Abstract
Group A rotavirus (RVA) infections cause severe economic losses in intensively reared livestock animals, particularly in herds of swine and cattle. RVA strains are antigenically heterogeneous, and are classified in multiple G and P types defined by the two outer capsid proteins, VP7 and VP4, respectively. This study summarizes published literature on the genetic and antigenic diversity of porcine and bovine RVA strains published over the last 3 decades. The single most prevalent genotype combination among porcine RVA strains was G5P[7], whereas the predominant genotype combination among bovine RVA strains was G6P[5], although spatiotemporal differences in RVA strain distribution were observed. These data provide important baseline data on epidemiologically important RVA strains in swine and cattle and may guide the development of more effective vaccines for veterinary use.
Keywords: Surveillance, Epidemiology, Vaccination, Zoonosis, Porcine, Bovine
1. Introduction
Rotavirus is a major pathogen associated with acute gastroenteritis in animals and humans. The disease is usually seen in young animals and the susceptibility to disease decreases as the age progress, most likely due to changes in animal physiology and/or acquired immunity due to previous exposures (Estes and Kapikian, 2007).
The Rotavirus genus is divided into at least 7 distinct genetic groups or serogroups (A–G; Estes and Kapikian, 2007). Of these, rotavirus A (RVA) is genetically and antigenically the most diverse species within the genus (Matthijnssens and Desselberger, 2012), although more recent data also show a significant diversity in VP7 of rotavirus B and C strains in pigs (Martella et al., 2007b, Marthaler et al., 2012). In addition, RVAs are the most important due to their high prevalence and pathogenicity in humans and a variety of animals. The genome of RVA consists of 11 segments of double-stranded RNA enclosed in a triple-layered virus particle, and encodes six structural (VP1–VP4, VP6, and VP7) and five or six nonstructural proteins (NSP1–NSP6). A binary classification system reminiscent of the one used to classify influenza viruses has been established to characterize the two outer capsid proteins, VP4 and VP7, which independently elicit neutralizing antibodies. Thus, RVA strains are classified into VP4 or P types (for protease-sensitive) and VP7 or G types (for glycoprotein) (Estes and Kapikian, 2007). Thus far, at least 27 G types and 35 P types have been described, many of these identified in the last 5 years (Matthijnssens et al., 2011).
The diversity of RVA strains is mainly increased by accumulation of point mutations leading to genetic/antigenic drift and reassortment of cognate genes leading to genetic/antigenic shift (Matthijnssens and Desselberger, 2012). An additional important evolutionary mechanism is interspecies transmission, occurring when a RVA strain is able to infect a heterologous host species. This is often coupled with reassortment of cognate genes (Martella et al., 2010). In cattle, RVA strains belonging to at least 12 G types (G1–G3, G5, G6, G8, G10, G11, G15, G17, G21, and G24) and 11 P types (P[1], P[3], P[5–7], P[11], P[14], P[17], P[21], P[29], and P[33]) have been identified. Bovine RVA strains belonging to G6, G8, and G10, in association with P[1], P[5], and P[11], are commonly found in cattle, though strains belonging to G1–G3, G5, and G11 and P[3], P[6], P[7], and P[14] have been detected sporadically. An unusual G17P[17] avian-like bovine RVA strain (Bo/993/83) has also been isolated from a calf, which is presumably the result of an interspecies transmission event from a bird RVA strain to a cow. In addition, bovine RVA strains with novel VP7 genotypes (G15, G21, and G24) and VP4 genotypes (P[21], P[29], and P[33]) have recently been identified (Matthijnssens et al., 2011). The most prevalent RVA strains found in pigs are G3, G4, G5, G9, and G11 in association with P[6] or P[7]. In addition, several VP7 types (G1, G2, G6, G8, G10, G12, and G26) and VP4 types (P[5], P[8], P[11], P[13], P[14], P[19], P[23], P[26], P[27], P[32], and P[34]) have been detected sporadically in pigs, bringing the total G and P types detected in swine to 12 and 13, respectively (Matthijnssens et al., 2011). Interestingly, several G and P types are shared between RVAs of these livestock animals and other host species, as indicated by molecular analysis of several RVA strains detected in horses, small ruminants or even birds (Martella et al., 2010). In addition, genotyping and phylogenetic analyses of rare human RVA strains have demonstrated on multiple occasions a close genetic relatedness to animal RVA strains (Gentsch et al., 2005, Matthijnssens et al., 2009a), and a comprehensive genetic analysis of all 11 genome segments revealed that the two major human genotype constellations, Wa-like and DS-1-like genogroups, have a common origin with porcine and bovine RVA strains, respectively (Matthijnssens et al., 2009b).
Given the natural history of RVA infection and the close relationship of swine and cattle with humans, porcine and bovine RVA strains are a large potential genetic pool for novel human RVA strains. In addition, porcine and bovine RVA strains are considered important pathogens in swine and cattle due to their economic impact on the swine and cattle industry. Therefore, understanding the molecular diversity of porcine and bovine RVA strains is important. In this study, we summarize the literature available on the prevalence and distribution of RVA G and P types until August 2011 in the two globally most important livestock animals, pigs and cattle, in order to provide baseline data following the concept delineated in a recent systematic review of human RVA strains (Bányai et al., 2012). This semiquantitative assessment of the dynamics of swine and cattle RVA strains may help better understand the evolution and ecology of RVA and also might help formulize better vaccines by the selection of more adequate antigens that evoke stronger and wider cross immunity, and better match the co-circulating RVA strains in a particular region.
