: 94 °C for 20 s, and 55 °C for 45 s for 40 cycles. Analysis was performed with the MXpro software. The viral RNA concentration (TCID50 equivalents per ml) of each individual sample was calculated using the standard curve.
2.9. PRRSV serology
PRRSV-specific antibodies in serum samples were tested with an antibody ELISA (HerdChek PRRS X3, IDEXX Laboratories), according to the manufacturer's instructions. A sample-to-positive (S/P) ratio of equal or greater than 0.4 was considered positive.
To detect virus neutralizing antibodies against PRRSV, serum samples from day 33 p.i. were heat-treated for 30 min at 56 °C and serial 2-fold dilutions (50 μl volumes) of the test serum were then made in cRPMI. An equal volume of the homologous PRRSV strains containing 102 TCID50 in cRPMI were added to each serum dilution and serum–virus mixtures were incubated at 37 °C for 60 min. Finally, 2.5 × 105 PAM cells were added to each well and plates were incubated for 3 days at 37 °C before IPMA staining of virus positive cells as described above.
2.10. ADV serology
ADV-specific IgG1 antibody responses were detected using an indirect ELISA (Kimman et al., 1992). The titer of a sample was expressed as log10 of the reciprocal of the highest dilution yielding an S/P ratio >0.4.
2.11. IFN-γ ELISPOT assay
The number of antigen-specific IFN-γ-secreting cells per 2.5 × 105 PBMC was determined using a ELISPOT assay according to the method described by the manufacturer of the used MultiScreenHTS-IP Filter plates (Millipore). Briefly, the plates were pre-coated overnight with 10 μg/ml of anti-pig IFN-γ mAb (BD). PBMC (2.5 × 105 PBMC in 50 μl/well) were plated in DMEM GlutaMAX™ medium supplemented with 4.5 g/l glucose, 25 mM HEPES, 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin and 5 μM β-mercaptoethanol (all from Gibco, Invitrogen). PBMC were stimulated for 20–24 h in triplicate wells by addition of homologous PRRSV at a multiplicity of infection (MOI) of 0.01. Concanavalin A (20 μg/ml [Sigma]) was used in triplicate wells as a positive control, and culture medium in triplicate wells as negative control (all 50 μl/well). Plates were incubated with 100 μl of anti-pig IFN-γ biotin-labeled mAb (BD) at a concentration of 0.17 μg/ml. The reaction was revealed by sequential incubation with streptavidin–alkaline phosphatase enzyme conjugate (R&D Systems) and BCIP/NBT substrate solution (R&D systems). The number of specific IFN-γ secreting cells, as determined using an ImmunoSpot® S4 Analyzer (Cellular Technology Ltd.), were calculated as the average number of spots in the triplicate PBMC cultures stimulated with virus, minus the number of spots in triplicate PBMC cultures exposed to culture medium only. The data were expressed as the background corrected number of IFN-γ-secreting cells per 2.5 × 105 PBMC.
2.12. Immunophenotyping of PBMC by flow cytometry
Isolated PBMC were transferred to microtiter plates (1 × 106 cells in a volume of 100 μl per well) and centrifuged for 3 min at 350 × g. Cells were triple stained with mAbs directed to porcine SWC1, CD172a and SWC8 for identification of leukocyte populations or CD3, CD4 and CD8 for identification of T-cell subpopulations. The staining with primary antibodies was followed by a combination of PerCP, FITC and PE labeled secondary antibodies. Primary antibodies used were: mouse anti-porcine-CD3: clone PPT3, IgG1 (Kirkham et al., 1996); mouse anti-porcine-CD4: clone 74-12-4, IgG2b (Pescovitz et al., 1984), mouse anti-porcine-CD8α: clone SL2 (11/295/33) IgG2a (Jonjić and Koszinowski, 1984), mouse anti-porcine-SWC1a, recently identified as CD52 (Leitner et al., 2012) clone K263.3D7, IgG1 (AbD Serotec), mouse anti-porcine-CD172a: clone BL1H7, IgG2b (VMRD), mouse anti-porcine-SWC8: clone MIL3, IgM (AbD Serotec). Secondary antibodies used were: rat anti-mouse IgG1 PerCP (BD), goat anti-mouse IgG2b FITC (Southern Biotech) and goat anti-mouse IgG2a PE (Southern Biotech) and goat anti-mouse IgM PE (Southern Biotech).
Flow cytometry analyses were performed on a constant event number of 50,000 cells, acquired on a FACScan flow cytometer using CellQuest Software (both BD biosciences). Monocytes were identified as SWC1+CD172a+SWC8−, and B cells as SWC1−CD172a−SWC8high (Summerfield et al., 2001). The cytotoxic T cell sub-population was identified as CD3+CD4−CD8high, naïve T helper cells as CD3+CD4+CD8− (T helper cells), memory T helper cells as CD3+CD4+CD8+ NK cells as CD3−CD4−CD8+ cells, and γδ T cells as CD3+CD4−CD8− (Nielsen et al., 2003, Gerner et al., 2009). Although the majority of γδ T cells are CD3+CD4−CD8−, they can also exist as CD8+ cells (Gerner et al., 2009). We, therefore, refer to this CD3+CD4−CD8− population as CD8− γδ T cells. The numbers of each cell population per μl blood were calculated using cell counts from EDTA stabilized blood samples with the Medonic® CA620 cell counter (Boule Medical AB).
2.13. Quantitative RT-PCR for the detection of cytokine RNA
The concentration of IL-1β, IL-10, IL-12, IFNα, IFN-γ, and TNF-α mRNA in RNA extracted from peripheral blood samples was determined by qRT-PCR as described by Wichgers Schreur et al. (2011) with slight modifications. Briefly, 200 ng of total RNA was reverse transcribed using random primers and Superscript III (Promega). An appropriate dilution of cDNA was amplified in a PCR reaction mix (20 μl) containing 5 μM of forward and reverse primer (2.5 μl of each primer), and a concentration of 2× SYBR Green Mix (Applied Biosystems). The following primers were used (5′–3′): IL-12: forward primer ATTGCATGCTTTTCCTGCTGTT, reverse primer GCTGAGTTATGCACTA- TGCACGTTA; IFN-γ forward primer GAGCATGGATGTGATCAAGCAA, reverse primer CATTCAGTT- TCCCAGAGCTACCA; 18S forward primer AGAGTCTCGTTCGTTATCGGAATT, reverse primer TGCATGGCCGTTCTTAGTTG. Primer sequences of IL-1β were obtained from Wichgers Schreur et al. (2011), IL-10 from Chaung et al. (2010), TNF-α from De Greeff et al. (2010), and IFN-α from Durand et al. (2009). Serial dilutions of pGem-T Easy plasmids containing the PCR fragment of interest were used as internal standards. C t values for the tested cytokines in each sample were expressed as cDNA quantity (ng) using the internal standards. Subsequently, the cytokine levels were normalized with the ng levels of the house keeping gene 18S rRNA. The data were expressed as the fold induction compared to the control group.
