Abstract
A longitudinal study was carried out in five French farrow-to-finish herds differently affected by respiratory diseases to describe the carrying and infection patterns of batches of sows to various respiratory pathogens during gestation and lactation. An entire batch of sows was followed during two successive reproduction cycles. Nasal, tonsillar and oro-pharyngeal swabs and blood samples were taken from each sow 9 and 4 weeks before farrowing and 1 and 4 weeks after farrowing. Mycoplasma hyopneumoniae, Actinobacillus pleuropneumoniae, Pasteurella multocida, Haemophilus parasuis and Streptococcus suis were detected from swab samples using PCR assays. Blood samples were tested for antibodies against M. hyopneumoniae, A. pleuropneumoniae serotypes 1-9-11 and 2, Porcine Circovirus type-2 (PCV-2) and Porcine Reproductive and Respiratory Syndrome virus (PRRSV) by ELISA tests. Antibodies against H1N1, H1N2 and H3N2 Swine Influenza Viruses (SIV) of European lineages were tested by hemagglutination inhibition assay. The results indicated that S. suis is widespread among sows (67.1% of PCR-positive sows). A. pleuropneumoniae, P. multocida, and H. parasuis were detected by PCR in 30.9%, 24.6% and 23.4% of the sows, respectively. Antibodies against M. hyopneumoniae were recovered from more than 55% of the sows in all herds whereas the micro-organism was detected in 2.4% of the sows. Although PCV-2 and SIV infections were highly prevalent, the PRRSV infection patterns ranged from no infection in farms mildly affected by respiratory diseases to active circulation in more severely affected herds. The sow population thus constitutes a reservoir for a continuous circulation of respiratory pathogens and needs to be properly considered in control strategies.
Keywords: Respiratory diseases, Sows, Pathogen detection, Infection dynamics
1. Introduction
Respiratory diseases are some of the most common and costly diseases affecting growing-finishing pigs reared under intensive confined conditions. Several bacterial and viral agents are involved in the pathogenesis of these diseases. Mycoplasma hyopneumoniae, most frequently in combination with other bacteria like Pasteurella multocida, Haemophilus parasuis, Streptococcus suis and Actinobacillus pleuropneumoniae, or viruses such as Swine Influenza Virus (SIV), Porcine Reproductive and Respiratory Syndrome virus (PRRSV), Porcine Circovirus type-2 (PCV-2) and Porcine Respiratory Coronavirus (PRCV) are the most important micro-organisms responsible for lung diseases (Sorensen et al., 2006). Together they are involved in the Porcine Respiratory Disease Complex (PRDC).
Some studies have indicated that disease outcome may be influenced by the infection pattern and infectious pressure of the pathogens in growing-finishing pigs, particularly for M. hyopneumoniae (Sibila et al., 2004, Fano et al., 2007). Indeed, the infection pressure had to be at a very low level throughout the pigs’ lifetime in order to reduce disease severity. Particular attention to critical transmission points is required to reduce pathogen transmission between animals. For many of the micro-organisms involved in respiratory diseases vertical transmission, relying on sow to piglet contamination, is suspected to occur in endemically infected herds during the suckling phase (Clark et al., 1991, Amass et al., 1996, Vigre et al., 2002, Zimmerman et al., 2006). Sows can thus serve as a reservoir for the continuous circulation of infectious pathogens especially in farrow-to-finish systems, where both populations, (i.e., the breeding herd and the growing-finishing pigs), are reared on the same site (Calsamiglia and Pijoan, 2000, Sorensen et al., 2006). Reducing the infection pressure in the sow herd may help to decrease spreading in the offspring. However, at present, little is known about the shedding and infection dynamics of respiratory pathogens, especially bacteria, in sow populations. Such information would be helpful to (i) properly design epidemiological studies for assessing/understanding infection dynamics within a herd and detecting the risk factors of respiratory diseases, (ii) provide accurate information for modeling studies, i.e. parameterization and validation of epidemiological models, and (iii) implement adequate control strategies. The aim of the present study was therefore to describe the carrying and infection patterns of batches of sows to various respiratory pathogens during gestation and lactation in herds differently affected by respiratory disorders.
2. Materials and methods
2.1. Herds selection
A longitudinal study was carried out in five single site farrow-to-finish herds located in Brittany (Western France). The farms were included on the basis of the veterinarian's knowledge of the farm and the results of an initial screening of 12 herds. Five herds were selected for different patterns regarding their respiratory health status in growing-finishing pigs and to tally with the most common herd health patterns, housing and herd management practices encountered in the field at that time. They were also selected for convenience as the farmer had to agree with our protocol. The screening visit took place 2–3 months before the present longitudinal field study was started. Clinical signs of respiratory disorders in the farrowing, post-weaning and finishing sections were recorded. Nasal, tonsillar and oro-pharyngeal swabs and blood samples were obtained from 30 pigs: 10 in the post-weaning section, 10 at the beginning and 10 at the end of the finishing phase. Lungs from at least 30 of the sampled finishing pigs were collected at the slaughterhouse. Macroscopic lung lesions of pneumonia were scored (Madec and Kobisch, 1982) and samples of lung lesions were taken. Swabs and lung samples were examined by PCR in our laboratory to detect M. hyopneumoniae, P. multocida, A. pleuropneumoniae, H. parasuis and S. suis. All sera were analysed for the detection of antibodies to PRRSV and SIV (H1N1, H3N2 and H1N2).
The retained farms differed in terms of clinical severity, infection profile, and the type and extent of lung lesions (Table 1 ). Disease severity and complexity showed a gradual increase from farms A to E. Herd A was mildly affected by respiratory diseases, according to clinical diagnosis and macroscopic lung lesions examination. The growing-finishing pigs were free from PRRSV and SIV. The lowest level of lung lesions in the five herds, was recorded in herd A. Herds B and C were endemically infected by PRRSV and differed in lung lesion severity, the finishing pigs in farm C being more affected by pneumonia. The last 2 farms (herds D and E) were the most severely affected by lung lesions as pneumonia and pleuritis were detected in more than 90% and 25% of the pigs, respectively. The finishing pigs were PRRSV and SIV positive, whereas the pigs from herd D tested positive for H1N1 antibodies, the growing pigs from farm E had antibodies against both SIV subtypes (H1N1 and H1N2).
Table 1.
Clinical, viral and bacteriological profiles and lung lesions of growing and finishing batches in the five selected farrow-to-finish pig herds (nasal, tonsillar, oro-pharyngeal swabs and blood samples of 30 growing-finishing pigs, lung examination of 30 slaughtered pigs, findings of the screening visit performed 2–3 months before the start of the longitudinal study).
| Herd | Clinical signs (cough)a | Pathogen detection by PCR in swab samples (number of positive pigs) |
Viral statusb |
Lung lesions |
Pathogen detection by PCR in lung tissuesb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mhp | Pm | App | Hps | Ssuis | PRRS | SIV | Pneumonia mean score (/28) (σ)c | % pleuritis | Mhp | Pm | App | Hps | Ssuis | ||
| A | Fin. | 1 | 9 | 0 | 28 | 30 | − | − | 0.9 (2.5) | 5.1 | + | − | − | − | + |
| B | Fin. | 0 | 16 | 0 | 30 | 2 | + | − | 2.1 (3.7) | 15 | + | + | − | − | − |
| Cd | Fin. | 0 | 20 | 0 | 17 | 17 | + | − | 9.6 (4.2) | 0 | + | − | + | + | − |
| De | Far./Fin. | 3 | 26 | 0 | 30 | 18 | + | H1N1 | 10.1 (6.3) | 38.5 | + | + | − | − | − |
| E | PW/Fin. | 0 | 4 | 2 | 29 | 13 | + | H1N1, H1N2 | 6.4 (5.1) | 28.6 | + | − | − | + | − |
Fin.: finishing; Far.: farrowing; PW: post-weaning.