2. Methods
During August 2011 we conducted a systematic search through PubMed using the terms “rotavirus” in combination with “porcine”, “swine”, “pig”, “piglet”, “Sus scrofa”, “hog”, “bovine”, “calf”, “Bos taurus”, or “cattle”. Searching for additional studies cited in the identified publications was also conducted. Data referring to porcine and bovine RVA strains were analyzed separately. Studies reported from the same country were cross-referenced by authors, location and time period to ensure the data were not duplicated. No stringent exclusion criteria were defined regarding sampling practice, sample size, length of study period, or typing method, however, we intended to keep only those studies which provided insight into the epidemiologic context.
For each study, the following information was inserted in a Microsoft Office Excel database: first author, manuscript title, journal name, year of publication, volume and page numbers, country of study, study period, sample size, typing method, type-specific RV prevalence. G and P type specificities were used as the primary endpoint to describe RVA strain prevalence and any possible shifts in their epidemiologic dynamics (Bányai et al., 2012).
3. Results
3.1. RVA strain prevalence in swine
A total of 763 original articles published between 1976 and 2011 were identified using PubMed searches. After 565 references were excluded as non-primary studies, the abstract of 198 papers were screened for relevance. Among these, 166 articles were further screened for eligibility, out of which 111 papers were excluded in the lack of G and/or P typing data and one additional article was excluded because it could not be obtained. Fifty-five articles were included in the final analysis. In total, 6968 fecal samples (including pooled samples) were collected in those studies, of which 1672 (24%) were found positive for RVA. Of these, 1149 strains were G and/or P-typed (Supplementary Table plus references 1–55).
Among the G typeable RVA strains (83%), at least 11 different specificities were identified, with G5 (45.8%) being by far the most prevalent genotype, followed by G3 (11.2%), and G4 (9.6%) (Fig. 1 ). The proportion of samples containing multiple G types or G untypeable RVA strains was 1.4% and 15.8%, respectively. Among the P typeable strains (76%), 17 P type specificities were found. The most prevalent P types were P[7] (47.4%), followed by P[6] (15.9%), and P[13] (3.2%). The rest of the P types represented individually <3% of the totals (Fig. 1). Mixed P types and P untypeable RVA strains were found in 3.2% and 21.1% of the tested cases, respectively.
Fig. 1.
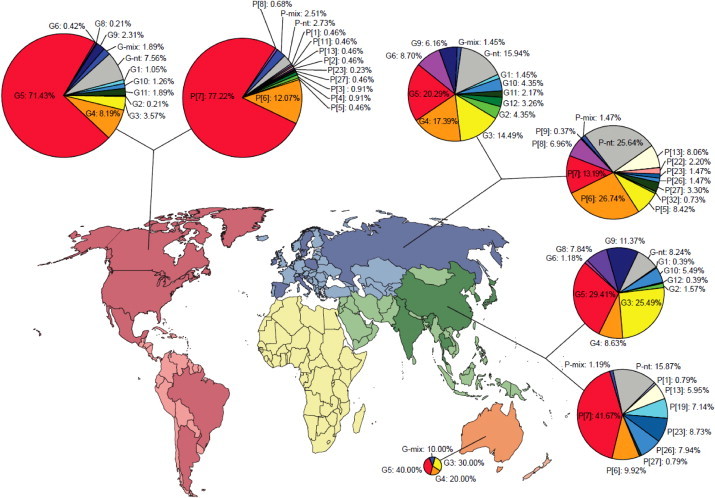
Geographical variation in the distribution of epizootically major and minor RVA strains in pigs. Continents are highlighted by various colors; dark shade shows countries providing data from any given region. ‘-mix’ and ‘-nt’ refers to samples containing multiple type specificities and non-typeable strains, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
When the G and P antigen specificities were combined to define the most prevalent porcine RVA strains, 47 individual G/P combinations were identified, but G5P[7] (37.3%) was the single most prevalent combination. Of interest, none of the remaining 46 G/P type combinations reached more than 6% prevalence (Fig. 2 ).
Fig. 2.
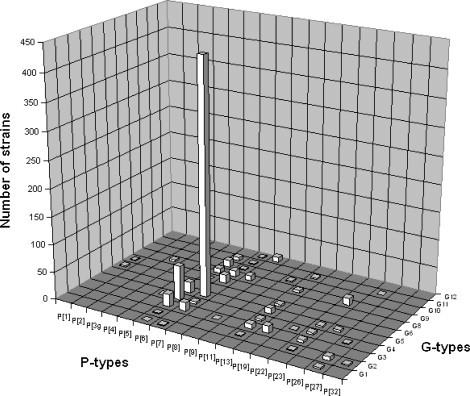
Relative importance of individual RVA G/P combinations in pigs. The numbers of porcine RVA strains identified with particular G/P combination are indicated in the graph. Percentile values are referred to in the text.
