Abstract
Calf diarrhea is a major economic burden for the US cattle industry. A variety of infectious agents are implicated in calf diarrhea and co-infection of multiple pathogens is not uncommon in diarrheic calves. A case–control study was conducted to assess infectious etiologies associated with calf diarrhea in Midwest cattle farms. A total of 199 and 245 fecal samples were obtained from diarrheic and healthy calves, respectively, from 165 cattle farms. Samples were tested by a panel of multiplex PCR assays for 11 enteric pathogens: bovine rotavirus group A (BRV-A), bovine coronavirus (BCoV), bovine viral diarrhea virus (BVDV), bovine enterovirus (BEV), bovine norovirus (BNoV), Nebovirus, bovine torovirus (BToV) Salmonella spp. (Salmonella), Escherichia coli (E. coli) K99+, Clostridium perfringens with β toxin gene and Cryptosporidium parvum (C. parvum). The association between diarrhea and detection of each pathogen was analyzed using a multivariate logistic regression model. More than a half of the fecal samples from the diarrheic calves had multiple pathogens. Statistically, BRV-A, BCoV, BNoV, Nebovirus, Salmonella, E. coli K99+, and C. parvum were significantly associated with calf diarrhea (p < 0.05). Among them, C. parvum and BRV-A were considered to be the most common enteric pathogens for calf diarrhea with high detection frequency (33.7% and 27.1%) and strong odds ratio (173 and 79.9). Unexpectedly BNoV (OR = 2.0) and Nebovirus (OR = 16.7) were identified with high frequency in diarrheic calves, suggesting these viruses may have a significant contribution to calf diarrhea.
Keywords: Calf diarrhea, Case–control study, Enteric pathogens, Multiplex PCR detection
1. Introduction
Calf diarrhea is a major cause of economic loss with high morbidity and mortality in the cattle industry worldwide (Bartels et al., 2010, de la Fuente et al., 1999, Kelling et al., 2002, Uhde et al., 2008, United, 2007). Many factors are known to contribute to calf diarrhea. Historically, calf diarrhea has been commonly attributed to bovine rotavirus group A (BRV-A), bovine coronavirus (BCoV), bovine viral diarrhea virus (BVDV), Salmonella spp. (Salmonella), Escherichia coli (E. coli) K99+, and Clostridium perfringens (C. perfringens) type C and Cryptosporidium parvum (C. parvum) (Acha et al., 2004, Reynolds et al., 1986, Saif and Smith, 1985, Snodgrass et al., 1986). The specific etiology of many field cases of calf diarrhea still remain undiagnosed (Milnes et al., 2007). Recently, bovine norovirus (BNoV), Nebovirus, bovine enterovirus (BEV) and bovine torovirus (BToV) have been identified as potential causes of calf diarrhea (Blas-Machado et al., 2007, Haschek et al., 2006, Hoet et al., 2003a, Kaplon et al., 2011, Otto et al., 2011, Park et al., 2007, Park et al., 2008a, Park et al., 2008b). Some of these agents (i.e., BNoV, BEV and BToV) have also been found in feces from clinically healthy calves (Haschek et al., 2006, Jimenez-Clavero et al., 2005, Mijovski et al., 2010, Shanks et al., 2008) and many of previous epidemiological studies for BNoV and BToV have been focused only on diarrheic calves (Hoet et al., 2003b, Milnes et al., 2007, Park et al., 2007, Park et al., 2008b). Their role in calf diarrhea still remains to be evaluated.
Various laboratory methods have been applied for the detection of infectious agents in feces. Historically, virus isolation, electron microscopy, enzyme-linked immunosorbent assay, latex agglutination test, bacterial culture, direct microscopy of fecal smear (acid-fast stain), and/or fecal flotation have been commonly used to test fecal samples for enteric pathogens (Cho et al., 2010). These procedures are reliable; however, they are time-consuming and require specialized knowledge. Recently, nucleic acid based tests, such as polymerase chain reaction (PCR) assays, have become popular for rapid and sensitive detection of infectious agents (Albini et al., 2008, Cho et al., 2010). Multiplex real-time PCR panels have been proven to be a useful diagnostic tool for concurrent detection of several target enteric pathogens with high sensitivity and specificity (Albini et al., 2008, Cho et al., 2010), which decreases bias in diagnostic outcome due to testing method.
The following case–control study was conducted to: (a) assess the prevalence of 11 infectious agents consisting of 7 common [BRV-A, BCoV, BVDV, Salmonella, E. coli K99+, C. perfringens with β toxin gene (Cpt β) and C. parvum] and 4 emerging enteric pathogens (BNoV, Nebovirus, BEV and BToV) in fecal samples from healthy and diarrheic calves in the Midwest by using a panel of PCR assays; and (b) determine their association with diarrhea as well as investigate their potential interactions in expression of disease.