(+) positive; (−) negative.
10 growers and 10 market-aged pigs.
Nasal swabs.
2.2. Animals and sampling scheme
The herds were managed all-in all-out by room in the farrowing, post-weaning and fattening sections with a 3-week batch interval between farrowing for 3 farms, 4-week interval for herd A and 1-week interval for herd B. The main features of the five herds are detailed in Table 2 . After checking for pregnancy, the sows in each herd were housed in a gestation room and were then moved to farrowing rooms about 1 week before the expected farrowing time. The piglets were weaned around 4 weeks after farrowing except in farms A and C where the piglets were weaned at 3 weeks of age. During the gestation and lactation periods, the sows were housed in closed facilities with mechanical ventilation and slatted floor. The sows in the selected herds were not vaccinated against M. hyopneumoniae during the study. Whether the sows were vaccinated earlier in life in the multiplier herd was unknown. The sows in herd C were vaccinated against A. pleuropneumoniae. The breeding sows in herd B were vaccinated against PRRSV and SIV. The same commercial SIV vaccine (antigens A/New Jersey/8/76 (H1N1) and A/Port Chalmers/1/73 (H3N2)) had previously been used in herd A but no SIV vaccine was used in this herd during the study. An inactivated PRRSV vaccine was used for the gilts in herd E. All farms except E vaccinated the sows and gilts against atrophic rhinitis.
Table 2.
Main characteristics of the five farrow-to-finish herds under study.
| Herd | Sow herd features |
Husbandry |
Technical performances |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of sows | Breed | % gilts | Vaccination scheme/respiratory diseases | Batch interval | Weaning age (weeks) | ADWGa (g/day; 7–25 kg) | FCRb (7–25 kg) | ADWGa (g/day; 25–105 kg) | FCRb (25–105 kg) | Age at 105 kg live weight (days) | % mortality from weaning to slaughter | |
| A | 130 | Naïma | 22 | Atrophic rhinitis | 4 weeks | 3 | 492 | 1.55 | 777 | 2.67 | 164 | 5.1 |
| B | 700 | LW × LR | 30 | Atrophic rhinitis, PRRSV, SIV | 1 week | 4 | 458 | 1.59 | 789 | 2.61 | 165 | 9.5 |
| C | 170 | Naïma + LW × LR | 31 | Atrophic rhinitis, App | 3 weeks | 3 | 501 | 1.64 | 817 | 2.62 | 160 | 7.0 |
| D | 220 | Naïma | 19 | Atrophic rhinitis | 3 weeks | 4 | 426 | 1.60 | 728 | 2.93 | 168 | 6.5 |
| E | 190 | LW × LR | 22 | PRRSV (gilts) | 3 weeks | 4 | nac | na | na | na | na | na |
Average daily weight gain.
Feed conversion ratio.
Not available.
All farms were visited 8 times. An entire batch of pregnant sows in each herd was followed during two successive gestation and lactation periods. The sows were sampled twice during gestation, 9 and 4 weeks before the expected farrowing date, and twice during the farrowing phase, then 1 and 4 weeks after farrowing. Prior to the first visit, the sows were individually identified (ear tag) by the farmer. Clinical observations and samplings were made by four specially trained operators from our laboratory throughout the study. To reduce the bias due to the observer, each operator was specialized in an activity: animal restraining (1), sow's mouth opening (1), swabbing (1) and blood sampling and clinical observations (1).
2.3. Sample collection
At each visit, nasal, tonsillar and oro-pharyngeal swabs and blood samples were taken from each sow. The animals were restrained by placing a conventional cable snare over the maxilla. The animal's mouth was held open with a gag to obtain samples from the tonsillar and oral–pharyngeal areas. The surface of the tonsils was swabbed with “CytoBrushs” (VWR International, Fontenay-sous-Bois, France). Oral–pharyngeal samples were obtained by swabbing the surface of the oral–pharyngeal cavity thoroughly but gently with a brush protected by a catheter (Ori Endometrial BrushTM, Orifice Medical AB, Ystad, Sweden). For nasal sampling, both nasal cavities were swabbed with “CytoBrushs” (VWR International), inserted into the nostrils and rotated to reach deeply into the turbinates. All samples were placed in 2 ml of Buffered Peptone Water Broth. They were individually identified and delivered to the laboratory for processing on the day of collection (Initial Suspension, IS).
Blood samples were collected by jugular vein puncture, using evacuated Tubes (Vacuette, Dutscher SAS, Brumath, France) without additive. The samples were individually identified, and delivered to the laboratory, where they were processed. Serum was obtained by centrifugation for 10 min at 3500 × g and stored at −20 °C until subsequent analysis.
2.4. Laboratory analyses
2.4.1. PCRs on swab samples for bacterial pathogen DNA detection
Briefly, 1 ml of each IS was centrifuged (12,000 × g, 20 min) and the pellets were resuspended in 800 μl of lysis solution. The remaining IS was stored at −70 °C with glycerol (20%) for future studies. Lysates were incubated for 1 h at 60 °C, 10 min at 95 °C and then kept at −20 °C until PCR analysis. All swabs were tested by PCR, as previously described, for DNA detection of M. hyopneumoniae (Marois et al., 2007), A. pleuropneumoniae (Schaller et al., 2001), P. multocida (Marois et al., 2008), S. suis (Marois et al., 2004) and H. parasuis (Oliveira et al., 2001). When an inhibition of the PCR reaction was observed, DNA was re-extracted from the lysates with phenol/chloroform/isoamyl(ic) alcohol (25/24/1) (Marois et al., 2004). A sample was considered positive to a pathogen when the specific PCR tested positive.
2.4.2. Serological analyses
All sera were tested for M. hyopneumoniae antibodies with a blocking enzyme-linked immunosorbent assay (ELISA), according to the manufacturer's instructions (DAKO ELISA, Kitvia, Labarthe-Inard, France). The inhibition percentage (IP) for each serum was calculated with the following formula: IP = 100 − [100 × (sample mean OD/buffer control mean OD)]. A sample was classified as positive if the IP value was >50%. The sensitivity and specificity of the ELISA were previously reported to be 100% and 100%, respectively, in experimental trials (Sørensen et al., 1997). Sera were also analysed to detect A. pleuropneumoniae antibodies serotype 2 and serogroup 1-9-11 using 2 ELISA commercial kits (Swinecheck App2 and Swinecheck App1911, Biovet, AES Laboratoire, Combourg, France). The positive threshold was fixed at an optical density (OD) of 0.5 and 0.55 for serotype 2 and serogroup 1-9-11, respectively. The sensitivity and specificity of the two ELISA tests were 77.8% and 97.4% and 89.5% and 95.1%, respectively, according to the manufacturer.