To identify any potential spatial trends in the porcine RVA strain distribution, the studies were analyzed and divided by geographical regions. The 55 eligible studies were performed in 19 countries (6 studies each in Europe, Americas, and Asia, and a single study in Australia) (Fig. 1, Supplementary Table). In general, G5 was the most common porcine RVA genotype in all 4 continents (20.3–71.4%). The second and third most common porcine VP7 specificities of RVA strains were G4 (17.4%) and G3 (14.5%) in Europe, G4 (8.2%) and G11 (1.9%) in the Americas, G3 (25.5%) and G9 (11.4%) in Asia, and G3 (30.0%) and G4 (20.0%) in Australia. Concerning the P type specificities, P[7] was the most common in the Americas (77.2%) and Asia (41.7%), while this genotype was less common in Europe (13.2%). In contrast, genotypes P[6] (26.7%) and P[13] (13.2%) were more common in Europe. The P[6] genotype was the second most common P type in the Americas (12.1%) and Asia (9.9%). In addition, P[23] was relatively common in Asia (8.7%).
Studies conducted before and after the mid-1990s alike reported G5 and P[7] as the globally most common VP7 and VP4 porcine RVA types, respectively (data not shown). To evaluate country-specific temporal trends in RVA strain prevalence, we analyzed data from countries representing different geographic areas and reporting relevant information over time. In this analysis Canada, Spain, and Thailand were selected based on availability of data. Fig. 3 clearly indicates the fluctuation in the G and P type prevalence in each country having reported relevant information over time, although it was not possible to breakdown data on annual or biennial bases. In addition to the temporal fluctuations, several new porcine VP7 and VP4 genotype specificities were reported in recent years (e.g., P[26], P[27], P[32]).
Fig. 3.
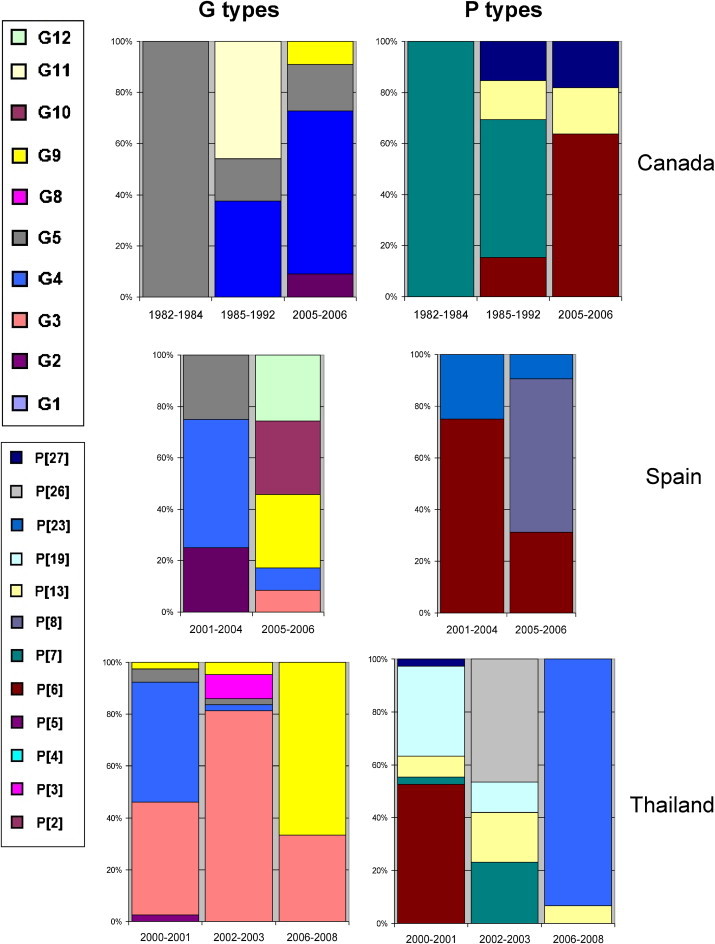
Temporal shift in the distribution of epizootically major and minor porcine RVA strains in selected countries. Relative prevalence of individual G and P types of porcine RVAs reported from Canada, Spain and Thailand over time is shown.
3.2. RVA strain prevalence in cattle
A total of 1480 original records were identified in the primary PubMed search. After excluding 1164 non-primary studies and screening 316 abstracts, 229 full articles were further screened for eligibility. Of these, 150 studies reported no G and/or P type data and 4 were no accessible for us. At the end data from 75 articles were analyzed. The total number of samples collected for RVA diagnosis in cattle was 14,076 of which 4749 samples (33.7%) were positive for RVA. Of these, 3204 samples were subjected to sero- or genotyping (Supplementary Table plus References 7,10,27,29,33,56–127).
Among the 11 VP7 specificities identified in this study, the most common types were G6 (56.7%), G10 (20.6%), and G8 (3.5%). The respective frequencies of the other G types found in cattle were <1% (Fig. 4, Fig. 5 ). Samples with multiple G types and infections due to untypeable strains were 5.9% and 11.3%, respectively. The predominating VP4 specificity among bovine RVA strains was P[5] (25.9%), followed by P[11] (21.5%) and P[1] (2.1%). Multiple P types were found in 6.2% of all samples, while 43.6% of RVA strains remained untypeable (Fig. 4).
Fig. 4.