2.14. Statistical analysis
Statistical analyses to compare flow cytometry data and ADV serology between groups were performed at each sampling point using a one-way ANOVA. This test was used because of the homogeneity of variances between groups. Statistical analyses to compare the virus load, PRRSV serology, IFN-γ ELISPOT and cytokine RNA levels between groups were performed at each sampling point using a non-parametric Kruskal–Wallis test followed by Mann–Whitney U tests for multiple comparisons. Statistical analysis to compare data between the ADV-immunized and non-immunized pigs within each group were performed using a Student's t-test. Calculations were performed with SPSS 19 (SPSS Inc.). Differences were considered statistically significant at p < 0.05.
3. Results
In general, there were no differences between the PRRSV-infected animals that were ADV-immunized or those non-immunized for fever, clinical signs, viral RNA load, PRRSV antibody levels, IFN-γ secreting cells or frequency of immune cells in blood. Therefore, in the results section, no distinction was made for the described parameters between the immunized and non-immunized pigs.
3.1. Clinical signs
Differences were observed in clinical signs and body temperature between the pigs infected with the Lena strain and pigs infected with the Belgium A or LV strain (Fig. 1 A and B). All Lena-infected pigs showed fever between days 2 and 15 p.i, one Belgium A-infected pig (40.1–40.4 °C) between days 0 and 3 p.i. and none of the LV-infected pigs (Fig. 1A). Clinical signs were observed in Lena-infected pigs between days 3 and 17 p.i. (Fig. 1B), with animals displaying reduced liveliness and appetite, an increased breathing frequency and coughing between days 3 and 17 p.i. In the Belgium A-infected group, one pig developed a red coloration of the skin of the ears between days 14 and 16 p.i. but did not show fever or other clinical signs related to the PRRSV infection. In the group of LV-infected pigs, one pig showed a clinical score of 6 at day 8 p.i. and was found dead the next day. Post-mortem examination showed that the cause of death was an intra-abdominal testicular torsion with infarction, thus the mortality was not PRRSV related and the clinical sign score of this pig on day 8 p.i. was removed from the analysis.
Fig. 1.
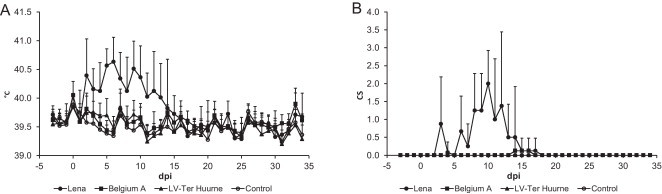
(A) Average body temperature and (B) clinical scores of pigs infected with different strains of PRRSV. Data points from d.p.i. 0–3 represents the average of sixteen pigs ±S.D., from.d.p.i.4 to 7 of twelve pigs and from d.p.i. 8 to 34 of seven or eight pigs.
3.2. PRRSV titers in serum
Virus isolation on PAM cells was performed on serum from all inoculated pigs, and showed that infectious virus was detected in the serum of almost all PRRSV inoculated pigs at day 3 p.i. (Table 2 ). From day 21 p.i., 50% or less of the pigs infected with the Belgium A and LV strain were still viraemic. This is in contrast to the Lena-infected pigs, with 7/8 animals viraemic at day 26 p.i. At day 33 p.i. most pigs had cleared the infection from blood and only one Lena-infected and one Belgium A infected pig remained viraemic. From the four non-ADV immunized pigs that remained in the experiment until day 33 p.i, the serum samples were also titrated. The virus titration assay showed that peak virus titers of the Lena-infected pigs, 105.4 TCID50/ml, were higher than pigs infected with the Belgium A or LV strain, 103.8 TCID50/ml and 104.0 TCID50/ml, respectively (Fig. 2 A). The qRT-PCR performed on serum from all inoculated animals revealed data comparable to the virus titrations, although the level of viral RNA in serum of LV-infected pigs (102.4 TCID50/ml) was higher than in serum from Belgium A-infected pigs (101.5 TCID50/ml) (Fig. 2B).
Table 2.
Proportion of PRRSV-positive animals by viral isolation on PAM cells.
| Treatment | d.p.i. |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 5 | 7 | 10 | 14 | 21 | 26 | 33 | |
| Lena | 0/16 | 16/16 | 12/12 | 12/12 | 8/8 | 8/8 | 7/8 | 7/8 | 1/8 |
| Belgium A | 0/16 | 12/16 | 12/12 | 12/12 | 8/8 | 8/8 | 8/8 | 4/8 | 1/8 |
| LV-TerHuurne | 0/16 | 13/16 | 11/12 | 11/12 | 7/7 | 7/7 | 6/7 | 2/7 | 0/7 |
| Control | 0/16 | 0/16 | 0/12 | 0/12 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 |
Fig. 2.
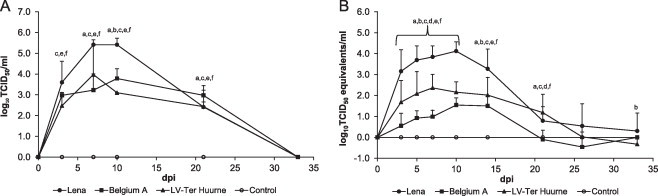
PRRSV levels in serum of pigs infected with different strains of PRRSV. (A) Infectious PRSV isolated from PRRSV inoculated pigs not immunized with an inactivated Aujeszky's disease (ADV) vaccine. Each data point represents the average of three or four pigs ±S.D. (B) PRRSV RNA levels in serum as determined by qRT-PCR. Data points from d.p.i. 0 to 3 represents the average of sixteen pigs ±S.D., from.d.p.i. 4 to 7 of twelve pigs and from d.p.i. 8 to 33 of seven or eight pigs. “a” denotes a significant difference (p < 0.05) between Lena and Belgium A-infected pigs; “b” between Lena and LV-infected pigs; “c” between Lena and control pigs; “d” between Belgium A and LV-infected pigs; “e” between Belgium A and control pigs; and “f” between LV and control pigs.