Sera of all herds, except herd A, were tested for PCV-2 antibodies using an ORF-2 protein based ELISA test (Blanchard et al., 2003) at 4 sampling dates: 1 and 4 weeks post-partum at the first reproduction cycle and 9 and 4 weeks before farrowing in the second cycle. An OD ratio of 1.5 served as positivity cut-off. The sensitivity and specificity of the PCV-2 ELISA were 98.2% and 94.5%, respectively. PRRS virus antibodies were tested (ELISA test, IDDEX Laboratory, Eragny, sur Oise, France) in sera of every herd at 3 sampling dates: 9 weeks before farrowing in the first reproduction cycle and 4 weeks after farrowing in both cycles. The sensitivity and specificity of the PRRS ELISA were 97.4% and 99.6%, respectively. During the first reproduction cycle of each herd, random samples of sera obtained 1-week post-partum from 12 sows were tested with an hemagglutination inhibition (HI) test (Van Reeth et al., 2008) for antibodies against European SIVs, using strains A/Sw/Finistere/2899/82 (H1N1), A/Sw/Gent/1/84 (H3N2) and A/Sw/Scotland/410440/94 (H1N2) as antigens. An animal was considered positive against a given SIV subtype when the HI antibody titre was 20 or more (Van Reeth et al., 2008).
2.5. Statistical analysis
The contamination and infection status was considered at the sow level. At each sampling date, an animal was considered contaminated by a bacterial pathogen when at least one out of the three samples tested positive by specific PCR assay. Generalized linear models using the Generalized Estimation Equations (GEE) method were used to assess the influence of parity, farm, physiological stage and cycle on the sow contamination status determined by PCR assays and the infection status determined by serological assays (M. hyopneumoniae and A. pleuropneumoniae) (proc genmod, SAS 9.1, SAS Inst Inc.). The influence of the sampling site on the sow contamination status was also assessed using GEE models. Since the data consisted in repeated measurements on the same animals over time and space, observations could not be considered as independent. Sow was taken as a repeated statement to assess the relationships between the dependent variable and parity, farm and sampling site whereas a farm cluster effect was considered in models assessing the influence of the physiological stage and cycle on the dependent variable. The score statistics for type-3 GEE analysis was used to test the significance of main effects (p < 0.05). GEE models were also used to estimate the frequency of positive animals by PCR and serological assays as regards bacterial pathogens throughout the survey period. Within-herd seroprevalences were adjusted according to sensitivity and specificity of ELISA assays.
3. Results
3.1. PCR analyses for bacterial pathogen detection
In total, 3453 swabs from 184 sows were tested by PCR assays aimed to detect bacterial pathogen genomes. 109 of these 184 sows were followed throughout the entire sampling period due to the culling/replacement process in-between cycles. On average, 29 sows (σ = 6.7) were sampled per batch at each sampling time and sows were sampled 6.9, 6.0, 6.6, 6.5, and 5.4 times in herds A, B, C, D and E, respectively. Parity distributions at each reproduction cycle are given for each herd in Table 3 . Sows from herd C were on average younger than those from the other herds since new breeding stock was introduced when moving from a 3-week to a 4-week batch interval. The high average parity in herd D was related to the breeding sows management policy (long lasting time of the sows).
Table 3.
Parity mean distribution during two successive reproduction cycles of five batches of sows from farrow-to-finish herds.
| Herd | Cycle 1 |
Cycle 2 |
||
|---|---|---|---|---|
| Mean (σ) | n | Mean (σ) | n | |
| A | 4.2 (2.4) | 23 | 3.8 (2.5) | 25 |
| B | 4.0 (2.6) | 35 | 3.0 (2.1) | 36 |
| C | 2.7 (1.7) | 39 | 2.3 (1.3) | 41 |
| D | 5.6 (3.1) | 27 | 4.7 (3.1) | 26 |
| E | 3.6 (2.0) | 25 | 2.9 (1.9) | 25 |
DNA of M. hyopneumoniae, H. parasuis, P. multocida, A. pleuropneumoniae and S. suis were detected from at least one out of the three swab samples in 2.4% (confidence interval at 95% (C.I.95%) (1.7; 3.3)), 23.4% (C.I.95%: (20.6;26.4)), 24.6% (C.I.95%: (21.0;27.5)), 30.9% (C.I.95%: (28.3;33.7)), and 67.1% (C.I.95%: (63.8;70.2)) of the sows, respectively. Whatever the micro-organism, the model-based estimated frequency of positive animals varied from farm to farm (Table 4 ). In all five herds, all the pathogens, except M. hyopneumoniae were detected at least once in the upper respiratory tract of the sows. S. suis was the pathogen the most frequently recovered in all herds, with more than 40% of positive sows. To a lesser extent A. pleuropneumoniae, P. multocida and H. parasuis were also frequently detected in almost all herds with herd levels ranging from 11.6% to 48%, 8.4% to 45% and 4.7% to 50% of positive sows, respectively.
Table 4.
Estimated frequencies (with 95% confidence interval) of M. hyopneumoniae, A. pleuropneumoniae, P. multocida, H. parasuis and S. suis detection in breeding sows followed in five farrow-to-finish herds (184 sows, 8 sampling times, nasal, tonsillar and oro-pharyngeal swabs, PCR assays, Generalized Estimation Equations models with an autoregressive correlation matrix).
| Herd | Mycoplasma hyopneumoniae | Actinobacillus pleuropneumoniae | Pasteurella multocida | Haemophilus parasuis | Streptococcus suis |
|---|---|---|---|---|---|
| A | 0.6b (0.0;37.3) | 48.0c (34.6;61.7) | 8.4a (3.6;18.5) | 21.6b (13.6;32.3) | 45.9a (30.3;62.3) |
| B | 0.4b (0.1;2.6) | 11.6a (9.3;14.4) | 20.1c (16.2;24.6) | 19.1b (15.7;23.1) | 71.2b,c (65.7;76.1) |
| C | 4.1a (0.1;68.2) | 29.6b (19.2;42.7) | 22.5c (13.4;35.3) | 4.7a (2.2;9.6) | 76.4b (63.4;85.8) |
| D | 5.7a (0.1;75.5) | 34.2b (23.6;46.7) | 45.0b (30.9;60.0) | 50.2c (37.0;63.3) | 54.5a (39.3;68.9) |
| E | 0b (0.0;0.0) | 41.2b,c (27.7;56.3) | 26.9c (15.8;41.9) | 30.1d (19.1;44.0) | 79.1c (66.2;88.0) |
Herds with different subscripts (a, b, c and d) have a significantly different PCR-prevalence (p < 0.05, Generalized Estimation Equations model).