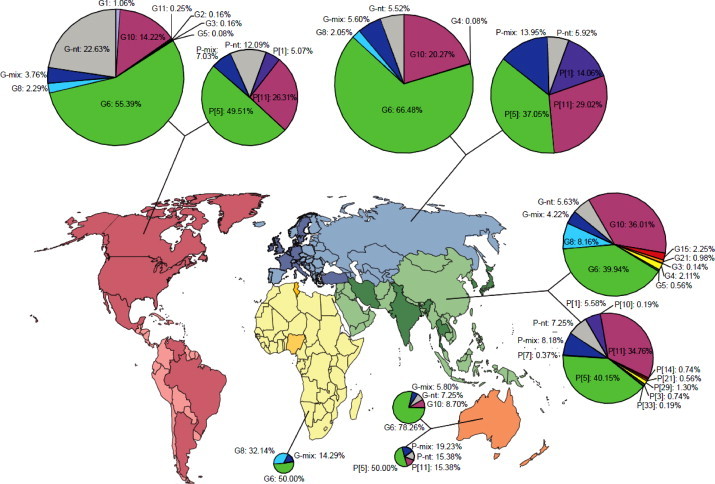
Geographical variation in the distribution of epizootically major and minor RVA strains in calves. Continents are highlighted by various colors; dark shade shows countries providing data from any given region. ‘-mix’ and ‘-nt’ refers to samples containing multiple type specificities and non-typeable strains, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
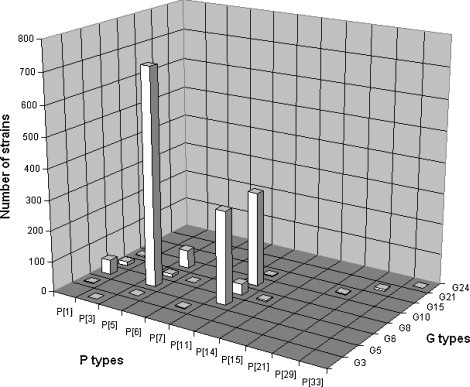
Relative importance of individual RVA G/P combinations in calves. The numbers of bovine RVA strains identified with particular G/P combinations are indicated in the graph. Percentile values are referred to in the text.
Nineteen individual G and P combinations were identified and three combinations, G6P[5], G6P[11], and G10P[11], represented 40.4% of all RVA strains. The frequency of other G and P combinations was <2% per individual G and P specificities (Fig. 5).
In total, 24 countries from 5 continents (9 in Europe, 6 in the Americas, 5 in Asia, 2 in Africa, and 2 in Australia-Oceania) (Fig. 4, Supplementary Table) provided relevant data to enable an assessment of the geographical distribution of bovine RVA strains. Serotype/genotype G6 strains were predominating in all 5 continents (range, 39.8–78.3%) followed by G10 in Americas, Europe, Asia, and Australia, and G8 in Africa. Regarding the P type specificities, genotype P[5] RVA strains were found most prevalent in Europe, the Americas, Asia, and Australia (range, 37.1–50.0%); this was followed by P[11] strains (range, 15.4–34.8%), and P[1] mainly in Europe, America, Australia, and Asia.
When published articles were analyzed by time frame of sample collection, the G6 and P[5] specificities were found to be globally predominant both in the period preceding the mid-1990s and the period thereafter (data not shown). Several countries reported data from different periods; Fig. 6 summarizes the temporal changes in strain prevalence in selected countries (Brazil, Italy, Japan) that periodically reported relevant information. Although the number of different specificities was relatively low in these countries, it is clear that distribution of different G and P types of bovine RVA strains changed over time.
Fig. 6.
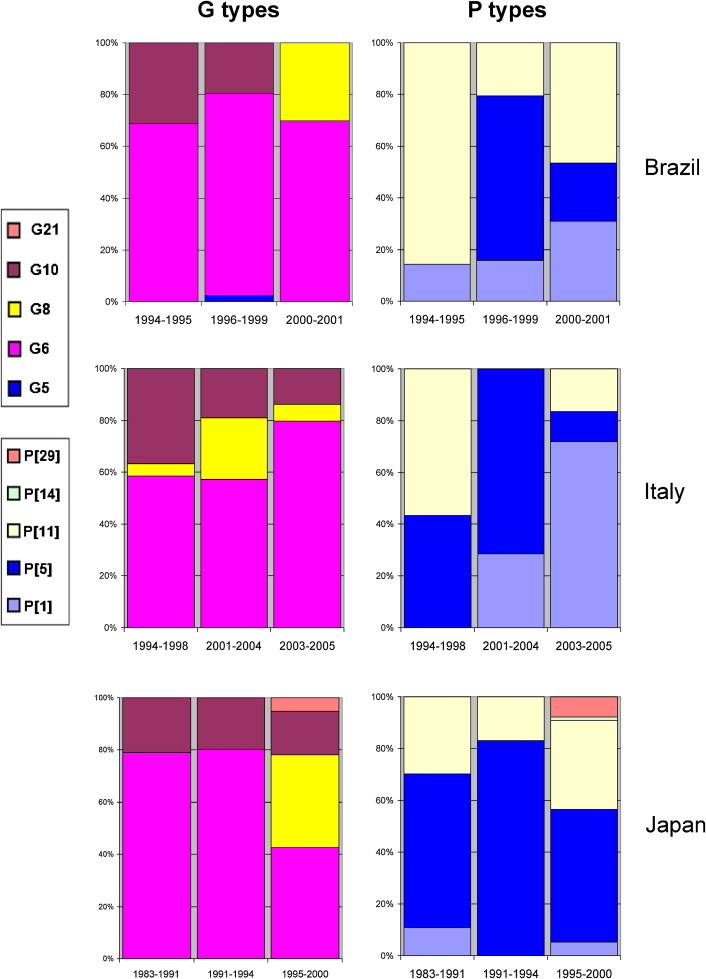
Temporal shift in the distribution of epizootically major and minor bovine RVA strains in selected countries. Relative prevalence of individual G and P types of bovine RVAs reported from Brazil, Italy and Japan over time is shown.