2. Materials and methods
2.1. Animals and samples
All fecal samples used in the study were originated from clinically diarrheic and healthy calves during year 2010–2011. A total of 199 fecal samples from diarrheic calves were procured from submissions to the Iowa State University Veterinary Diagnostic Laboratory (ISUVDL) and used as cases. The samples were from 140 cattle farms with the most of the samples (99%) originated in the Midwest [Iowa (78%), Minnesota (8%), Wisconsin (4%), Missouri (3%), Ohio (3%), Illinois (1%), South Dakota (1%) and Nebraska (1%)]. No more than 4 samples were randomly selected from the same farm if a large number of samples were submitted. A vast majority of the samples tested were from sick animals before treatment begun according to referring veterinarians. Approximately 41% and 42% of the samples were from dairy and beef breeds, respectively. The remaining 18.5% of the samples were submitted without breed identification. Physical appearance of first 99 of the 199 fecal samples was recorded as ‘watery’ or ‘semi-solid’ upon receiving as fresh samples were available to the investigators before freezing.
A total of 245 fecal samples were collected from clinically healthy (i.e., no diarrhea) calves in 25 different beef or dairy farms which were evenly distributed across the State of Iowa and used as controls. These farms were pre-selected to be part of other field-based study in which on-going health monitoring was required including use of any medication. Samples were collected twice from each farm at approximately 2-week intervals with continuous monitoring of health status to ensure lack of diarrhea among animals on each farm. At each time of sample collection, 5 calves were randomly selected for sampling.
Most of the source farms were similar in overall farm management, including vaccination and medication, and nutritional status. Most (96.4%) of the calves tested were less than 6 months old in age. Two third of the control calves were less than 3 months of age while 80% of the case calves were less than 3 months of age. Only 1and 7 cases were submitted from a 7-month-old diarrheic calf and clinically healthy yearlings or older cattle, respectively.
2.2. Detection of pathogens
All fecal samples were examined for 11 different microorganisms (i.e., BRV-A, BCoV, BVDV, BEV, BNoV, BToV, Nebovirus, Salmonella, E. coli K99+, C. parvum and Cpt β) using a panel of polymerase chain reaction (PCR) based assays. All except BEV have been reported as pathogens implicated in calf diarrhea.
Before PCR testing, each fecal sample was suspended in 0.01 M phosphate-buffered saline (pH 7.4) to make 30% fecal homogenates and then centrifuged for 1 min at 100 × g for clarification as previously described (Cho et al., 2010). The supernatant was then used for viral and bacterial nucleic acid extraction using MagMax™ Total Nucleic Acid Isolation Kit (Applied Biosystems, Austin, TX) according to the manufacturer's instruction. The extraction procedure was performed using Kingfisher® 96 Magnetic Particle Processor (Thermo Fisher Scientific Inc., Waltham, MA). All extracts were stored at −80 °C until tested.
Probe-based real-time PCR (rtPCR) assays for all pathogens except BToV and Nebovirus were performed in a duplex or singleplex PCR format with Path-ID™ Multiplex One-Step RT-PCR Kit (Applied Biosystems, Austin, TX) and AgPath-ID™ One-Step RT-PCR Kit (Applied Biosystems, Austin, TX), respectively. For BToV, a SYBR Green rtPCR assay was used with QuantiTest™ SYBR® Green PCR Kit (QIAGEN, Valencia, CA).
For rtPCR set-up, 7 μl of template and 18 μl of the reaction mixture for the duplex PCRs (Table 1 , real-time PCR set 1, 2, 5 and 6) and 5 μl of template and 20 μl of the reaction mixture for singleplex PCRs (Table 1, real-time PCR set 3 and 4) were used. All reaction mixtures contained 400 nM of each primer, 120 nM of the probe except BToV, RT-PCR buffer, RT-PCR enzyme mix, and nuclease-free water. The volume of each reagent added to a reaction mixture was as per manufacturer's instruction. The sequence information of primers and probes used for specific detection of each pathogen is summarized in Table 1.
Table 1.
Oligonucleotide sequence of primers and probe used in PCR to detect each target enteric pathogen.
| PCR format | Target pathogen [primer/probe sequence (5′–3′)] | References |
|---|---|---|
| Real-time PCR (set 1) | BCoV-fwd: CTAGTAACCAGGCTGATGTCAATACC BCoV-rev: GGCGGAAACCTAGTCGGAATA BCoV-probe: (FAM/MGB) CGGCTGACATTCTCGATC |