3.3. PRRSV specific antibody and T cell IFN-γ responses
Antibodies against PRRSV were detected in all infected groups from day 10 p.i. (Fig. 3 A). No antibodies were detected in the control group. There were no significant differences in antibody levels between the strains. At day 33 p.i. no neutralizing antibodies were detected in any of the three infected groups (data not shown).
Fig. 3.
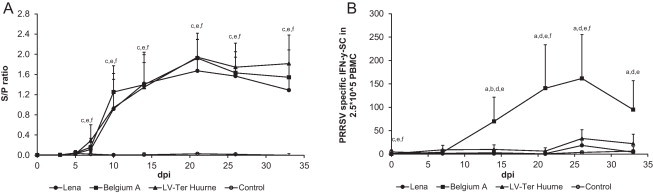
Virus specific antibody and T cell IFN-γ responses of pigs infected with different strains of PRRSV. (A) PRRSV antibody titers as determined by ELISA. Data points from d.p.i. 0 to 3 represents the average of sixteen pigs ±S.D., from.d.p.i. 4 to 7 of twelve pigs and from d.p.i. 8 to 33 of seven or eight pigs. (B) Background corrected virus-specific IFN-γ secreting cells as determined by ELISPOT assay. Each data point represents the average of seven or eight pigs ±S.D. “a” denotes a significant difference (p < 0.05) between Lena and Belgium A-infected pigs; “b” between Lena and LV-infected pigs; “c” between Lena and control pigs; “d” between Belgium A and LV-infected pigs; “e” between Belgium A and control pigs; and “f” between LV and control pigs.
The highest numbers of IFN-γ secreting cells after homologous PRRSV stimulation were observed in pigs infected with the Belgium A strain (peaking at a mean of 162 IFN-γ SC/2.5 × 105 PBMC) (Fig. 3B). The lowest numbers of IFN-γ secreting cells were observed for control pigs and Lena-infected pigs (maximum mean of 6 and 19 IFN-γ SC/2.5 × 105 PBMC, respectively) and these groups were not significantly different. The pigs infected with the LV strain had a significantly higher number of IFN-γ secreting cells than the control pigs at days 21 and 26 p.i. (mean of 6 IFN-γ SC/2.5 × 105 PBMC at day 21 p.i., and 33 IFN-γ SC/2.5 × 105 PBMC at day 26, compared to 1 IFN-γ SC/2.5 × 105 PBMC at day 21 p.i, and 4 IFN-γ SC/2.5 × 105 PBMC at day 26 p.i. for the control pigs).
3.4. Immunophentoyping peripheral blood leukocytes
The effect of infection on the number of immune cells in blood was most pronounced in Lena-infected pigs (Fig. 4 ). The number of T helper cells and CD8−γδ T cells were significantly lower in the Lena-infected pigs than in the control pigs. This was also observed for the Belgium A-infected pigs, although the level was not decreased to the same extent as in the Lena-infected pigs. The memory T helper cell level was also lower in the Lena-infected pigs compared to the control pigs, but was followed by an increase. This was also observed for the Belgium A strain, but the effect was less pronounced. The LV-infected pigs did not show a decrease, but only an increase of memory T helper cell at the end of the experiment. The number of NK cells of all infected groups increased after inoculation. It was significantly higher than the control pigs for LV-infected pigs at days 10 and 33 p.i. and for Belgium A-infected pigs at day 33 p.i. Only the LV-infected pigs showed an increase in cytotoxic T cells at the end of the experiment, compared to the control pigs. Initially, a significant decrease was observed in the number of B cells of all infected groups compared to the control pigs. Then, the number of B cells increased to a significantly higher level for the LV and Belgium A-infected pigs at the end of the experiment. All strains showed a significant increase in monocytes at day 21 p.i. compared to the control pigs.
Fig. 4.
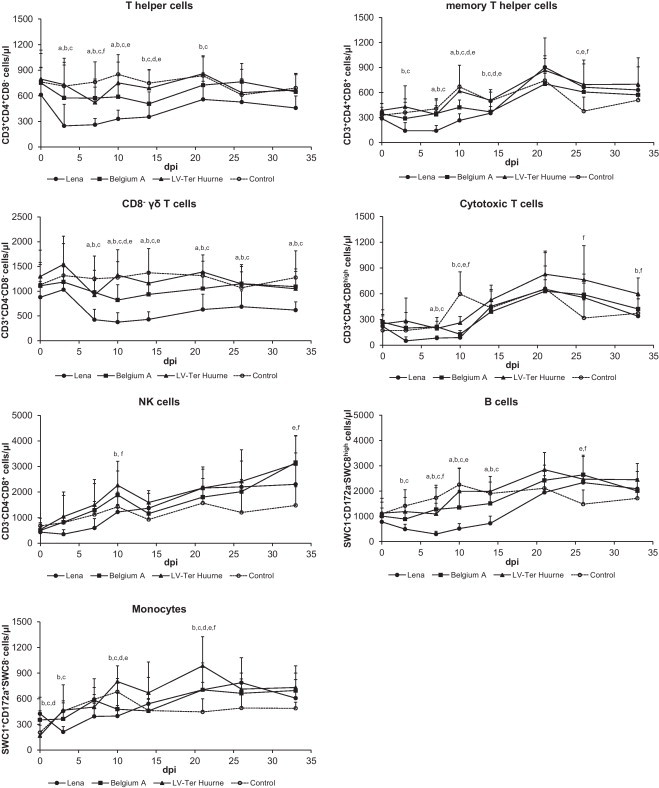
Frequency of immune cells in blood of pigs infected with different PRRSV strains. Each data point represents the average of seven or eight pigs ±S.D. “a” denotes a significant difference (p < 0.05) between Lena and Belgium A-infected pigs; “b” between Lena and LV-infected pigs; “c” between Lena and control pigs; “d” between Belgium A and LV-infected pigs; “e” between Belgium A and control pigs; and “f” between LV and control pigs.