4. Discussion
RVA is considered a major gastroenteritis pathogen in cattle and swine, being responsible for significant economic losses due to increased mortality, treatment costs, and reduced weight gain. Co- and super-infections may worsen the outcome of primary RVA infections (Martella et al., 2007a).
Veterinary RVA vaccines, either live or inactivated, have been developed for prevention of neonatal calf diarrhea (e.g., Guardian®, Intervet/Merck Animal Health) and post-weaning enteritis of pigs (e.g., ProSystems RCE, Intervet/Merck Animal Health), although vaccination is not performed routinely. ProSystems RCE is a bivalent live oral RVA vaccine which is commercially available for swine in the US; this vaccine contains strains OSU (G5P[7]) and Gottfried (G4P[6]), but recently, a third porcine RVA strain, A2 (G9P[7]) was also isolated from the vaccine (the vaccine also contains four major Escherichia coli pilus antigens (K88, K99, F41, and 987P) and Clostridium perfringens type C toxoid) (Saif and Fernandez, 1996). Guardian® is a multiple antigen vaccine, which includes a cell-free extract of K99 pilus type of E. coli, a unique combination of two inactivated coronaviruses, two bovine RVA strains, NCDV (G6P[1]) and UK (G6P[5]), and a bacterin-toxoid from C. perfringens types C and D (Saif and Fernandez, 1996). There are other veterinary rotavirus vaccines licensed for use in other geographic regions and many of these share the antigen composition with the above listed combination vaccines. For example, in Italy, Pfizer distributes combination vaccines, such as Scourguard 3 that includes rotavirus, coronavirus and K99 pilus type E. coli strains, or Scourguard 4KC that includes rotavirus, coronavirus, K99 pilus type E. coli strains and C. perfringens type C., while Merial offers Trivaction 6, which has been developed to confer protection simultaneously against rotavirus, coronavirus and various E. coli strains. However, little or no additional information is available about their usage and effectiveness against RVA in the field. Nonetheless commercial RVA vaccines are administered parenterally to cows and sows during the late stage of gestation, in order to elicit a strong maternal immunity that is readily conferred to newborn animals (Saif and Fernandez, 1996). Some studies have demonstrated vaccine failure or breakthroughs that have been related to a number of factors, including inadequate managing conditions of animals or antigenic differences between vaccine and field RVA strains, even if vaccine and field strains shared partially their surface antigen specificities (Saif and Fernandez, 1996; Supplementary Table plus References 62,99,116). Therefore, assessing precisely the prevalence of various VP7 and VP4 type specificities is required to evaluate adequately vaccine effectiveness and to understand whether or not it may be necessary to construct polyvalent RVA vaccines for livestock animals.
Typical cattle and swine RVA strains may be able to infect other species through interspecies transmission (Martella et al., 2010). In addition, several studies have demonstrated that reassortment of genome segments between porcine and bovine RVA strains does occur. For example, porcine-like RVA G5P[7] strains were found in Korean cattle herds and vice versa, bovine-like RVA G6P[1] strains were sporadically detected in some Argentinean pig herds (Lorenzetti et al., 2011; Supplementary Table plus References 37,52). Interestingly, reassortant bovine-porcine RVAs with advantageous genetic configurations have been demonstrated to retain the ability to infect and cause disease in the heterologous host (Park et al., 2011, Kim et al., 2012).
From a public health perspective, the anthropozoonotic potential of porcine and bovine RVA strains has been recognized previously (Matthijnssens et al., 2008b, Ghosh and Kobayashi, 2011), but we are just beginning to understand its potential magnitude as surveillance of human RVA strains in the vaccine era continuously intensifies. Several antigen combinations of RVA are shared between humans and animals and it has been demonstrated that the 2 major genogroups of human RVAs, Wa-like and DS1-like, have a common origin with porcine and bovine RVA strains, respectively (Martella et al., 2010, Matthijnssens et al., 2008a, Matthijnssens et al., 2009a, Bányai et al., 2012). In addition, although being widespread in pigs worldwide, only a single unique G5P[7] RVA strain has been identified in humans among >110,000 genotyped RVA strains (Esona et al., 2009a, Bányai et al., 2012). Also, a number of human RVA strains possessing a P[6] genotype closely resembling the VP4 of porcine RVA strains have been identified (Bányai et al., 2004, Bányai et al., 2009). On the other hand, bovine-like RVA G8 and G10 strains have been identified from humans on several occasions, which may reach an epidemiological relevance in some geographical areas (Esona et al., 2009b, Esona et al., 2011). In contrast, some G/P genotype combinations are rare in humans, cattle and several other host species as exemplified by G6P[14] and G8P[14] RVA strains (Matthijnssens et al., 2009b, Bányai et al., 2010, Iturriza-Gómara et al., 2011, Chitambar et al., 2011, El Sherif et al., 2011).