In the subsample of 109 sows which were swabbed during all eight visits, for all the sows found positive to M. hyopneumoniae, the DNA of the organism was detected at one sampling occasion, whereas genomes of P. multocida, H. parasuis, A. pleuropneumoniae and S. suis were detected on 2 sampling occasions or more for 60.4%, 66.3%, 75.5% and 98.1% of the positive sows, respectively. Whatever the micro-organism, the frequency of positive animals in the herd varied over time, without any significant effect of the physiological stage (Fig. 1 and Table 5 ) whereas a significant effect of the reproduction cycle was observed for S. suis and H. parasuis. A significant effect of the farm on the sow contamination status of the upper respiratory tract was identified for each micro-organism (p < 0.05, Table 5), except for M. hyopneumoniae which could not be estimated due to the absence of detection in some farms. The ability to detect contaminated sows was significantly influenced by the sampling site for four out of the five bacteria (Table 5). Swabbing the tonsils significantly increased the probability to detect sows contaminated by A. pleuropneumoniae and H. parasuis than sampling the nasal cavities or the oro-pharyngeal area (p < 0.05, Table 6 ). The detection of P. multocida PCR-positive sows was significantly improved when nasal swabs were used (p < 0.05, Table 6). The odds for a sow being PCR-positive to S. suis by oro-pharyngeal swabbing was significantly lower than by nasal or tonsillar sampling (p < 0.05, Table 6).
Fig. 1.
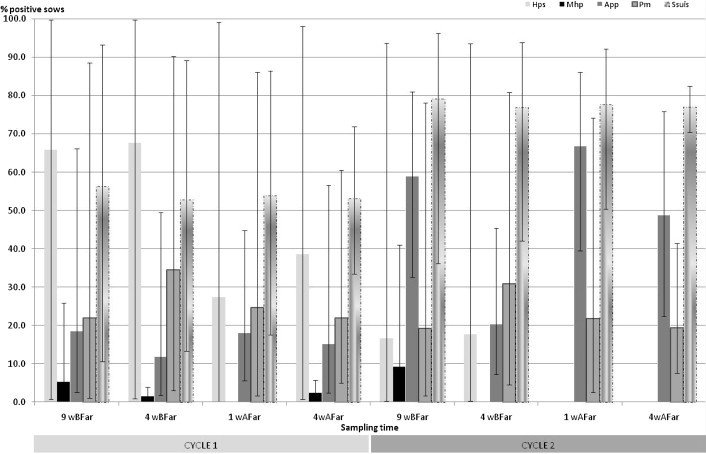
Estimated within herd pathogen detection frequency (and 95% confidence interval) of M. hyopneumoniae (Mhp), A. pleuropneumoniae (App), P. multocida (Pm), H. parasuis (Hps) and S. suis (Ssuis) during two successive gestation and lactation periods (cycles 1 and 2) (5 farrow-to-finish herds, 184 sows, 8 sampling times, nasal, tonsillar and oro-pharyngeal swabs, PCR assays, Generalized Estimation Equations with an exchangeable correlation matrix). 9wBFar: 9 weeks before farrowing; 4wBFar: 4 weeks before farrowing; 1wAFar: 1 week after farrowing; 4wAFar: 4 weeks after farrowing.
Table 5.
Results of the score statistics for type-3 Generalized Estimation Equations analysis to test the significance of herd, parity, physiological stage, cycle effects and sampling site on the sow contamination status determined by PCR assays and the infection status determined by serological assays.
| Dependent variable | Independent variable |
||||
|---|---|---|---|---|---|
| Herd | Parity number | Physiological stage | Cycle | Sampling site | |
| Mhp PCR-status | n.e.a | n.e. | n.e. | n.e. | 0.63 |
| App PCR-status | <0.001 | 0.77 | n.e. | n.e. | <0.001 |
| Pm PCR-status | <0.001 | 0.07 | 0.55 | 0.61 | 0.002 |
| Hps PCR-status | <0.001 | 0.35 | 0.31 | 0.04 | 0.002 |
| Ssuis PCR-status | <0.001 | 0.12 | 0.99 | 0.05 | <0.001 |
| Mhp serological status | 0.03 | 0.79 | 0.18 | 0.26 | – |
| App1-9-11 serological status | <0.001 | 0.005 | 0.19 | 0.11 | – |
| App2 serological status | <0.001 | 0.002 | 0.29 | 0.41 | – |
Shaded values are significant at p < 0.05.
Not estimable.
Table 6.
Results of the sampling site comparison to detect PCR-positive sows to M. hyopneumoniae, A. pleuropneumoniae, P. multocida, H. parasuis and S. suis (5 herds, 184 sows, nasal (NS), tonsillar (TS) and oro-pharyngeal (OPS) swabs, odds ratio (OR) with confidence interval at 95% (C.I.95%)).
| Sampling site comparison | Site |
M. hyopneumoniae |
A. pleuropneumoniae |
P. multocida |
H. parasuis |
S. suis |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p | OR | C.I.95% | p | OR | C.I.95% | p | OR | C.I.95% | p | OR | C.I.95% | p | OR | C.I.95% | ||
| NS vs. OPS | 0.74 | 0.17 | 0.002 | 0.049 | <0.001 | |||||||||||
| NS | 1.11 | 0.59;2.11 | 0.87 | 0.71;1.06 | 1.39 | 1.13;1.71 | 0.81 | 0.67;0.99 | 1.40 | 1.23;1.60 | ||||||
| OPS | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | ||||||
| TS vs. NS | 0.34 | <0.001 | <0.001 | <0.001 | 0.48 | |||||||||||
| TS | 0.72 | 0.37;1.4 | 1.65 | 1.35;2.02 | 0.70 | 0.58;0.86 | 1.43 | 1.16;1.77 | 1.05 | 0.92;1.18 | ||||||
| NS | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | ||||||
| TS vs. OPS | 0.58 | <0.001 | 0.87 | 0.046 | <0.001 | |||||||||||
| TS | 0.8 | 0.37;1.73 | 1.43 | 1.24;1.66 | 0.98 | 0.79;1.21 | 1.17 | 1.00;1.37 | 1.47 | 1.29;1.67 | ||||||
| OPS | 1 | – | 1 | – | 1 | – | 1 | – | 1 | – | ||||||
Bold values are values with a p < 0.05.
3.2. Serological analyses
3.2.1. Bacterial pathogens
Sows seropositive to M. hyopneumoniae, A. pleuropneumoniae serogroups 1-9-11 and 2 were detected in all five herds at different frequencies depending on the farm (Table 7 ). The odds of sows from herd E being M. hyopneumoniae seropositive were significantly lower than those of herds A, C and D (p < 0.05). The odds of being seropositive to A. pleuropneumoniae serogroup 1-9-11 were significantly higher in herds C and E than in the three other herds whereas for serotype 2 it was higher in herds A and C. The lowest proportions of seropositive sows to A. pleuropneumoniae serogroup 1-9-11 and serotype 2 were found in herds A, B, D and D, respectively.
Table 7.