Cho et al. (2010) |
| BRV-fwd1: TCAACATGGATGTCCTGTACTCCT BRV-fwd2: TCAACATGGATGTCCTGTATTCCT BRV-fwd3: TCAACATGGATGTCCTTTATTCCT BRV-rev1: TCCTCCAGTTTGGAACTCATT BRV-rev2: TCCCCCAGTTTGGAATTCATT BRV-rev3: CCCTCCAGTTTGGAATTCATT BRV-probe1: (VIC/MGB) TCAAAAACTCTTAAAGATGCTAG BRV-probe2: (VIC/MGB) TCAAAAACTCTTAAAGATGCAAG |
Cho et al. (2010) | |
| Real-time PCR (set 2) | BEV-fwd: GCCGTGAATGCTGCTAATCC BEV-rev: GTAGTCTGTTCCGCCYCYRACT BEV-probe: (FAM/BHQ1) CGCACAATCCAGTGTTGCTACGTCGTAAC |
Jimenez-Clavero et al. (2005) |
| BVDV-fwd: GGG NAG TCG TCA RTG GTT CG BVDV-rev: GTG CCA TGT ACA GCA GAG WTT TT BVDV probe: (Cy5/BHQ2) CTTGGTGTACCTCTATACTCA |
Mahlum et al. (2002) | |
| Real-time PCR (set 3) | BNoV-fwd: CGCTCCATGTTYGCBTGG BNoV-rev: TCAGTCATCTTCATTTACAAAATC BNoV-probe: (Fam/Zen/IABkFQ) TGTGGGAAGGTAGTCGCGACRYC |
Wolf et al. (2007) |
| SYBR green real-time PCR (set 4) | BToV-fwd: TTACTGGYTATTGGGCMYT BToV-rev: AAAGGRGTGCAGTGWAGCTT |
Hosmillo et al. (2010) |
| Real-time PCR (set 5) |
E. coli K99+-fwd: GCTATTAGTGGTCATGGCACTGTAG E. coli K99+-rev: TTTGTTTTCGCTAGGCAGTCATTA E. coli K99+-Probe: (FAM/BHQ1) ATTTTAAACTAAAACCAGCGCCCGGCA |
West et al. (2007) |
|
C. parvum-fwd: CAAATTGATACCGTTTGTCCTTCTGT C. parvum -rev: GGCATGTCGATTCTAATTCAGCT C. parvum -probe: (Cy5/BHQ2) TGCCATACATTGTTGTCCTGACAAATTGAA |
Guy et al. (2003) | |
| Real-time PCR (set 6) |
Samonella-fwd: GCCATGCTGTTCGATGAT Samonella -rev: GTTACCGATAGCGGGAAAGG Samonella -probe: (FAM/BHQ1) TTTTGCACCACMGCCAGCCC |
Moore and Feist (2007) |
|
C. perfringens β-fwd: TGGAGCGTGAAAGAAACTGTTATTA C. perfringens β-rev: GGTATCAAAAGCTAGCCTGGAATAGA C. perfringens β-probe:(Cy5/BHQ2) CTTAATTGGAATGGTGCTAACTGGGTAGGACAA |
Albini et al. (2008) | |
| Internal control | P1570: TGGCCCGCAGTATTCTGATT P1642: CAGCTGGGACAGCAGTTGAG P1591M: (Cy3/BHQ1) CCTCGAATCAAACGCCGTTGGAATG |
Cho et al. (2010) |
| Nested RT-PCR | Nebo-fwd: TTTCTAACYTATGGGGAYGAYG Nebo-rev: GTCACTCATGTTTCCTTCTCTAAT nNebo-fwd: CGCTCCGTGTGGGATCACGA nNebo-rev: GCACGGGCTTCTTCTAGAGA |
Kaplon et al. (2011) |
Amplification of the targeted genomic region was conducted using ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Austin, TX). Cycling conditions of the probe-based rtPCRs were as follows: (a) reverse transcription (RT) for 10 min at 48 °C (45 °C for singleplex); (b) activation of DNA polymerase at 95 °C for 15 min (10 min for singleplex); and (c) 40 cycles of denaturation at 94 °C for 10 s and annealing/extension at 60 °C for 60 s (45 s for singleplex). The RT step was applied only for viral targets. Running conditions of the SYBR Green rtPCR for BToV were: (a) RT step for 10 min at 50 °C; and (b) 40 cycles of denaturation at 95 °C and annealing/extension at 60 °C for 30. After 40 cycle reaction, the melting curve analysis was performed. Samples with cycle threshold (Ct) ≤ 35 for any given targets were considered positive for those pathogens.
For detection of Nebovirus, a gel-based nested RT-PCR was used as previously described (Jor et al., 2010). The PCR was conducted using OneStep RT-PCR Kit (QIAGEN, Valencia, CA) with QIAGEN® RNase inhibitor and HotStarTaq® DNA Polymerase Kit (QIAGEN, Valencia, CA) for RT-PCR and nested PCR, respectively, according to the manufacturer's instruction. Cycling conditions of the RT-PCR were: (a) RT step at 50 °C for 30 min; (b) DNA polymerase activation step at 95 °C for 15 min; (c) 40 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s and extension at 72 °C for 1 min; and (d) followed by a final cycle at 72 °C for 10 min. Cycling conditions of the nested PCR were: (a) activation step at 95 °C for 15 min; and (b) 35 cycles of denaturation at 94 °C for 30 s, annealing at 54 °C for 30 s and extension at 72 °C for 1 min; and (c) followed by a final cycle at 72 °C for 10 min.
2.3. Effect of test methods on detection frequency of enteric pathogens
Detection frequencies of selected enteric pathogens (i.e., BRV-A, BCoV and C. parvum) in calf diarrhea cases submitted to ISUVDL from 2003 to 2011 were compared based on laboratory methods used. Before 2008, antigen-capturing ELISA and fecal smear direct microscopy (acid-fast stain) tests were used to detect BRV-A/BCoV and C. parvum in feces, respectively. Since then, a bovine enteric panel (BEP) consisting of 2 multiplex rtPCR tests (Cho et al., 2010) was implemented for simultaneous detection of BRV-A, BCoV, and C. parvum in feces. All diagnostic data were retrieved from the ISUVDL laboratory information management system.