3.5. Cytokine mRNA production in blood
In general, significant inductions of cytokine mRNA in blood cells were detected in the Lena and Belgium A-infected pigs within the first week after inoculation (Fig. 5 ). No significant induction in cytokine mRNA levels was observed for the LV-infected pigs compared to the control pigs. A significant up regulation of IL-12 mRNA has been observed for the Lena (7.6 fold) and Belgium A strain (4 fold) at day 3 p.i. compared to the control. For IFN-α mRNA (Lena: 4.2 fold; Belgium A: 4.0 fold), IFN-γ (Lena: 8.9 fold; Belgium A: 9.2 fold), IL-1β (Lena: 6.7 fold; Belgium A: 5.4 fold), and IL-10 (Lena: 13.0 fold; Belgium A: 18.3 fold) an upregulation was observed at day 7 p.i. At day 7 p.i. there was also a significant increase of TNF-α mRNA in blood of the Lena-infected pigs (3.7 fold) compared to the control. Interestingly, at the end of the trial at day 33 p.i., there was an increase in IFN-γ mRNA in the blood of Belgium A (5.2 fold) and LV-infected pigs (4.7 fold), and not in blood of the Lena-infected pigs.
Fig. 5.
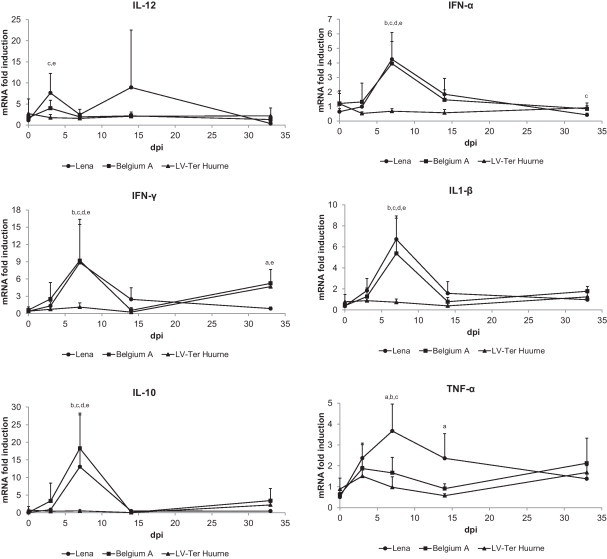
Peripheral blood cytokine responses of pigs infected with different PRRSV strains. Cytokine gene expression was assessed by qRT-PCR and expressed as fold induction compared to the control group ±S.D. “a” denotes a significant difference (p < 0.05) between Lena and Belgium A-infected pigs; “b” between Lena and LV-infected pigs; “c” between Lena and control pigs; “d” between Belgium A and LV-infected pigs; “e” between Belgium A and control pigs.
3.6. Humoral immune competence of pigs after PRRSV infection
Following ADV immunization, antibodies were detected in serum from pigs infected with the Belgium A or LV strain and control pigs from day 21 p.i. (Fig. 6 ). In serum from pigs infected with the Lena strain, antibodies were detected one sampling point later, at day 26 p.i. At this time, antibody titers from pigs infected with the Lena strain were significantly lower (0.3 log10) than the other groups (control pigs: 2.4 log10; pigs infected with: LV: 2.1 log10, Belgium A: 1.8 log10).
Fig. 6.
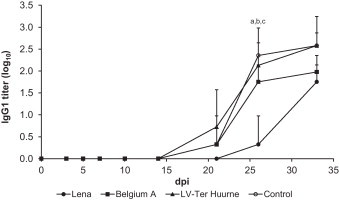
Aujeszky's disease (ADV) antibody titers as determined by ELISA of pigs infected with different PRRSV strains and subsequently immunized with an ADV vaccine. Each data point represents the average PRRSV levels of three or four pigs ±S.D. “a” denotes a significant difference (p < 0.05) between Lena and Belgium A-infected pigs; “b” between Lena and LV-infected pigs; “c” between Lena and control pigs.
4. Discussion
In this study, the pathogenicity of and immune response to three European type PRRSV strains were compared. Infection with the subtype 3 strain Lena resulted in clinical signs, in contrast to infection with the subtype 1 strains, which did not induce fever or clinical signs. In a previous study with this strain in 6-week-old conventional pigs, clinical signs were more severe than in the present study and 40% of the pigs died within 3 weeks post-inoculation (Karniychuk et al., 2010). The severity of clinical signs and death of the pigs in that study might have been due to secondary infections, since Arcanobacterium pyogenes and S. suis were isolated from the lungs of two pigs. We used pigs from a farm with a specific-pathogen-free status and pigs were kept under animal biosafety level 3 conditions, which would prevent introductions of infectious agents. This could explain the different clinical outcome between the studies, although it cannot be excluded that differences in severity of clinical signs were caused by other factors such as differences in pig breed (Lewis et al., 2007).
Fever was only observed in pigs infected with the Lena strain. These pigs had around 100 times higher virus loads during the peak of infection, and higher cytokine levels in blood, mainly TNF-α, compared to the other strains. A relationship between TNF-α production/response in blood and the pathogenicity of PRRSV strains has been demonstrated before (Liu et al., 2010). It was described that the highly pathogenic American genotype PRRSV strain HuN4, which induces fever and clinical signs, induced higher levels of TNF-α in blood early in infection than its derivative strain HuN4-F112 of lower virulence (Liu et al., 2010). The HuN4 also replicated faster and to higher titers than the HuN4-F112 strain. The higher virus replication most likely means more trigger of innate immune defences, more cytokine production and more inflammation, resulting in fever.
The level of IFN-γ secreting cells was used to measure the efficiency of the adaptive cellular immune response. The level of IFN-γ secreting cells remained low for pigs infected with the Lena strain, but for pigs infected with the Belgium A (from day 14 p.i) or LV strain (from day 21 p.i.), an increase in IFN-γ secreting cells was observed compared to the control pigs. This would indicate that the subtype 1 strains Belgium A and LV induce a cell-mediated immune response, in contrast to pigs infected with the Lena strain. Interestingly, the level of IFN-γ secreting cells of the Belgium A-infected pigs was high (maximum average of 647 IFN-γ secreting cells/106 PBMC), compared to the LV strain (maximum average of 132 IFN-γ secreting cells/106 PBMC). Most studies with both European and American genotype strains showed levels lower than 100 IFN-γ secreting cells/106 PBMC before day 33 p.i. (Diaz et al., 2005, Batista et al., 2004). It is not clear why the level of IFN-γ secreting cells of the Belgium A-infected pigs was that high. Besides a stronger induction of the IFN-γ response in vivo, it is also possible that the Belgium A strain has a stronger ability to stimulate cells in vitro in the ELISPOT assay. This has been observed for other strains previously by Díaz et al. (2012).