This systematic review on bovine and porcine RVA strain diversity aimed at determining the relative prevalence of G and P type specificities across geographic regions over time based on published data in the literature. These studies revealed a sharp difference in the surveillance activity in humans and livestock. While characterization of human RVA strains has been intensified recently worldwide with over 110,000 genotyped RVA strains from 1996 to 2008, in swine and cattle a considerably lower number of strains were genotyped over the last 3 decades (porcine, ~1100; bovine, ~3200). The number of countries reporting RVA strain prevalence data for swine and cattle was fairly low, yet numerous new genotypes have been identified, mainly by research groups strongly motivated to investigate animal RVA molecular epidemiology.
The major findings of this review were the followings: (i) RVA G and P type diversity in swine was higher than in cattle and was comparable to that seen in humans from recent reviews (Gentsch et al., 2005, Matthijnssens et al., 2009a, Bányai et al., 2012) even though considerably fewer RVA strains were characterized in swine. The existence of numerous individual G and P genotypes in both host species may serve as a basis for further strain diversity through reassortment, and the description of mixed antigen specificities in various studies suggests the background for such reassortment events are given. Regarding the less frequently isolated strains, mistyping can sometimes occur, probably due to variations in the nucleotide sequence within primer binding regions (Cashman et al., 2010, Garaicoechea et al., 2006). (ii) RVA strain prevalence changes over time in different locations in both pigs and calves and the recognized strain diversity increases as more efforts are implemented to characterize untypeable strains using sophisticated methods, primarily nucleotide sequencing. (iii) Although RVA strains are diverse in livestock animals, both pigs and calves had a particular predominant genotype combination: G5P[7] in pigs and G6P[5] in calves. The predominance of these strains was seen across continents over time although some fluctuation was also evident and in some locations the dominating strains were different from these two genotypes. (iv) Finally, given that G5P[7] and G6P[5] are shared with some vaccine strains used in swine and cattle, respectively, further studies are needed to elucidate more specifically the vaccine effectiveness in herds where these vaccines are used to confer passive immunity. It will be also important to determine if there are genetic or antigenic differences in the structure of surface antigens between vaccine strains and field strains causing disease in calves and pigs born to vaccinated cows and sows, respectively. However, as rotavirus vaccines may not be administered continuously in the majority of swine or cattle herds, assessment of strain specific effectiveness on either a herd or individual basis may be challenging.
Our study has several limitations partly inherent to the heterogeneity of the data, potential sampling artifacts, and studies analyzed in our search. For example, various detection and typing methods markedly differ in sensitivity and specificity, which might bias the result of certain studies reporting G and P type prevalence in pigs and cattle. These differences in typing methods are clearly illustrated by the relatively high rates of untypeable strains in both host species. Different animal breeds and different animal housing practices might further complicate the situation raising additional concerns regarding the conclusions of the delineated spatiotemporal dynamics of RVA strain prevalence. In several studies, RVA strains were characterized after isolation on cell culture; because various strains can be isolated at different efficiency, this could also have resulted in bias in the overall animal RVA strain prevalence and diversity. Although we tried to be liberal when we defined our study selection criteria some studies may have been overlooked during the review process lowering the totals of RVA strains with available type specificity data. RVA has both endemic and epidemic forms, particularly in swine herds; therefore, it is unclear whether detection of multiple strains may actually reflect strain diversity on a single farm when samples were pooled before laboratory process. Moreover, the number of countries providing relevant information was very low and most studies analyzed a selected small number of strains, thus an extrapolation of data to a wider geographic region may result in distortion in the true global strain prevalence in these animal populations. Similar pitfalls have been encountered during the review of human RVA strains (Bányai et al., 2012). Also, we found limited information on vaccine use in settings where the analyzed studies were reported from; thus, any possible vaccine-associated pressure on RVA strain prevalence remained hidden.
Nonetheless, this study is the first to report comprehensive baseline data of RVA strain prevalence in livestock animals using the methodology of systematic reviews. Despite the fact that relatively low number of countries have provided relevant data on bovine and porcine RVA strain prevalence and diversity, the extrapolations on the regional strain prevalence may be valid given that the same few G and P type specificities were identified to be dominating across countries and over decades. In particular, genotypes G5 and P[7] in swine, and G6, P[5], and P[11] in cattle, were found epizootically to be the most important. Synchronized RVA strain surveillance in humans and animals using standardized methods may provide important and relevant data on animal RVA strain prevalence to aid vaccination efforts and understand the health risk of human diseases caused by animal RVAs.
Conflict of interest
The authors declare that they have no competing interests.
Authors’ contribution
KB and VM created and designed the study; HP and BL collected the data; HP, BL, JM, MC, and BG performed the data analysis and drafted the illustrations; KB, HP, BL, FJ, SG, BG, MC, JM, and VM interpreted the data; HP, VM, and KB drafted the manuscript; FJ, BL, BG, SG, MC, and JM critically revised the manuscript. All authors approved the final manuscript.
Acknowledgements
KB was supported by the Hungarian Academy of Sciences (OTKA, PD76364; Momentum program). JM was supported by an FWO (‘Fondsvoor Wetenschappelijk Onderzoek’) post-doctoral fellowship. FJ received János Bolyai scholarship from the Hungarian Academy of Sciences. We are grateful to the anonymous reviewers of this journal for giving valuable suggestions.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vetmic.2013.03.020.
Contributor Information
Hajnalka Papp, Email: papp.hajnalka.1986@gmail.com.
Brigitta László, Email: laszlob83@gmail.com.