2.4. Statistics
The PCR results on each of the fecal samples were recorded as either positive or negative for each pathogen and categorized under disease status (i.e., diarrheic versus non-diarrheic) of each animal. The association between diarrhea and detection of each pathogen was determined using a multivariate logistic regression model. The probability of concurrent detection among pathogens was also analyzed in the same manner. The final model was built with stepwise selection using Firth's penalized likelihood method due to quasi-complete separation of the data. Odds ratios (ORs) with 95% confidence intervals were calculated to assess the likelihood of association.
The association between the severity of diarrhea (i.e., watery versus semi-solid) and the presence of each pathogen was also analyzed using multivariate logistic regression model with stepwise model selection.
Since BNoV and BCoV were detected in feces from both diarrheic and healthy calves at a relatively high frequency, Ct values of feces for BNoV and BCoV were analyzed by the non-parametric Wilcoxon rank-sum test to evaluate the quantitative difference in virus shedding between diarrheic and healthy calves.
All statistical analyses were conducted by using SAS 9.2 (SAS Institute, Cary, NC). For all analyses, a value of p < 0.05 was considered significant.
3. Results
3.1. Survey of calves in the Midwest USA for infection with enteric pathogens
A total of the 199 fecal samples from diarrheic calves and 245 fecal samples from healthy calves were tested for 11 putative enteric pathogens. PCR testing revealed that 80.4% and 27.8% of the diarrheic and normal fecal samples, respectively, were positive for at least one of these infectious agents.
As summarized in Table 2 , BNoV (44.7%), C. parvum (33.7%), BCoV (31.7%), BRV-A (27.1%), Nebovirus (21.6%) and Salmonella (9.0%) were commonly detected in feces from the diarrheic calves, while BVDV, BToV, E. coli K99+ and BEV were found at a much lower frequency (0.4–5%). BNoV (16.3%) and BCoV (12.2%) were also detected in the feces from healthy calves but at a lower frequency than that in diarrheic feces. While Nebovirus (1.6%) and BVDV (0.4%) were infrequently detected in the feces from healthy calves, C. parvum, BRV-A, E. coli K99+ and BToV were detected only in the feces from diarrheic calves. In contrast, BEV (32.7%) was much more frequently detected in the feces from healthy calves than those from diarrheic calves. C. perfringens with β toxin gene (i.e., C. perfringens type B or C) was not detected in any of the feces examined in this study.
Table 2.
Detection frequency of various bovine enteric pathogens among feces from diarrheic and healthy calves in the Midwest and association between positivity and calf diarrhea.
| Pathogens | Overall % positive | % positives among diarrheic calves | % positives among healthy calves | p-Value | Odds ratio |
|---|---|---|---|---|---|
| Bovine norovirus | 29.1 | 44.7 (89/199)a | 16.3 (40/245)a | 0.042 | 2.0 (1.002–3.9)b |
| Cryptosporidium parvumc | 15.1 | 33.7 (67/199) | 0.0 (0/245) | 0.0007 | 173.0 (8.9–3365.1) |
| Bovine coronavirus | 20.9 | 31.7 (63/199) | 12.2 (30/245) | 0.0034 | 2.7 (1.4–5.1) |
| Bovine rotavirus group A | 12.2 | 27.1 (54/199) | 0.0 (0/245) | 0.0025 | 79.9 (4.7–1369.5) |
| Nebovirus | 0.9 | 21.6 (43/199) | 1.6 (4/245) | 0.0001 | 16.7 (4.0–68.8) |
| Salmonella spp. | 4.1 | 9.0 (18/199) | 0.0 (0/245) | 0.0056 | 80.6 (3.6–1803.7) |
| Bovine enterovirus | 20.3 | 5.0 (10/199) | 32.7 (80/245) | <0.0001 | 0.113 (0.04–0.3) |
| Escherichia coliK99+ | 1.8 | 4.0 (8/199) | 0.0 (0/245) | 0.0143 | 98.4 (2.5–3859.9) |
| Bovine torovirus | 1.1 | 2.5 (5/199) | 0.0 (0/245) | 0.2404 | 10.4 (0.2–520.3) |
| Bovine viral diarrhea virus | 0.5 | 0.5 (1/199) | 0.4 (1/245) | – | – |
| Clostridium perfringens toxin β | 0.0 | 0.0 (0/199) | 0.0 (0/245) | – | – |
Numbers in the parenthesis show number of positive feces/number of samples tested.
Numbers in the parenthesis is 95% confidence interval of the estimated odds ratio.
The bold letters indicate microorganisms detected only in feces from diarrheic calves.
Although BNoV and BCoV were detected in feces from both diarrheic and healthy calves, the detection frequency and fecal shedding quantity of the viruses were significantly higher in the feces from diarrheic calves except for one healthy calf feces which showed the lowest Ct value (17.4) for BCoV (Fig. 1 ), as compared to those in the feces from healthy calves. The median (mean ± SE) Ct values of feces from diarrheic calves positive for BNoV and BCoV were 26.2 (26.4 ± 0.53) and 25.6 (24.3 ± 0.87) respectively, whereas those from healthy calves positive for BNoV and BCoV were 31.3 (31.0 ± 0.54) and 31.3 (30.2 ± 0.78) respectively.
Fig. 1.