Adjusted frequencies (with 95% confidence interval) of M. hyopneumoniae (Mhp), A. pleuropneumoniae serogroup 1-9-11 (App1-9-11) and serotype 2 (App2) detection in breeding sows followed in five farrow-to-finish herds (184 sows, 8 sampling times, ELISA assays, Generalized Estimation Equations models with an autoregressive correlation matrix).
| Herd | Mhp | App1-9-11 | App2 |
|---|---|---|---|
| A | 82.5a (51.6;95.4) | 36.9a (8.9;76.1) | 65.2a (22.9;100) |
| B | 69.3a,b (57.6;78.9) | 24.3a (13.9;37.6) | 31.6b,c (19.2;47.6) |
| C | 83.5a (57.1;95.1) | 100b (77.1;100) | 73.0a (33.6;100) |
| D | 79.9a (51.2;93.8) | 29.6a (5.4;69.2) | 14.4b (0.2;58.2) |
| E | 55.4b (24.8;82.4) | 65.7c (27.8;95.3) | 35.1c (10.2;75.5) |
Herds with different subscripts (a, b and c) have a significantly different PCR-prevalence (p < 0.05, Generalized Estimation Equations model).
Neither physiological stage nor cycle influenced the sow serological status to M. hyopneumoniae, A. pleuropneumoniae serogroup 1-9-11 or serotype 2 (p > 0.05, Fig. 2 and Table 5). In herds C and E, an increase of the percentage of A. pleuropneumoniae serotype 2-seropositive sows was detected between gestation and lactation in the first reproduction cycle (data not shown). The probability of young sows (parity number ≤2) having antibodies against A. pleuropneumoniae 1-9-11 and 2 (p < 0.05) was significantly lower. No parity effect was identified for M. hyopneumoniae (Table 5).
Fig. 2.
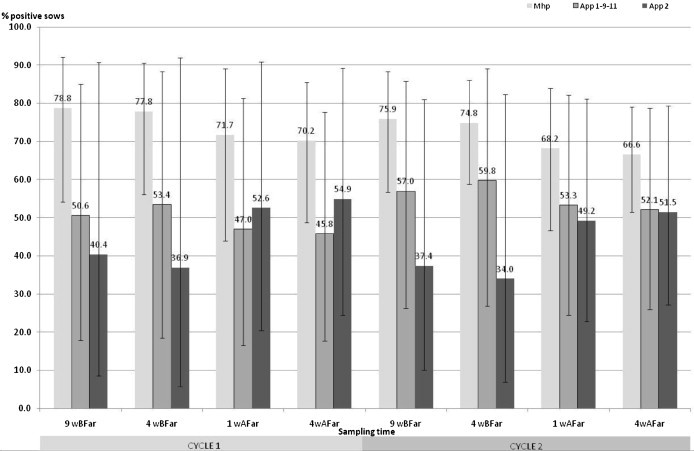
Adjusted within herd antibody detection frequency (with 95% confidence interval) of M. hyopneumoniae (Mhp), A. pleuropneumoniae serogroup 1-9-11 (App1-9-11) and serotype 2 (App2) during two successive gestation and lactation periods (cycles 1 and 2) (5 farrow-to-finish herds, 184 sows, 8 sampling times, ELISA assays, Generalized Estimation Equations with an exchangeable correlation matrix). 9wBFar: 9 weeks before farrowing; 4wBFar: 4 weeks before farrowing; 1wAFar: 1 week after farrowing; 4wAFar: 4 weeks after farrowing.
3.2.2. Viral pathogens
The viral infection patterns varied between farms. Sows from herd A were seronegative to PRRSV during the first gestation and lactation periods. Blood samples from the second reproduction cycle were not tested. However, no clinical signs of PRRSV infection were recorded in this farm during the second cycle. The percentage of PRRS-seropositive sows increased between the first gestation and second lactation periods in herd E whereas it decreased in herd C and remained high in herd D as shown in Fig. 3 .
Fig. 3.
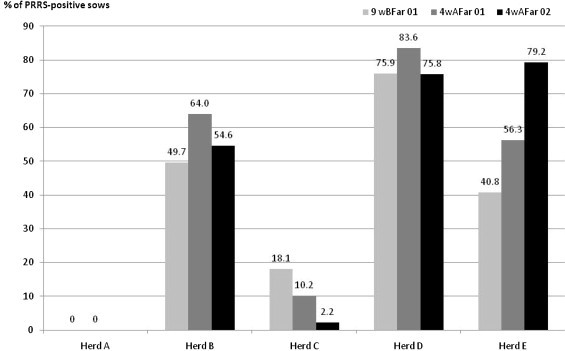
Frequency of PRRS-seropositive sows during gestation and lactation phases in five farrow-to-finish herds (184 sows, 3 sampling dates). wBFar01: 9 weeks before farrowing, replicate 01; 4wAFar01: 4 weeks after farrowing, replicate 01; 4wAFar02: 4 weeks before farrowing, replicate 02.
All herds tested positive for SIV by HI assays (Fig. 4 ). The highest frequencies of SIV-seropositive sows were found for subtype H1N2 in all herds, except in the SIV-vaccinated herd B, where 100% and 91.7% of the sows tested positive for subtypes H3N2 and H1N1, respectively. At the animal level, three herds (A, B and C) had sows with subtype H1N1-antibodies titres that were higher than those of the first more frequent subtype and were considered positive for H1N1. HI titres for subtype H3N2 equal to those of the most prevalent subtype were detected in herd A in which the sows used to be vaccinated before the study. Both the low frequency of positive sows and the low antibodies titres for the H3N2 subtype observed in herds D and E were considered to be due to cross-reactions in the HI test.
Fig. 4.
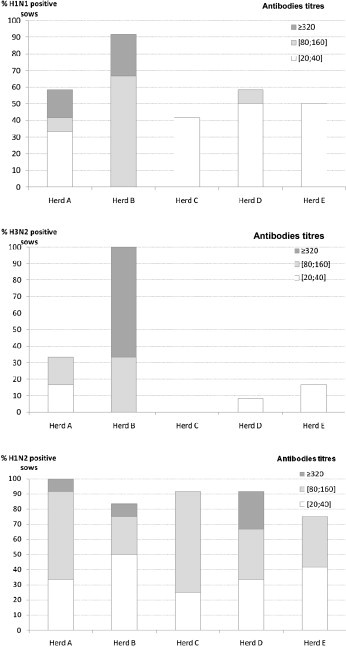
Distribution of the frequency of sows with SIV antibodies and antibodies titres for subtypes H1N1, H1N2 and H3N2 4 weeks after farrowing in five farrow-to-finish pig herds (12 sows/herd).
PCV-2 antibodies were detected in more than 75% of sows in the four tested herds at 1 and 4 weeks after farrowing for the first reproduction cycle and at 9 and 4 weeks before farrowing for the second cycle.
4. Discussion
The aim of the present study was to describe the carrying and infection patterns of batches of sows for various respiratory pathogens during two successive gestation and lactation periods. Three sampling sites in the upper respiratory tract, i.e., nasal cavities, tonsils, and oro-pharyngeal area, were chosen to describe the carrier state of the animals regarding five bacteria involved in PRDC. The sampling areas were selected according to the site where each bacteria was likely to be recovered (Møller et al., 1993, Pijoan, 1999, Rapp-Gabrielson, 1999, Marois et al., 2004, Fablet et al., 2010). On the other hand, as no test is perfect, sensitivity should be increased by sampling several sites. Therefore, for each bacterium, samples from all three sites were tested by PCR to enhance the likelihood of detecting positive animals. The carrier state was hence studied at the animal level.