For all strains, virus could be isolated from day 3 p.i. and peaked between days 7 and 14 p.i., then decreased strongly after day 14 p.i. During the decrease of viremia, an increase of NK cells was observed, mainly for the Belgium A and LV strain, which could be related to viral clearance. Beside the role of NK cells in virus clearing, cytotoxic T cells might have been involved in the clearing at the end of the experiment. Cytotoxic T cells were higher than in the control pigs for all infected groups at day 26 p.i., although only significantly higher for LV-infected pigs. This higher level of cytotoxic T cells coincided with a peak in IFN-γ secreting cells. The increase observed in the cytotoxic T cell level could (partly) be related to clonal expansion of PRRSV-specific T cells, although this increase could also be due to a homeostatic regulation of the infection and inflammation. A relationship between viral clearance and cytotoxic T cells has been previously observed in pigs infected with European and American genotype strains (Lamontagne et al., 2003, Gómez-Laguna et al., 2009). However, others failed to show that CD3+CD8high cells exert CTL activity toward LV-infected alveolar macrophages (Costers et al., 2009). Further studies should determine whether the PRRSV-specific cytotoxic T cell activity in vivo is involved in viral clearance.
It has been described for PRRSV that the polyclonal activation of B cells occurs mainly in lymphoid organs (Lamontagne et al., 2001, Diaz et al., 2005). The mobilization of these cells to tissue may explain the decrease in frequency of B cells in blood of the infected pigs. The activation of B cells in the lymph nodes could have resulted in the production of PRRSV-specific antibodies that were detected in blood from day 10 p.i. During the last week of the experiment at day 26 p.i., the level of B cells of the infected pigs was higher than the control pigs, at which point the level of PRRSV-specific antibodies was also at a peak. A similar course of CD21+ B cell levels in blood has been observed previously (Shi et al., 2008). Others, however, observed either an increase or a decrease in CD21+ cells for at least 4 weeks after infection with a European genotype strain (Diaz et al., 2005, Gómez-Laguna et al., 2009). Although in our study higher levels of B cells were observed in infected pigs than in the control pigs at the end of the experiment, this did not contribute to viral clearance by the production of neutralizing antibodies. The absence of neutralizing antibodies has been observed for infections with European genotype strains before (Diaz et al., 2005). However, others found neutralizing antibodies with the Lelystad strain around week 3–4 p.i. (Vanhee et al., 2010). The difference in results can be attributed to differences in the virus neutralization test method. Therefore, it cannot be ruled out that there were small amounts of neutralizing antibodies at day 33 p.i., but the level remained below the detection limit of the virus neutralization test used.
After many viral infections, such as swine influenza or porcine coronavirus (Van Reeth et al., 1999), a rapid production of type I IFN (IFN-α/β) by the innate immune response supresses initial viral replication and promotes the adaptive immune response (Kimman et al., 2009). For PRRSV, it has been shown in vitro that the type I IFN response is suppressed (Ait-Ali et al., 2011). There are, however, conflicting reports of PRRSV inhibiting or inducing a type I IFN response in vivo (Van Reeth et al., 1999, Gómez-Laguna et al., 2009). In the present study an increase in IFN-α mRNA in blood was observed at day 7 p.i. for the Lena and Belgium A-infected pigs, in parallel with the viral load. No increased level of IFN-α mRNA production was observed in the group of LV-infected pigs. This can be the result of differences between strains in IFN-α induction, as has been described to occur in in vitro studies with infection of porcine alveolar macrophages with different PRRSV strains (Lee et al., 2004), or different infection/replication kinetics of this strain in combination with the short-lived nature of mRNA in blood.
Pigs have large numbers of γδ T cells circulating in blood. These cells have been described to play a role in both innate and adaptive immune responses to viral or bacterial infections (Gerner et al., 2009). Gamma delta T cells can act as antigen presenting cells or induce cytotoxic responses. Regulatory functions have been demonstrated in mice and humans (Gerner et al., 2009). After infection with PRRSV, they can respond by proliferation and production of IFN-γ (Olin et al., 2005). In the present study, the level of CD8−γδ T cells decreased to a significantly lower level than the control pigs for the Lena and Belgium A-infected pigs at the peak of viremia. This level returned to normal for the Belgium A-infected pigs at day 21 p.i. A similar course as for the Belgium A-infected pigs in the level of γδ T cells has been observed after infection with an American genotype strain (Shi et al., 2008). However, for the Lena-infected pigs, the level of CD8−γδ T cells remained low until the end of the experiment at day 33 p.i. Because of the role of these cells in innate and adaptive responses, this could result in increased susceptibility or a decreased response to secondary infections, such as those observed for the lower response of Lena-infected pigs to immunization with ADV.
There is a general concept that PRRSV is an immunosuppressive agent. There are, however, contradicting results about the immunomodulating effect of PRRSV (De Bruin et al., 2000, Murtaugh et al., 2002, Suradhat et al., 2006, Jung et al., 2009). These contradicting results could, amongst other things, be caused by the differences in virus strains used. In the current study, ADV immunization was used to study the effect of the PRRSV strains on the humoral immune response to another antigen. It was shown that infection with Lena influenced the development of an antibody response against ADV. This was in contrast to infection with the LV or Belgium A strains. The results with the LV strain were in agreement with the results of De Bruin et al. (2000), who also showed that there was no effect of the LV strain infection on the antibody development against ADV. For more virulent strains like Lena, it has been shown before that infection can suppress the immune system. Infection with American genotype isolates that caused clinical signs resulted in classical swine fever virus vaccine failure or increased disease after a secondary infection with porcine respiratory coronavirus (Suradhat et al., 2006, Jung et al., 2009). In the present study, the mechanisms behind the delayed ADV antibody response of pigs infected with the Lena strain might be the lower level of T helper cells, CD8−γδ T cells and B cells in blood of these Lena infected pigs before the first immunization at day 7 p.i., possibly resulting in a lower level of effector cells.
The present study showed differences in the immune responses to three different European genotype PRRSV strains. However, the general picture was similar to other European genotype strains: a late adaptive immune response with no detectable neutralizing antibodies, and especially for the Lena strain, low levels of IFN-γ secreting cells (Diaz et al., 2005, Gómez-Laguna et al., 2009). The virulence of the strain seemed to be mostly affecting the cell populations and cytokine levels in blood. Although several mechanisms behind the difference in virulence have been discussed, further studies on cell populations and cytokine production in tissues should provide more insight in the mechanisms behind immune modulation by different virus strains. This knowledge on immune modulation by PRRSV should aid the development of intervention strategies such as improved vaccines or other therapeutic agents.