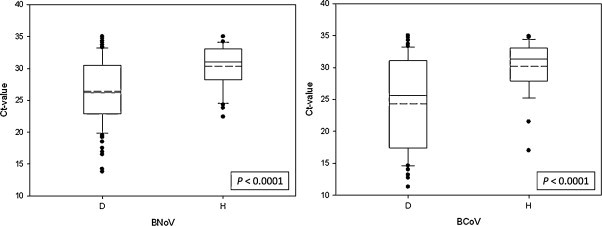
Quantitative comparison of bovine norovirus (BNoV) and bovine coronavirus (BCoV) shedding in fecal samples from diarrheic (D) and healthy (H) calves. Mean (solid line) and median (dotted line) are shown on a boxplot with 50 percentile distribution. The lower and upper whiskers represent 10th and 90th percentile plot, resepctively, and dots represent outlayers. Virus shedding level between the 2 groups was compared based on Ct values by the non-parametric Wilcoxon rank-sum test.
With respect to age distribution, many of fecal samples from diarrheic calves positive for BNoV, C. parvum, BCoV, BRV-A, Nebovirus, Salmonella, BToV, and E. coli K99+ were from calves at 0 to 4 weeks of age (Fig. 2 ). In particular, calves at 0–2 weeks of age were the most commonly positive for these pathogens.
Fig. 2.
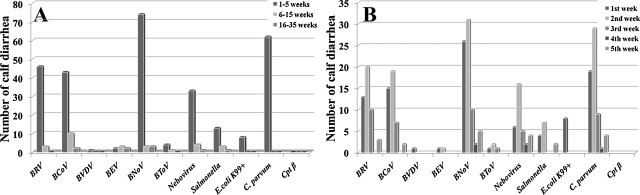
Age distribution of diarrheic calves whose feces were positive for one or more enteric pathogens. Animals are classified into 3 age groups: 0–4 weeks, 5–14 weeks and 15–34 weeks of age (A) based on the information provided by submitting veterinarians. Animals at 0–4 weeks of age are further broken down on the weekly basis after birth (B). BRV (bovine rotavirus), BCoV (bovine coronavirus), BVDV (bovine viral diarrhea virus), BEV (bovine enterovirus), BNoV (bovine norovirus), C. parvum (cryptosporidium parvum) and Cpt β (clostridium perfringens β toxin).
3.2. Assessment of the association of 11 enteric pathogens with diarrhea
As summarized in Table 2, the presence of C. parvum, E. coli K99+, Salmonella, BRV-A, Nebovirus, BCoV and BNoV in feces were significantly associated with calf diarrhea (p < 0.05). Among these pathogens, C. parvum, E. coli K99+, Salmonella, BRV-A and Nebovirus showed a stronger association with diarrhea (OR > 10.0). In contrast, detection of BEV was inversely correlated with diarrhea (OR = 0.113); therefore, BEV was not included in further statistical analyses.
No statistically significant association between the presence of BToV in feces and diarrhea was observed in this study even though the virus was detected only in the feces from diarrheic calves, probably due to a low frequency of detection (Table 2). The ORs could not be calculated for BVDV and Cpt β because of either extremely low frequency of detection or no detection; hence, statistical significance could not be determined. Bovine rotavirus group A was the only pathogen significantly (p = 0.013) associated with liquid form of diarrheic feces (Table 3 ).
Table 3.
Association of enteric pathogens with the severity of diarrhea (i.e., watery diarrhea) based on physical appearance of feces.
| Number of samples positive for each target | Physical appearance of fecesa |
p-Value | Odds ratio | ||
|---|---|---|---|---|---|
| Watery (n = 30) | Semisolid (n = 69) | ||||
| Bovine rotavirus group A | 26/99b (26.3%) | 13/30 (43.3%) | 13/69 (18.8%) | 0.013 | 3.3 (1.3–8.4)c |
| Bovine coronavirus | 30/99 (30.3%) | 10/30 (33.3%) | 20/69 (29.0%) | –d | – |
| Bovine viral diarrhea virus | 1/99 (1.0%) | 1/30 (3.3%) | 0/69 (0.0%) | – | – |
| Bovine norovirus | 42/99 (42.4%) | 15/30 (50.0%) | 27/69 (39.1%) | – | – |
| Bovine torovirus | 2/99 (2.0%) | 1/30 (3.3%) | 1/69 (1.4%) | – | – |
| Nebovirus | 23/99 (23.2%) | 10/30 (33.3%) | 13/69 (18.8%) | – | – |
| Salmonella spp. | 13/99 (13.1%) | 4/30 (13.3%) | 9/69 (13.0%) | – | – |
| Escherichia coli K99+ | 7/99 (7.1%) | 4/30 (13.3%) | 3/69 (4.3%) | – | – |
| Cryptosporidium parvum | 28/99 (28.3%) | 10/30 (33.3%) | 18/69 (26.1%) | – | – |
| Clostridium perfringens toxin β | 0/99 (0.0%) | 0/30 (0.0%) | 0/69 (0.0%) | – | – |
Physical appearance of feces was upon receiving of samples with clinical history of diarrhea.
Number of positives/number of samples tested.
Numbers in the parenthesis are 95% confidence interval of the estimated odds ratio.
No significant association was observed.