The herds included in this study were not selected at random and may not be representative of the population of French farrow-to finish pig herds. However, they were selected to tally with the different respiratory patterns in growing-finishing pigs most frequently encountered by veterinarians in the field. Their inclusion was based on the results of a preliminary screening step involving 12 herds which were clinically examined and tested for the different agents under investigation. Hence, they were supposed to represent the different categories of herds identified in this screening step as suffering from different levels of severity and complexity of respiratory diseases. As all sows from a batch, selected at random, were included in the study, the cohorts should be appropriate representatives of the herds under study and thereby provide a valid picture of the infection patterns and even their variability between herds. However, one limitation of the study relies on the kind of production system sampled which may not be representative of farming types in use in all other pig producing countries.
To the best of our knowledge, this is the first time that a comprehensive description of the microbial flora of the airways of naturally infected sows has been obtained under field conditions. We showed that the upper part of the airways of clinically healthy sows harboured a variety of respiratory pathogens such as S. suis, A. pleuropneumoniae, P. multocida and H. parasuis. This is in agreement with results of previous studies indicating that these bacteria are common inhabitants of the upper respiratory tract of asymptomatic pigs raised under commercial conditions (Dritz et al., 1996, Baele et al., 2001, MacInnes et al., 2008). Even if for H. parasuis, A.pleuropneumoniae, S.suis and P. multocida only a limited number of serotypes are known to be highly pathogenic, PCR assays detecting species of pathogens were used as general descriptors of the infectious status towards the main bacteria species involved in respiratory diseases. An overall low pathogen load in a herd being recommended, the aim was to assess, at the pathogen species level, whether different infection pressure may be identified and were related to the overall respiratory health status of the farm. The results of our study in sows indicated that whatever the pathogen, its prevalence varied between farms and over time within herds. As only a small percentage of sows (<10%) was consistently detected positive to a given pathogen at all 8 sampling times, it can be hypothesized that the pathogen might transiently be present in small quantities, i.e., under the detection threshold of the PCR assay in the upper respiratory tract of the sows, and that shedding was intermittent in most of the animals. While S. suis, A. pleuropneumoniae, P. multocida and H. parasuis were the most frequent pathogens identified in the upper part of the airways of the sows, these pathogens were seldom detected in lung tissues of slaughtered pigs during the recruitment step. M. hyopneumoniae was indeed the only pathogen detected at slaughter from lungs of finishing pigs in all five herds. Even if the slaughtered sampled pigs were not the offspring of the sows followed in the present study, we can assume that other factors than the bacterial carrier state of the upper part of the respiratory tract of the sows may influence the infectious status of the lung in growing-finishing pigs.
M. hyopneumoniae plays a central role in PRDC and is widespread in pig herds (Thacker, 2006). Interestingly, even if the M. hyopneumoniae genome was detected in four out of the five selected herds, the micro-organism was relatively seldom identified at the sow level. The detection threshold of the simple PCR was estimated to be 10–100-fold higher than that of nested-PCR (Marois et al., 2007). Therefore, this could have limited our capacity to detect sows carrying low numbers of M. hyopneumoniae (<1000 mycoplasma). The true prevalence of positive animals may in turn have been underestimated. On the other hand, when studying infection dynamics, the bacterial load carried by the animals is more important than a positive shedding status. Further studies using quantitative tests are needed to improve our knowledge on the course of M. hyopneumoniae infection under field conditions. Although M. hyopneumoniae DNA was not consistently detected, antibodies against the micro-organism were recovered in sows from all herds. Positive reactions in serum from sows may reflect previous exposure to M. hyopneumoniae, as vaccination was not carried out in the investigated herds. Interestingly, in the herd that showed the lowest number of seropositive sows, M. hyopneumoniae carriage was not detected at any sampling time. It can be speculated that M. hyopneumoniae infection dynamics within the sow population remained limited in this farm. The fact that M. hyopneumoniae was detected in the upper respiratory tract of seropositive sows, indicated, as already reported (Sibila et al., 2007), that circulating antibodies against M. hyopneumoniae did not prevent colonization of the airways. M. hyopneumoniae DNA was detected in sows from the first until the tenth parity suggesting that even the oldest sows may carry the organism and have the potential to spread the bacteria to susceptible animals, especially their offspring. In previous studies, relationships between sow parity and M. hyopneumoniae infection were inconsistent (Calsamiglia and Pijoan, 2000, Ruiz et al., 2003, grosse Beilage et al., 2009). Our study, being primarily descriptive, may lack sufficient power to find any significant association. Thus, further research seems to be required to conclude about the influence of sow age on M. hyopneumoniae infection, but, due to the expected low prevalence of carrier sows, altogether, these results emphasize the need to sample a large number of sows of different parity groups when assessing M. hyopneumoniae infection dynamics.
The odds for a sow being seropositive for A. pleuropneumoniae serogroup 1-9-11 and serotype 2 were significantly lower in first and second parity than in third or more parity sows. These findings indicate that seroconversion is more likely to occur during the first two reproduction cycles and suggest that young sows are exposed to the micro-organism following their introduction in the breeding herd. Antibodies to the two considered serotypes were found in all herds. While the high seropositivity levels observed in herd C could be related to the vaccination, the antibodies detected in the four non-vaccinated herds indicated that sows were naturally infected. This also suggested that the infection is widespread among clinically healthy sow herds as reported for A. pleuropneumoniae serotype 2 (Levonen et al., 1994). It should be noted that the infection pressure differed between farms, as revealed by the varying levels of seroprevalence between herds.
As expected, the cluster herd effect had a significant influence on infection status of the sows for all the examined pathogens which demonstrated variation between farms as a result of differences in farm layout and management practices. This is in line with a cross-sectional study showing that M. hyopneumoniae seropositivity in sows is influenced by various management factors (grosse Beilage et al., 2009). According to the results of the present study, the carrier state or serological status of the sows did not seem to be affected by whether they were in gestation or lactation. A decrease in the level of M. hyopneumoniae serum antibodies during the last month of pregnancy to the farrowing day was previously described (Wallgren et al., 1998). Since the last sampling time during pregnancy was performed 4 weeks before farrowing, our sampling scheme may have limited the ability to detect such a reduction. Whatever the micro-organism, a specific physiological stage with a maximum prevalence of carrier sows was not identified. Therefore, sampling sows at different gestation and lactation stages should be recommended, especially in cross-sectional studies looking for prevalence. On the other hand, the carrying patterns of the same batches of sows, especially for H. parasuis and S. suis, were different from the first to the second reproduction cycle suggesting a time effect on the level of carriage for a given batch. This tended to indicate that a batch effect is important to take into account in epidemiological studies as well as in diagnostic.