Acknowledgements
The authors thank Hans van Dasler from Pfizer Animal Health Weesp for providing the ADV vaccine, and Hans Nauwynck for provision of PRRSV strain Belgium A and Lena. The authors thank Helmi Fijten, Bernie Moonen and Esther Willems for their technical assistance. Jean-Pierre Frossard is thanked for critical reading of the manuscript. The animal caretakers are thanked for collection of all samples. This work was supported by the PoRRSCon Project of the European Union Seventh Framework Programme (Grant Agreement # 245141).
References
- Ait-Ali T., Wilson A.D., Carre W., Westcott D.G., Frossard J.P., Mellencamp M.A., Mouzaki D., Matika O., Waddington D., Drew T.W., Bishop S.C., Archibald A.L. Host inhibits replication of European porcine reproductive and respiratory syndrome virus in macrophages by altering differential regulation of type-I interferon transcriptional response. Immunogenetics. 2011;63:437–448. doi: 10.1007/s00251-011-0518-8. [DOI] [PubMed] [Google Scholar]
- Batista L., Pijoan C., Dee S., Olin M., Molitor T., Joo H.S., Xiao Z., Murtaugh M. Virological and immunological responses to porcine reproductive and respiratory syndrome virus in a large population of gilts. Can. J. Vet. Res. 2004;68:267–273. [PMC free article] [PubMed] [Google Scholar]
- Chaung H.C., Chen C.W., Hsieh B.L., Chung W.B. Toll-like receptor expressions in porcine alveolar macrophages and dendritic cells in responding to poly IC stimulation and porcine reproductive and respiratory syndrome virus (PRRSV) infection. Comp. Immunol. Microbiol. Infect. Dis. 2010;33:197–213. doi: 10.1016/j.cimid.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Collins J.E., Benfield D.A., Christianson W.T., Harris L., Hennings J.C., Shaw D.P., Goyal S.M., McCullough S., Morrison R.B., Joo H.S., Gorcyca D., Chladek D. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Invest. 1992;4:117–126. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- Costers S., Lefebvre D.J., Goddeeris B., Delputte P.L., Nauwynck H.J. Functional impairment of PRRSV-specific peripheral CD3 + CD8 high cells. Vet. Res. 2009;40:46. doi: 10.1051/vetres/2009029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwich L., Díaz I., Mateu E. Certainties, doubts and hypotheses in porcine reproductive and respiratory syndrome virus immunobiology. Virus Res. 2010;154:123–132. doi: 10.1016/j.virusres.2010.07.017. [DOI] [PubMed] [Google Scholar]
- De Bruin M.G., Samsom J.N., Voermans J.J., Van Rooij E.M., De Visser Y.E., Bianchi A.T. Effects of a porcine reproductive and respiratory syndrome virus infection on the development of the immune response against pseudorabies virus. Vet. Immunol. Immunopathol. 2000;76:125–135. doi: 10.1016/S0165-2427(00)00208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Greeff A., Benga L., Wichgers Schreur P.J., Valentin-Weigand P., Rebel J.M., Smith H.E. Involvement of NF-κB and MAP-kinases in the transcriptional response of alveolar macrophages to Streptococcus suis. Vet. Microbiol. 2010;141:59–67. doi: 10.1016/j.vetmic.2009.07.031. [DOI] [PubMed] [Google Scholar]
- Diaz I., Darwich L., Pappaterra G., Pujols J., Mateu E. Immune responses of pigs after experimental infection with a European strain of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2005;86:1943–1951. doi: 10.1099/vir.0.80959-0. [DOI] [PubMed] [Google Scholar]
- Diaz I., Darwich L., Pappaterra G., Pujols J., Mateu E. Different European-type vaccines against porcine reproductive and respiratory syndrome virus have different immunological properties and confer different protection to pigs. Virology. 2006;351:249–259. doi: 10.1016/j.virol.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Díaz I., Gimeno M., Darwich L., Navarro N., Kuzemtseva L., López S., Galindo I., Segalés J., Martín M., Pujols J., Mateu E. Characterization of homologous and heterologous adaptive immune responses in porcine reproductive and respiratory syndrome virus infection. Vet. Res. 2012;43:30. doi: 10.1186/1297-9716-43-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr P.M., Gebreyes W.A., Almond G.W. Porcine reproductive and respiratory syndrome virus: age and management system disease modeling for pathogenic co-infection. 2007. J. Swine Health. Prod. 2007;15:258–263. [Google Scholar]
- Durand S.V., Hulst M.M., De Wit A.A., Mastebroek L., Loeffen W.L. Activation and modulation of antiviral and apoptotic genes in pigs infected with classical swine fever viruses of high, moderate or low virulence. Arch. Virol. 2009;154:1417–1431. doi: 10.1007/s00705-009-0460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney D.J. Charles Griffin & Company Ltd.; London: 1978. Statistical Methods in Biological Assay. pp. 394–401. [Google Scholar]
- Frossard J.P., Fearnley C., Naidu B., Errington J., Westcott D.G., Drew T.W. Porcine reproductive and respiratory syndrome virus: antigenic and molecular diversity of British isolates and implications for diagnosis. Vet. Microbiol. 2012 doi: 10.1016/j.vetmic.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Gerner W., Käser T., Saalmüller A. Porcine T lymphocytes and NK cells—an update. Dev. Comp. Immunol. 2009;33:310–320. doi: 10.1016/j.dci.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Gómez-Laguna J., Salguero F.J., De Marco M.F., Pallarés F.J., Bernabé A., Carrasco L. Changes in lymphocyte subsets and cytokines during European porcine reproductive and respiratory syndrome: increased expression of IL-12 and IL-10 and proliferation of CD4(−) CD8(high) Viral Immunol. 2009;22:261–271. doi: 10.1089/vim.2009.0003. [DOI] [PubMed] [Google Scholar]