3.3. Concurrent infection of enteric pathogens for calf diarrhea
While 55% of the diarrheic fecal samples had more than 1 enteric pathogen detected, only 3% of the fecal samples from healthy calves had multiple pathogens (Fig. 3 ). In the diarrheic fecal samples, the presence of 2 different pathogens (31%) was the most commonly seen and 1% of the samples even had up to 6 different pathogens concurrently.
Fig. 3.
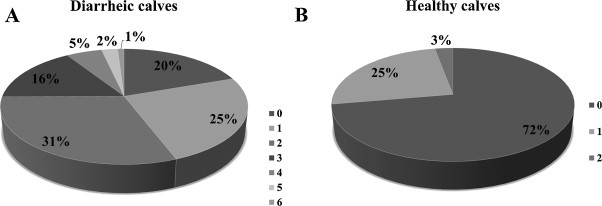
Frequency (%) of concurrent infection in diarrheic (A) and healthy calves (B). Numbers (0–6) represent the number of pathogens concurrently detected within each fecal sample. Bovine enterovirus is not included in assessment.
The probability of detecting certain agents together is summarized in Table 4 . Bovine norovirus, BCoV, Salmonella, and C. parvum were commonly detected in feces which were also positive for BRV-A. Nebovirus was commonly detected in feces also positive for BCoV, C. parvum or BToV. BNoV presence was significantly correlated with C. parvum presence in addition to BRV-A. While many of the pathogens were concurrently detected with more than 2 other pathogens, BToV and Salmonella were identified only with Nebovirus and BRV-A, respectively. The concurrent presence of BToV and Nebovirus was much stronger [13.4 ≤ OR ≤ 15.7 (2.2–114.5)] as compared to other mixed infections. Statistically significant synergistic interaction between pathogens for causing the diarrhea or exacerbating the severity of diarrhea was not observed.
Table 4.
Concurrent detection of enteric pathogens in feces from diarrheic calves and their association strength.
| Reference pathogens | Associated pathogens | p-Value | Odds ratio |
|---|---|---|---|
| Bovine norovirus | BRV-A | <0.0001 | 3.6 (1.9–6.8)a |
| (BNoV) | C. parvum | <0.0001 | 4.2 (2.4–7.4) |
| Bovine coronavirus | BRV-A | <0.0001 | 3.7 (2.0–6.8) |
| (BCoV) | Nebovirus | 0.0232 | 2.2 (1.1–4.3) |
| Bovine rotavirus group A | BNoV | 0.0005 | 3.2 (1.7–6.0) |
| (BRV-A) | BCoV | <0.0001 | 3.6 (1.9–6.9) |
| Salmonella spp. | 0.0012 | 5.9 (2.0–17.1) | |
| C. parvum | 0.0008 | 3.3 (1.6–6.7) | |
| Nebovirus | BCoV | 0.0496 | 2.1 (1.0–4.2) |
| BToV | 0.0066 | 15.7 (2.2–114.5) | |
| C. parvum | <0.0001 | 9.6 (4.9–18.9) | |
| Bovine torovirus | Nebovirus | 0.005 | 13.5 (2.2–82.8) |
| (BToV) | |||
| Salmonella spp. | BRV-A | 0.0013 | 5.1 (1.9–13.9) |
| Cryptosporidium parvum | BNoV | <0.001 | 3.6 (2.0–6.5) |
| (C. parvum) | BRV-A | 0.0057 | 2.7 (1.3–5.6) |
| Nebovirus | <0.001 | 7.1 (3.5–14.2) | |
Numbers in the parenthesis is 95% confidence interval of the estimated odds ratio.
When the pathogens were sorted based on their taxonomical property (i.e., virus, bacteria and protozoa) and compared for their detection frequency between diarrheic and healthy calves, virus only (36.2%) or virus/C. parvum co-infection (28.1%) was the most commonly observed in the diarrheic calves. In comparison, virus only (28.0%) was common in the healthy calves (Table 5 ). BNoV and BCoV were the pathogens that were the most commonly detected in the feces from healthy calves.
Table 5.
Frequency of viral, bacterial and/or protozoan infections in diarrheic and healthy calves. Viral pathogens included for testing are group A bovine rotavirus, bovine coronavirus, bovine torovirus, bovine norovirus, Nebovirus and bovine viral diarrhea virus. Bacterial pathogens included for testing are Escherichia coli K99+, Salmonella spp. and Clostridium perfringens with β toxin. Cryptosporidium parvum (C. parvum) is the only pathogen 3 representing the protozoa group.
| Pathogens | Diarrheic calves (%) | Healthy claves (%) |
|---|---|---|
| Viral pathogens infection | 36.2 | 27.3 |
| Bacterial pathogens infection | 4.0 | 0 |
| C. parvum infection | 4.5 | 0 |
| Viral and bacterial pathogens co-infection | 7.5 | 0 |
| Viral pathogens and C. parvum co-infection | 28.1 | 0 |
| Bacterial pathogens and C. parvum co-infection | 1.5 | 0 |
| Viral, bacterial pathogens and C. parvum co-infection | 1.5 | 0 |
3.4. Influence of laboratory methods on the detection frequency
The mean detection frequency of BRV-A, BCoV and C. parvum in diarrhea cases during year 2003–2007 were 24.6%, 11.9% and 8.7%, respectively, when antigen-capturing ELISAs and direct microscopy (acid-fast stain) were the main laboratory methods for detection of these pathogens at ISUVDL. After implementation of a PCR panel for the major calf diarrhea pathogens, the mean detection frequency of BRV-A, BCoV and C. parvum were 37.2%, 29.2% and 38.3%, respectively, during year 2008–2011 (Table 6 ).