The seroprevalence of PCV-2 infection was high in the four herds we tested, hence confirming the widespread occurrence of PCV-2 in French sow herds as described earlier (Rose et al., 2003). Serological investigations also showed that SIV antibodies were highly prevalent in the sows which is consistent with the fact that viruses of H1N1 and H1N2 European subtypes are widespread in pig populations in France (Kuntz-Simon and Madec, 2009). HI tests revealed infections with H1N1 and/or H1N2 viruses, while H3N2 antibodies could only be detected in vaccinated sows or as a result of cross-reactions in the HI test as already reported (Van Reeth et al., 2006). The infection pattern of the sow batches towards PRRSV differed between the five herds in which the growing pigs were experiencing different levels of severity of respiratory disorders. While in herd A, being the less affected by respiratory diseases, PRRSV infection was not detected in the sows during the study, an increase in the frequency of PRRSV-seropositive sows was found in the herd the most severely affected by lung diseases in finishing pigs (herd E). This increase in seropositivity frequency indicated that PRRSV was actively circulating in the sow herd. A higher and stable frequency of PRRSV infection was recovered in the second herd with growing pigs severely affected by respiratory diseases. Conversely a lower and decreasing rate of infected sows was described in herd C, suggesting that the virus was no longer actively spreading in the sow population. In the latter herd, the growing-finishing pigs which were tested remained sero-negative throughout the course of the study (data not shown). In herd B, despite the use of a commercial inactivated vaccine against PRRSV, from 50% to 64% of the sows tested positive by serology. Under experimental and field conditions, some variations in seropositivity rates following vaccination with a killed vaccine have been described (Reynaud et al., 1999). In the same line, Papatsiros et al. (2006) found that the humoral response of sows following two vaccinations was short, lasting 40 days after the booster vaccination. In the present study, the antibodies detected 72 days after the vaccination were probably related to active infection. In addition, the PRRSV antibodies detected in growing pigs confirmed that this herd was not virus-free.
In conclusion, the results of the present study indicate that in conventional commercial farrow-to-finish herds, the relevant infectious agents involved in PRDC are present in the sow herd, with limited consequences on the health status of the sows, and thus confirms that the reproductive herd constitutes a reservoir for the continuous circulation of respiratory pathogens. Therefore, these results suggest that the sow population needs to be properly considered in the measures taken to control respiratory diseases. However, further studies may help to know more about the factors that influence the infection process in sow herds and to evaluate the consequences on the infection dynamics in their offspring.
Acknowledgements
The authors gratefully acknowledge the farmers involved in the study for their very efficient participation and the related farm organizations for their help as well as Nathalie Malledan. They are also indebted to the Regional Council of Brittany, the “Comité Régional Porcin” and Boehringer Ingelheim Animal Health France, Fort Dodge Animal Health, Intervet S.A., Pfizer Animal Health and Schering-Plough Animal Health for their financial support.
References
- Amass S.F., Clark L.K., Knox K., Ching Wu C., Hill M.A. Streptococcus suis colonization of piglets during parturition. Swine Health and Production. 1996;4:269–272. [Google Scholar]
- Baele M., Chiers K., Devriese L.A., Smith H.E., Wisselink H.J., Vaneechoutte M., Haesebrouck F. The Gram-positive tonsillar and nasal flora of piglets before and after weaning. Journal of Applied Microbiology. 2001;91:997–1003. doi: 10.1046/j.1365-2672.2001.01463.x. [DOI] [PubMed] [Google Scholar]
- Blanchard P., Mahé D., Cariolet R., Truong C., Le Dimna M., Arnauld C., Rose N., Eveno E., Albina E., Madec F., Jestin A. An ORF2 protein-based ELISA for porcine circovirus type 2 antibodies in post-weaning multisystemic wasting syndrome. Veterinary Microbiology. 2003;94:183–194. doi: 10.1016/S0378-1135(03)00131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calsamiglia M., Pijoan C. Colonisation state and colostral immunity to Mycoplasma hyopneumoniae of different parity sows. Veterinary Record. 2000;146:530–532. doi: 10.1136/vr.146.18.530. [DOI] [PubMed] [Google Scholar]
- Clark L., Freeman M., Scheidt A., Knox K. Investigating the transmission of Mycoplasma hyopneumoniae in a swine herd with enzootic pneumonia. Veterinary Medicine. 1991;86:543–550. [Google Scholar]
- Dritz S.S., Chengappa M.M., Nelssen J.L., Tokach M.D., Goodband R.D., Nietfeld J.C., Staats J.J. Growth and microbial flora of non medicated segregated, early weaned pigs from a commercial swine operation. Journal of Veterinary Medical Association. 1996;208:711–715. [PubMed] [Google Scholar]
- Fablet C, Marois C., Kobisch M., Madec F., Rose N. Estimation of the sensitivity of four sampling methods for Mycoplasma hyopneumoniae detection in live pigs using a Bayesian approach. Veterinary Microbiology. 2010;143:238–245. doi: 10.1016/j.vetmic.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Fano E., Pijoan C., Dee S., Deen J. Effect of Mycoplasma hyopneumoniae colonization at weaning on disease severity in growing pigs. Canadian Journal of Veterinary Research. 2007;71:195–200. [PMC free article] [PubMed] [Google Scholar]
- grosse Beilage E., Rohde N., Krieter J. Seroprevalence and risk factors associated with seropositivity in sows from 67 herds in north-west Germany infected with Mycoplasma hyopneumoniae. Preventive Veterinary Medicine. 2009;88:255–263. doi: 10.1016/j.prevetmed.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Kuntz-Simon G., Madec F. Genetic and antigenic evolution of swine influenza viruses in Europe and evaluation of their zoonotic potential. Zoonoses and Public Health. 2009;56:310–325. doi: 10.1111/j.1863-2378.2009.01236.x. [DOI] [PubMed] [Google Scholar]
- Levonen K., Veijalainen P., Seppänen J. Actinobacillus pleuropneumoniae serotype-2 antibodies in sow colostrum in Finnish pig-health-scheme herds. Zentralblatt fur Veterinarmedizin B. 1994;41:567–573. doi: 10.1111/j.1439-0450.1994.tb00265.x. [DOI] [PubMed] [Google Scholar]