Ferenc Jakab, Email: jakabf@gamma.ttk.pte.hu.
Balasubramanian Ganesh, Email: ganeshvirologist@yahoo.co.in.
Simona De Grazia, Email: simona.degrazia@unipa.it.
Jelle Matthijnssens, Email: jelle.matthijnssens@uzleuven.be.
Max Ciarlet, Email: max.ciarlet@novartis.com.
Vito Martella, Email: vito.martella@uniba.it.
Krisztián Bányai, Email: bkrota@hotmail.com.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Bányai K., Martella V., Jakab F., Melegh B., Szűcs G. Sequencing and phylogenetic analysis of human genotype P[6] rotavirus strains detected in Hungary provides evidence for genetic heterogeneity within the P[6] VP4 gene. J. Clin. Microbiol. 2004;42:4338–4343. doi: 10.1128/JCM.42.9.4338-4343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bányai K., Esona M.D., Kerin T.K., Hull J.J., Mijatovic S., Vásconez N., Torres C., de Filippis A.M., Foytich K.R., Gentsch J.R. Molecular characterization of a rare, human-porcine reassortant rotavirus strain, G11P[6], from Ecuador. Arch. Virol. 2009;154:1823–1829. doi: 10.1007/s00705-009-0499-1. [DOI] [PubMed] [Google Scholar]
- Bányai K., Papp H., Dandár E., Molnár P., Mihály I., Van Ranst M., Martella V., Matthijnssens J. Whole genome sequencing and phylogenetic analysis of a zoonotic human G8P[14] rotavirus strain. Infect. Genet. Evol. 2010;10:1140–1144. doi: 10.1016/j.meegid.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Bányai K., László B., Duque J., Steele A.D., Nelson E.A., Gentsch J.R., Parashar U.D. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30(Suppl. 1):A122–A130. doi: 10.1016/j.vaccine.2011.09.111. [DOI] [PubMed] [Google Scholar]
- Cashman O., Lennon G., Sleator R.D., Power E., Fanning S., O'Shea H. Changing profile of the bovine rotavirus G6 population in the south of Ireland from 2002 to 2009. Vet. Microbiol. 2010;146:238–244. doi: 10.1016/j.vetmic.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Chitambar S.D., Arora R., Kolpe A.B., Yadav M.M., Raut C.G. Molecular characterization of unusual bovine group A rotavirus G8P[14] strains identified in western India: emergence of P[14] genotype. Vet. Microbiol. 2011;148:384–388. doi: 10.1016/j.vetmic.2010.08.027. [DOI] [PubMed] [Google Scholar]
- El Sherif M., Esona M.D., Wang Y., Gentsch J.R., Jiang B., Glass R.I., Abou Baker S., Klena J.D. Detection of the first G6P[14] human rotavirus strain from a child with diarrhea in Egypt. Infect. Genet. Evol. 2011;11:1436–1442. doi: 10.1016/j.meegid.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Esona M.D., Geyer A., Banyai K., Page N., Aminu M., Armah G.E., Hull J., Steele D.A., Glass R.I., Gentsch J.R. Novel human rotavirus genotype G5P[7] from child with diarrhea Cameroon. Emerg. Infect. Dis. 2009;15:83–86. doi: 10.3201/eid1501.080899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esona M.D., Geyer A., Page N., Trabelsi A., Fodha I., Aminu M., Agbaya V.A., Tsion B., Kerin T.K., Armah G.E., Steele A.D., Glass R.I., Gentsch J.R. Genomic characterization of human rotavirus G8 strains from the African rotavirus network: relationship to animal rotaviruses. J. Med. Virol. 2009;81:937–951. doi: 10.1002/jmv.21468. [DOI] [PubMed] [Google Scholar]
- Esona M.D., Banyai K., Foytich K., Freeman M., Mijatovic-Rustempasic S., Hull J., Kerin T., Steele A.D., Armah G.E., Geyer A., Page N., Agbaya V.A., Forbi J.C., Aminu M., Gautam R., Seheri L.M., Nyangao J., Glass R., Bowen M.D., Gentsch J.R. Genomic characterization of human rotavirus G10 strains from the African rotavirus network: relationship to animal rotaviruses. Infect. Genet. Evol. 2011;11:237–241. doi: 10.1016/j.meegid.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Estes M.K., Kapikian A.Z. Rotaviruses. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. 5th edn. Vol. 2. Lippincott Williams & Wilkins/Wolters Kluwer; Philadelphia: 2007. pp. 1917–1974. (Fields Virology). [Google Scholar]
- Garaicoechea L., Bok K., Jones L.R., Combessies G., Odeón A., Fernandez F., Parreño V. Molecular characterization of bovine rotavirus circulating in beef and dairy herds in Argentina during a 10-year period (1994–2003) Vet. Microbiol. 2006;118:1–11. doi: 10.1016/j.vetmic.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Gentsch J.R., Laird A.R., Bielfelt B., Griffin D.D., Banyai K., Ramachandran M., Jain V., Cunliffe N.A., Nakagomi O., Kirkwood C.D., Fischer T.K., Parashar U.D., Bresee J.S., Jiang B., Glass R.I. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J. Infect. Dis. 2005;192(Suppl. 1):S146–S159. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Kobayashi N. Whole-genomic analysis of rotavirus strains: current status and future prospects. Future Microbiol. 2011;6:1049–1065. doi: 10.2217/fmb.11.90. [DOI] [PubMed] [Google Scholar]