- Grau-Roma L., Fraile L., Segalés J. Recent advances in the epidemiology, diagnosis and control of diseases caused by porcine circovirus type 2. Vet. J. 2011;187:23–32. doi: 10.1016/j.tvjl.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Haynes J.S., Halbur P.G., Sirinarumitr T., Paul P.S., Meng X.J., Huffman E.L. Temporal and morphologic characterization of the distribution of porcine reproductive and respiratory syndrome virus (PRRSV) by in situ hybridization in pigs infected with isolates of PRRSV that differ in virulence. Vet. Pathol. 1997;34:39–43. doi: 10.1177/030098589703400106. [DOI] [PubMed] [Google Scholar]
- Johnson W., Roof M., Vaughn E., Christopher-Hennings J., Johnson C.R., Murtaugh M.P. Pathogenic and humoral immune responses to porcine reproductive and respiratory syndrome virus (PRRSV) are related to viral load in acute infection. Vet. Immunol. Immunopathol. 2004;102:147–233. doi: 10.1016/j.vetimm.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Jonjić S., Koszinowski U.H. Monoclonal antibodies reactive with swine lymphocytes. I. Antibodies to membrane structures that define the cytolytic T lymphocyte subset in the swine. J. Immunol. 1984;133:647–652. [PubMed] [Google Scholar]
- Jung K., Renukaradhya G.J., Alekseev K.P., Fang Y., Tang Y., Saif L.J. Porcine reproductive and respiratory syndrome virus modifies innate immunity and alters disease outcome in pigs subsequently infected with porcine respiratory coronavirus: implications for respiratory viral co-infections. J. Gen. Virol. 2009;90:2713–2723. doi: 10.1099/vir.0.014001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniychuk U.U., Geldhof M., Vanhee M., Van Doorsselaere J., Saveleva T.A., Nauwynck H.J. Pathogenesis and antigenic characterization of a new East European subtype 3 porcine reproductive and respiratory syndrome virus isolate. BMC Vet. Res. 2010;6:30. doi: 10.1186/1746-6148-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimman T.G., Brouwers R.A.M., Daus F.J., Van Oirschot J.T., Van Zaane D. Measurement of isotype-specific antibody responses to Aujeszky's disease virus in sera and mucosal secretions of pigs. Vet. Immunol. Immunopathol. 1992;31:95–113. doi: 10.1016/0165-2427(92)90089-9. [DOI] [PubMed] [Google Scholar]
- Kimman T.G., Cornelissen L.A., Moormann R.J., Rebel J.M., Stockhofe-Zurwieden N. Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine. 2009;27:3704–3718. doi: 10.1016/j.vaccine.2009.04.022. [DOI] [PubMed] [Google Scholar]
- Kirkham P.A., Takamatsu H., Yang H., Parkhouse R.M. Porcine CD3 epsilon: its characterization, expression and involvement in activation of porcine T lymphocytes. Immunology. 1996;87:616–623. doi: 10.1046/j.1365-2567.1996.498566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge K.L., Vaughn E.M., Roof M.B., Bautista E.M., Murtaugh M.P. Age-dependent resistance to porcine reproductive and respiratory syndrome virus replication in swine. Virol. J. 2009;6:177. doi: 10.1186/1743-422X-6-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarque G., Reeth K.V., Nauwynck H., Drexler C., Van Gucht S., Pensaert M. Impact of genetic diversity of European-type porcine reproductive and respiratory syndrome virus strains on vaccine efficacy. Vaccine. 2004;22:4183–4190. doi: 10.1016/j.vaccine.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Lamontagne L., Page C., Larochelle R., Longtin D., Magar R. Polyclonal activation of B cells occurs in lymphoid organs from porcine reproductive and respiratory syndrome virus (PRRSV)-infected pigs. Vet. Immunol. Immunopathol. 2001;82:165–182. doi: 10.1016/s0165-2427(01)00335-x. [DOI] [PubMed] [Google Scholar]
- Lamontagne L., Pagé C., Larochelle R., Magar R. Porcine reproductive and respiratory syndrome virus persistence in blood, spleen, lymph nodes, and tonsils of experimentally infected pigs depends on the level of CD8high T cells. Viral Immunol. 2003;16:395–406. doi: 10.1089/088282403322396181. [DOI] [PubMed] [Google Scholar]
- Lee S.M., Schommer S.K., Kleiboeker S.B. Porcine reproductive and respiratory syndrome virus field isolates differ in in vitro interferon phenotypes. Vet. Immunol. Immunopathol. 2004;102:217–231. doi: 10.1016/j.vetimm.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J., Reutner K., Essler S.E., Popow I., Gerner W., Steinberger P., Saalmüller A. Porcine SWC1 is CD52 – final determination by the use of a retroviral cDNA expression library. Vet. Immunol. Immunopathol. 2012;146:27–34. doi: 10.1016/j.vetimm.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C.R., Ait-Ali T., Clapperton M., Archibald A.L., Bishop S. Genetic perspectives on host responses to porcine reproductive and respiratory syndrome (PRRS) Viral Immunol. 2007;20:343–358. doi: 10.1089/vim.2007.0024. [DOI] [PubMed] [Google Scholar]
- Liu Y., Shi W., Zhou E., Wang S., Hu S., Cai X., Rong F., Wu J., Xu M., Xu M., Li L. Dynamic changes in inflammatory cytokines in pigs infected with highly pathogenic porcine reproductive and respiratory syndrome virus. Clin. Vaccine Immunol. 2010;17:1439–1445. doi: 10.1128/CVI.00517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loving C.L., Brockmeier S.L., Vincent A.L., Lager K.M., Sacco R.E. Differences in clinical disease and immune response of pigs challenged with a high-dose versus low-dose inoculum of porcine reproductive and respiratory syndrome virus. Viral Immunol. 2008;21:315–325. doi: 10.1089/vim.2008.0038. [DOI] [PubMed] [Google Scholar]
- Meier W.A., Galeota J., Osorio F.A., Husmann R.J., Schnitzlein W.M., Zuckermann F.A. Gradual development of the interferon-gamma response of swine to porcine reproductive and respiratory syndrome virus infection or vaccination. Virology. 2003;309:18–31. doi: 10.1016/s0042-6822(03)00009-6. [DOI] [PubMed] [Google Scholar]
- Meulenberg J.J., Hulst M.M., De Meijer E.J., Moonen P.L., Den Besten A., De Kluyver E.P., Wensvoort G., Moormann R.J. Lelystad virus belongs to a new virus family, comprising lactate dehydrogenase-elevating virus, equine arteritis virus, and simian hemorrhagic fever virus. Arch. Virol. Suppl. 1994;9(9):441–448. doi: 10.1007/978-3-7091-9326-6_43. [DOI] [PubMed] [Google Scholar]