Table 6.
Comparison of the detection frequency of bovine rotavirus group A (BRV-A), bovine coronavirus (BCoV) and Cryptosporidium parvum (C. parvum) in feces from diarrheic calves before/after use of a PCR-based bovine enteric panel (BEP) in Iowa State University Veterinary Diagnostic Laboratory.
| Year | BRV-A | BCoV | C. parvum | |
|---|---|---|---|---|
| ELISA | ELISA | Direct microscopya | ||
| Before BEP | 2003 | 29.8% (131/440)b | 12.4% (50/405) | 12.2% (15/123) |
| 2004 | 25.8% (102/396) | 11.8% (46/391) | 12.7% (13/102) | |
| 2005 | 25.6% (103/402) | 9.8% (41/418) | 8.4% (12/143) | |
| 2006 | 18.6% (67/361) | 17.0% (24/141) | 5.7% (7/123) | |
| 2007 | 22.7% (123/542) | – | 4.5% (5/111) | |
| Average | 24.6% (123/2142) | 11.9% (161/1355) | 8.6% (52/602) |
| Year | BRV-A |
BCoV |
C. parvum |
|
|---|---|---|---|---|
| Real-time PCR | ||||
| After BEP | 2008 | 40.7% (198/487) | 36.8% (179/487) | 42.5% (207/487) |
| 2009 | 39.5% (213/539) | 27.8% (150/539) | 42.3% (228/539) | |
| 2010 | 40.1 (242/603) | 28.4% (171/603) | 38.1% (230/603) | |
| 2011 | 29.2% (176/602) | 25.1% (151/602) | 31.6% (190/602) | |
| Average | 37.2% (829/2231) | 29.2% (651/2231) | 38.3% (855/2231) | |
Acid-fast staining was used.
% positive (number of positives/number of total cases).
4. Discussion
In this study, we investigated the prevalence of 11 calf enteric pathogens consisting of 7 common (BRV-A, BCoV, BVDV, Salmonella, E. coli K99+, Cpt β and C. parvum) and 4 emerging pathogens (BNoV, Nebovirus, BEV and BToV) and then evaluated two aspects; their clinical significance in calf diarrhea and co-infection between them. Not unexpectedly, 80% of diarrheic calves tested were positive for at least one of the target enteric pathogens, suggesting that the infectious factor is still a major cause of calf diarrhea. More than 50% of the diarrheic calves tested were concurrently infected with more than one pathogen. Co-infection with 2 pathogens was the most common finding (31%) with up to 6 pathogens detected in 1% of the fecal samples from diarrheic calves. The majority of diarrheic cases were identified among 0- to 4-week-old calves and concentrated among calves at 0–2 weeks of age, which is similar to previous reports by other investigators (Bartels et al., 2010, de la Fuente et al., 1999, McDonough et al., 1994). High frequency of co-infection by multiple pathogens in young animals emphasizes that interventions for calf diarrhea should be focused on husbandry and management strategies, including assurance of colostrum intake, hygiene, reduction of population density, or modified components of the Sandhills calving system (Larson and Tyler, 2005). Twenty percent of the diarrheic calves were negative for all of the 11 pathogens in this study. While low sensitivity of the test might be accounted for the negative result, the role of non-infectious factors (e.g., cold weather, impaired uptake of colostrum, or poor sanitation) in calf diarrhea cannot be discounted. In addition, the possibility of other pathogens (e.g., rotavirus B or C; coccidia; C. perfringens type A, D or E; and other pathogroups of E. coli) or previously unrecognized agent(s) involved in diarrhea remains to be further studied.
Viral infections (36.1%) or combination of viruses and C. parvum (28.1%) were the most commonly detected etiology in feces from diarrheic calves, which is similar to previous reports on calf diarrhea (de la Fuente et al., 1998, Garcia et al., 2000, McDonough et al., 1994). In contrast, the proportion of bacteria-positive samples was relatively small. Of three target bacterial pathogens, Salmonella (9%) was the most commonly detected in the diarrhea feces examined. Interestingly, none of the fecal samples from both diarrheic and healthy calves was positive for Cpt β which is contained in either C. perfringens type B or C. This was an unexpected observation since C. perfringens type C has been postulated as the main type causing calf diarrhea. Although the PCR results were not confirmed by anaerobic bacterial culture, it should be noted that our observation is in agreement with previous reports by other investigators describing no (Albini et al., 2008, Ferrarezi et al., 2008, Sting, 2009) or very low detection of Cpt β (Gurjar et al., 2008) in diarrheic calves, suggesting that C. perfringens type C is rarely involved in outbreaks of calf diarrhea or is simply an opportunistic bacterium causing acute enterotoxemia under certain favorable conditions. As it was suggested that all types of C. perfringens should be considered as a calf diarrhea etiology (Ferrarezi et al., 2008), involvement of other types of C. perfringens in diarrhea cases may be necessary.