- Iturriza-Gómara M., Dallman T., Bányai K., Böttiger B., Buesa J., Diedrich S., Fiore L., Johansen K., Koopmans M., Korsun N., Koukou D., Kroneman A., László B., Lappalainen M., Maunula L., Marques A.M., Matthijnssens J., Midgley S., Mladenova Z., Nawaz S., Poljsak-Prijatelj M., Pothier P., Ruggeri F.M., Sanchez-Fauquier A., Steyer A., Sidaraviciute-Ivaskeviciene I., Syriopoulou V., Tran A.N., Usonis V., Van Ranst M., De Rougemont A., Gray J. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiol. Infect. 2011;139:895–909. doi: 10.1017/S0950268810001810. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Park J.G., Alfajaro M.M., Kim D.S., Hosmillo M., Son K.Y., Lee J.H., Bae Y.C., Park S.I., Kang M.I., Cho K.O. Pathogenicity characterization of a bovine triple reassortant rotavirus in calves and piglets. Vet. Microbiol. 2012;159:11–22. doi: 10.1016/j.vetmic.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Lorenzetti E., da Silva Medeiros T.N., Alfieri A.F., Alfieri A.A. Genetic heterogeneity of wild-type G4P[6] porcine rotavirus strains detected in a diarrhea outbreak in a regularly vaccinated pig herd. Vet. Microbiol. 2011;154:191–196. doi: 10.1016/j.vetmic.2011.06.026. [DOI] [PubMed] [Google Scholar]
- Martella V., Bányai K., Lorusso E., Bellacicco A.L., Decaro N., Camero M., Bozzo G., Moschidou P., Arista S., Pezzotti G., Lavazza A., Buonavoglia C. Prevalence of group C rotaviruses in weaning and post-weaning pigs with enteritis. Vet. Microbiol. 2007;123:26–33. doi: 10.1016/j.vetmic.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Martella V., Bányai K., Lorusso E., Decaro N., Bellacicco A., Desario C., Corrente M., Greco G., Moschidou P., Tempesta M., Arista S., Ciarlet M., Lavazza A., Buonavoglia C. Genetic heterogeneity in the VP7 of group C rotaviruses. Virology. 2007;367:358–366. doi: 10.1016/j.virol.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Martella V., Bányai K., Matthijnssens J., Buonavoglia C., Ciarlet M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010;140:246–255. doi: 10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- Marthaler D., Rossow K., Gramer M., Collins J., Tsunemitsu H., Kuga K., Suzuki T., Ciarlet M., Matthijnssens J. Detection of substantial porcine group B rotavirus genetic diversity in the United States, resulting in a modified classification proposal for G genotypes. Virology. 2012;433:85–96. doi: 10.1016/j.virol.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Ciarlet M., Heiman E., Arijs I., Delbeke T., McDonald S.M., Palombo E.A., Iturriza-Gómara M., Maes P., Patton J.T., Rahman M., Van Ranst M. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 2008;82:3204–3219. doi: 10.1128/JVI.02257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Rahman M., Ciarlet M., Van Ranst M. Emerging human rotavirus genotypes. In: Palombo E.A., Kirkwood C.D., editors. Viruses in the Environment. Tribandrum; India: 2008. pp. 171–219. [Google Scholar]
- Matthijnssens J., Bilcke J., Ciarlet M., Martella V., Bányai K., Rahman M., Zeller M., Beutels P., Van Damme P., Van Ranst M. Rotavirus disease and vaccination: impact on genotype diversity. Future Microbiol. 2009;4:1303–1316. doi: 10.2217/fmb.09.96. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J., Potgieter C.A., Ciarlet M., Parreño V., Martella V., Bányai K., Garaicoechea L., Palombo E.A., Novo L., Zeller M., Arista S., Gerna G., Rahman M., Van Ranst M. Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? J. Virol. 2009;83:2917–2929. doi: 10.1128/JVI.02246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Ciarlet M., McDonald S.M., Attoui H., Bányai K., Brister J.R., Buesa J., Esona M.D., Estes M.K., Gentsch J.R., Iturriza-Gómara M., Johne R., Kirkwood C.D., Martella V., Mertens P.P., Nakagomi O., Parreño V., Rahman M., Ruggeri F.M., Saif L.J., Santos N., Steyer A., Taniguchi K., Patton J.T., Desselberger U., Van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch. Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Desselberger U. Genome diversity and evolution of rotaviruses. In: Hacker J., Dobrindt U., Kurth R., editors. Genome Plasticity and Infectious Diseases. ASM Press; Washington, D.C.: 2012. pp. 219–241. [Google Scholar]
- Park S.I., Matthijnssens J., Saif L.J., Kim H.J., Park J.G., Alfajaro M.M., Kim D.S., Son K.Y., Yang D.K., Hyun B.H., Kang M.I., Cho K.O. Reassortment among bovine, porcine and human rotavirus strains results in G8P[7] and G6P[7] strains isolated from cattle in South Korea. Vet. Microbiol. 2011;152:55–66. doi: 10.1016/j.vetmic.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Saif L.J., Fernandez F.M. Group A rotavirus veterinary vaccines. J. Infect. Dis. 1996;174(Suppl. 1):S98–S106. doi: 10.1093/infdis/174.Supplement_1.S98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