- MacInnes J.I., Gottschalk M., Lone A.G., Metcalf D.S., Ojha S., Rosendal T., Watson S.B., Friendship R.M. Prevalence of Actinobacillus pleuropneumoniae, Actinobacillus suis, Haemophilus parasuis, Pasteurella multocida, and Streptococcus suis in representative Ontario swine herds. Canadian Journal of Veterinary Research. 2008;72:242–248. [PMC free article] [PubMed] [Google Scholar]
- Madec F., Kobisch M. Bilan lesionnel des poumons de porcs charcutiers à l’abattoir. Journées de la Recherche Porcine, Paris, France. 1982;14:405–412. [Google Scholar]
- Marois C., Bougeard S., Gottschalk M., Kobisch M. Multiplex PCR assay for detection of Streptococcus suis species and serotypes 2 and 1/2 in tonsils of live and dead pigs. Journal of Clinical Microbiology. 2004;42:3169–3175. doi: 10.1128/JCM.42.7.3169-3175.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marois C., Cariolet R., Morvan H., Kobisch M. Transmission of pathogenic respiratory bacteria to specific pathogen free pigs at slaughter. Veterinary Microbiology. 2008;129:325–332. doi: 10.1016/j.vetmic.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Marois C., Le Devendec L., Fablet C., Dorenlor V., Eono F., Eveno E., Jolly J.P., Madec F., Kobisch M. Comparaison de la PCR simple et de la PCR nichée pour détecter Mycoplasma hyopneumoniae chez le porc vivant. Journées de la Recherche Porcine, Paris, France. 2007;39:435–436. [Google Scholar]
- Møller K., Andersen L.V., Christensen G., Kilian M. Optimalization of the detection of NAD dependent Pasteurellaceae from the respiratory tract of slaughterhouse pigs. Veterinary Microbiology. 1993;36:261–271. doi: 10.1016/0378-1135(93)90093-m. [DOI] [PubMed] [Google Scholar]
- Oliveira S., Galina L., Pijoan C. Development of a PCR test to diagnose Haemophilus parasuis infections. Journal of Veterinary Diagnostic Investigation. 2001;13:495–501. doi: 10.1177/104063870101300607. [DOI] [PubMed] [Google Scholar]
- Papatsiros V.G., Alexopoulos C., Kritas S.K., Koptopoulos G., Nauwynck H.J., Pensaert M.B., Kyriakis S.C. Long-term administration of a commercial porcine reproductive and respiratory syndrome virus (PRRSV)-inactivated vaccine in PRRSV-endemically infected sows. Journal of Veterinary Medicine Series B. 2006;53:266–272. doi: 10.1111/j.1439-0450.2006.00965.x. [DOI] [PubMed] [Google Scholar]
- Pijoan C. Pneumonic pasteurellosis. In: Straw B.E., Mengelng W.L., D’Allaire S., Taylor D.J., editors. Diseases of Swine. Iowa State University Press; Iowa: 1999. pp. 511–520. [Google Scholar]
- Rapp-Gabrielson V.J. Haemophilus parasuis. In: Straw B.E., D’Allaire S., Mengeling W.L., Taylor D.J., editors. Diseases of Swine. Iowa University Press; Iowa, USA: 1999. pp. 475–481. [Google Scholar]
- Reynaud G., Chevallier S., Brun A., Herin J.B., Albina E., Charreyre C. Evaluation in the field of serological and virological efficacy of an inactivated PRRS vaccine in sows. Proceedings of 3rd International Symposium on PRRS and Aujeszky; Ploufragan, France, 21–24 June; 1999. pp. 281–282. [Google Scholar]
- Rose N., Larour G., Le Diguerher G., Eveno E., Jolly J.P., Blanchard P., Oger A., Le Dimna M., Jestin A., Madec F. Risk factors for porcine post-weaning multisystemic wasting syndrome (PMWS) in 149 French farrow-to-finish herds. Preventive Veterinary Medicine. 2003;61:209–225. doi: 10.1016/j.prevetmed.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Ruiz A.R., Utrera V., Pijoan C. Effect of Mycoplasma hyopneumoniae sow vaccination on piglet colonisation at weaning. Journal of Swine Health and Production. 2003;11:131–135. [Google Scholar]
- Schaller A., Djordjevic S.P., Eamens G.J., Forbes W.A., Kuhn R., Kuhnert P., Gottschalk M., Nicolet J., Frey J. Identification and detection of Actinobacillus pleuropneumoniae by PCR based on the gene apxIVA. Veterinary Microbiology. 2001;79:47–62. doi: 10.1016/s0378-1135(00)00345-x. [DOI] [PubMed] [Google Scholar]
- Sibila M., Calsamiglia M., Vidal D., Badiella L., Aldaz A., Jensen J.C. Dynamics of Mycoplasma hyopneumoniae infection in 12 farms with different production systems. Canadian Journal of Veterinary Research. 2004;68:12–18. [PMC free article] [PubMed] [Google Scholar]
- Sibila M., Nofrarias M., Lopez-Soria S., Segales J., Riera P., Llopart D., Calsamiglia M. Exploratory field study on Mycoplasma hyopneumoniae infection in suckling pigs. Veterinary Microbiology. 2007;121:352–356. doi: 10.1016/j.vetmic.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Sørensen V., Ahrens P., Barfod K., Feenstra A.A., Feld N.C., Friis N.F., Bille-Hansen V., Jensen N.E., Pedersen M.W. Mycoplasma hyopneumoniae infection in pigs: duration of the disease and evaluation of four diagnostic assays. Veterinary Microbiology. 1997;54:23–34. doi: 10.1016/s0378-1135(96)01266-7. [DOI] [PubMed] [Google Scholar]
- Sorensen V., Jorsal S.E., Mousing J. Diseases of the respiratory system. In: Straw B., Zimmermann W., D’Allaire S., Taylor D.J., editors. Diseases of Swine. 9th edition. Iowa State University Press; Ames, Iowa: 2006. pp. 149–177. [Google Scholar]
- Thacker E. Mycoplasmal disease. In: Straw B.E., Zimmerman J.J., D’Allaire S., Taylor D.J., editors. Diseases of Swine. Iowa State University Press; Ames: 2006. pp. 701–717. [Google Scholar]
- Van Reeth K., Brown I.H., Dürrwald R., Foni E., Labarque G., Lenihan P., Maldonado J., Markowska-Daniel I., Pensaert M., Pospisil Z., Koch G. Seroprevalence of H1N1, H3N2 and H1N2 influenza viruses in pigs in seven European countries in 2002–2003. Influenza and Other Respiratory Viruses. 2008;2:99–105. doi: 10.1111/j.1750-2659.2008.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Reeth K., Labarque G., Pensaert M. Serological profiles after consecutive experimental infections of pigs with European H1N1, H3N2, and H1N2 swine influenza viruses. Viral Immunology. 2006;19:373–382. doi: 10.1089/vim.2006.19.373. [DOI] [PubMed] [Google Scholar]
- Vigre H., Angen Ø., Barfod K., Lavritsen D.T., Sørensen V. Transmission of Actinobacillus pleuropneumoniae in pigs under field-like conditions: emphasis on tonsillar colonisation and passively acquired colostral antibodies. Veterinary Microbiology. 2002;89:151–159. doi: 10.1016/s0378-1135(02)00149-9. [DOI] [PubMed] [Google Scholar]
- Wallgren P., Bölske G., Gustafsson S., Mattsson J.G., Fossum C. Humoral immune response to Mycoplsama hyopneumoniae in sows and offspring following an outbreak of mycoplasmosis. Veterinary Microbiology. 1998;60:193–205. doi: 10.1016/s0378-1135(98)00155-2. [DOI] [PubMed] [Google Scholar]
- Zimmerman J., Benfield D.A., Murtaugh M.P., Osorio F., Stevenson G.W., Torremorell M. Porcine reproductive and respiratory syndrome virus (Porcine arterivirus) In: Straw B.E., Zimmerman J.J., D’Allaire S., Taylor D.J., editors. Diseases of Swine. Iowa State University Press; Ames: 2006. pp. 387–418. [Google Scholar]