- Mittelholzer C., Moser C., Tratschin J., Hofmann M.A. Analysis of classical swine fever virus replication kinetics allows differentiation of highly virulent from avirulent strains. Vet. Microbiol. 2000;74:293–308. doi: 10.1016/s0378-1135(00)00195-4. [DOI] [PubMed] [Google Scholar]
- Murtaugh M., Genzow P.M. Immunological solutions for treatment and prevention of porcine reproductive and respiratory syndrome (PRRS) Vaccine. 2011;29:8192–8204. doi: 10.1016/j.vaccine.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Murtaugh M., Xiao Z., Zuckermann F. Immunological responses of swine to porcine reproductive and respiratory syndrome virus infection. Viral Immunol. 2002;15:533–547. doi: 10.1089/088282402320914485. [DOI] [PubMed] [Google Scholar]
- Neumann E.J., Kliebenstein J.B., Johnson C.D., Mabry J.W., Bush E.J., Seitzinger A.H., Green A.L., Zimmerman J.J. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 2005;227:385–392. doi: 10.2460/javma.2005.227.385. [DOI] [PubMed] [Google Scholar]
- Nielsen J., Vincent I.E., Bøtner A., Ladekaer-Mikkelsen A.S., Allan G., Summerfield A., McCullough K.C. Association of lymphopenia with porcine circovirus type 2 induced postweaning multisystemic wasting syndrome (PMWS) Vet. Immunol. Immunopathol. 2003;92:97–111. doi: 10.1016/s0165-2427(03)00031-x. [DOI] [PubMed] [Google Scholar]
- Olin M.R., Batista L., Xiao Z., Dee S.A., Murtaugh M.P., Pijoan C.C., Molitor T.W. Gammadelta lymphocyte response to porcine reproductive and respiratory syndrome virus. Viral Immunol. 2005;18:490–499. doi: 10.1089/vim.2005.18.490. [DOI] [PubMed] [Google Scholar]
- Pescovitz M.D., Lunney J.K., Sachs D.H. Preparation and characterization of monoclonal antibodies reactive with porcine PBL. J. Immunol. 1984;133:368–375. [PubMed] [Google Scholar]
- Shi K., Li H., Guo X., Ge X., Jia H., Zheng S., Yang H. Changes in peripheral blood leukocyte subpopulations in piglets co-infected experimentally with porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Vet. Microbiol. 2008;129:367–377. doi: 10.1016/j.vetmic.2007.11.020. [DOI] [PubMed] [Google Scholar]
- Stadejek T., Oleksiewicz M.B., Scherbakov A.V., Timina A.M., Krabbe J.S., Chabros K., Potapchuk D. Definition of subtypes in the European genotype of porcine reproductive and respiratory syndrome virus: nucleocapsid characteristics and geographical distribution in Europe. Arch. Virol. 2008;153:1479–1488. doi: 10.1007/s00705-008-0146-2. [DOI] [PubMed] [Google Scholar]
- Summerfield A., McNeilly F., Walker I., Allan G., Knoetig S.M., McCullough K.C. Depletion of CD4(+) and CD8(high+) T-cells before the onset of viraemia during classical swine fever. Vet. Immunol. Immunopathol. 2001;78:3–19. doi: 10.1016/s0165-2427(00)00248-8. [DOI] [PubMed] [Google Scholar]
- Suradhat S., Kesdangsakonwut S., Sada W., Buranapraditkun S., Wongsawang S., Thanawongnuwech R. Negative impact of porcine reproductive and respiratory syndrome virus infection on the efficacy of classical swine fever vaccine. Vaccine. 2006;24:2634–2642. doi: 10.1016/j.vaccine.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Van Breedam W., Delputte P.L., Van Gorp H., Misinzo G., Vanderheijden N., Duan X., Nauwynck H.J. Porcine reproductive and respiratory syndrome virus entry into the porcine macrophage. J. Gen. Virol. 2010;91:1659–1667. doi: 10.1099/vir.0.020503-0. [DOI] [PubMed] [Google Scholar]
- Vanhee M., Costers S., Van Breedam W., Geldhof M.F., Van Doorsselaere J., Nauwynck H.J. A variable region in GP4 of European-type porcine reproductive and respiratory syndrome virus induces neutralizing antibodies against homologous but not heterologous virus strains. Viral Immunol. 2010;23:403–413. doi: 10.1089/vim.2010.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Linden I.F., Voermans J.J., Van der Linde-Bril E.M., Bianchi A.T., Steverink P.J. Virological kinetics and immunological responses to a porcine reproductive and respiratory syndrome virus infection of pigs at different ages. Vaccine. 2003;21:1952–1957. [PubMed] [Google Scholar]
- Van Reeth K., Labarque G., Nauwynck H., Pensaert M. Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res. Vet. Sci. 1999;67:47–52. doi: 10.1053/rvsc.1998.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Song T., Yu Y., Liu Y., Shi W., Wang S., Rong F., Dong J., Liu H., Cai X., Zhou E.M. Immune responses in piglets infected with highly pathogenic porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 2011;142:170–178. doi: 10.1016/j.vetimm.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Wensvoort G., Terpstra C., Pol J.M., Ter Laak E.A., Bloemraad M., De Kluyver E.P., Kragten C., Van Buiten L., Den Besten A., Wagenaar F., Broekhuijsen J.M., Moonen P.L.J.M., Zetstra T., De Boer E.A., Tibben H.J., De Jong M.F., Van’t Veld P., Groenland G.J.R., Van Gennip J.A., Voets M.T., Verheijden J.H.M., Braamskamp J. Mystery swine disease in the Netherlands: the isolation of Lelystad virus. Vet. Q. 1991;13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- Wichgers Schreur P.J., Rebel J.M., Smits M.A., Van Putten J.P., Smith H.E. Lgt processing is an essential step in Streptococcus suis lipoprotein mediated innate immune activation. PLoS ONE. 2011;6:e22299. doi: 10.1371/journal.pone.0022299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckermann F.A., Garcia E.A., Luque I.D., Christopher-Hennings J., Doster A., Brito M., Osorio F. Assessment of the efficacy of commercial porcine reproductive and respiratory syndrome virus (PRRSV) vaccines based on measurement of serologic response, frequency of gamma-IFN-producing cells and virological parameters of protection upon challenge. Vet. Microbiol. 2007;123:69–85. doi: 10.1016/j.vetmic.2007.02.009. [DOI] [PubMed] [Google Scholar]