C. parvum was frequently (33.7%) detected in calf diarrhea cases, which is in agreement with previous reports (Bartels et al., 2010, Izzo et al., 2011, McDonough et al., 1994, Uhde et al., 2008). It may imply the difficulty with C. parvum control in the field due to autoinfection, environment resistance of oocysts and lack of effective treatment and vaccine (Joachim et al., 2003). Preventative measures for C. parvum in cow-calf operations should be focused on keeping good herd sanitation and sick animals segregated from healthy ones (Trotz-Williams et al., 2007). Co-infection with viruses (28.1%), particularly BRV-A (OR = 2.7), BNoV (OR = 3.6) and Nebovirus (OR = 7.1), was much more common than with bacteria in our study. While co-infection of BRV-A and C. parvum in diarrheic calves has been frequently reported (Bartels et al., 2010, Bjorkman et al., 2003, de la Fuente et al., 1999, Garcia et al., 2000, Uhde et al., 2008), common association of Nebovirus and C. parvum in diarrheic animals is a new observation. It has been reported that viral infections, such as porcine circovirus type 2 and human immunodeficiency virus, can increase the susceptibility of pigs and humans, respectively, to C. parvum (Nunez et al., 2003, Putignani and Menichella, 2010), suggesting that immunosuppressive viruses can predispose animals or humans to C. parvum. In the absence of effective treatment options for C. parvum, it may be prudent to rely on management practices and specific aids in prevention of viral infections to reduce clinical problems with C. parvum infections.
Bovine rotavirus A was found solely in many of the diarrhea cases (27.1%) and positively correlated with the severity (i.e., liquid feces) of diarrhea (OR = 3.3). This observation is similar to reports of human rotavirus infection being highly associated with acute watery diarrhea (Olesen et al., 2005, Wilhelmi et al., 2003). A high correlation between BRV-A detection and diarrhea (OR = 79.9) and a wide range of association with other pathogens (BNoV, BCoV, Salmonella and C. parvum) may be an evidence that BRV-A is a primary major bovine enteric pathogen of calf diarrhea, which is also in agreement with previous reports describing the primary role of BRV-A in neonatal calf diarrhea (Bartels et al., 2010, Garcia et al., 2000, Uhde et al., 2008). Our and others’ observations raise concerns regarding vaccination practices on farms and the efficacy of current licensed BRV-A vaccines since vaccination has been a main tool for prevention of BRV-A associated diarrhea in neonates. Implementation of a regular vaccination program for BRV-A can be easily achieved through enhancing the awareness of the high frequency of rotavirus-associated calf diarrhea in the field, but continuing efficacy of BRV-A vaccines may require frequent surveillance and further characterization of rotaviruses circulating in the field. Surveillance is warranted since antigenic variation of rotavirus due to frequent mutation and recombination is of a great concern for emerging a variant or new serotype (Martella et al., 2010).
New and emerging viruses with pathogenic potential for calf diarrhea (i.e., BNoV, Nebovirus and BToV) were also studied together with historically well-known major enteric pathogens (i.e., BRV-A, BCoV, BVDV, C. parvum, Salmonella and E. coli K99+ and Cpt β). The most noteworthy observations from our study were the significant association of BNoV (OR = 2.0) and Nebovirus (OR = 16.7) with calf diarrhea and their frequent detection (44.7% and 21.6%, respectively) in calf diarrhea cases, suggesting that bovine caliciviruses may play a more significant role in calf diarrhea than what was believed. It is an unexpected observation that Nebovirus was detected in diarrheic animals at a much higher rate than what was previously reported from France (Kaplon et al., 2011). A high frequency of BNoV detection is, on the other hand, not a surprise since many other investigators have previously reported a high prevalence of BNoV infection in the studied bovine populations (Cho et al., 2011, Di Bartolo et al., 2011, Jor et al., 2010, Kaplon et al., 2011, Mijovski et al., 2010, Park et al., 2007, Reuter et al., 2009, van der Poel et al., 2003, Wise et al., 2004, Yilmaz et al., 2011). Clinical significance of BNoV infection has not been clear in the field because the virus has also been found in clinically healthy calves (Jor et al., 2010, Mijovski et al., 2010) as also shown in our study. Recently an animal study has demonstrated that BNoV is pathogenic to naïve calves (Otto et al., 2011). In our study, which is the first case–control study evaluating BNoV as bovine enteric pathogen for calf diarrhea, a significant quantitative difference in the virus amount between fecal samples from diarrheic and healthy calves was detected, suggesting that disease progression may depend upon the initial exposure dose of the virus or factors contributing to BNoV replication to a high titer. While such a difference in replication ability did not appear to be due to unique genetic profile of BNoV's polymerase gene (data not shown), further study remains to redefine the pathogenicity of bovine caliciviruses and to determine the correlation between virus amount and the pathogenicity and identify contributing factors.
Unlike bovine caliciviruses, it was difficult to judge the role that BToV may play in calf diarrhea because the virus was detected in a relatively small number of the fecal samples examined (1.1%). Such a detection frequency of BToV in our study is similar to what was previously reported from Korea (2.9%) and Austria (5.2%) but different from that reported in USA (36.4%) and Japan (18%) (Duckmanton et al., 1998, Haschek et al., 2006, Kirisawa et al., 2007, Park et al., 2008b). Although a statistically significant association between BToV and diarrhea could not be demonstrated due to a low prevalence, it must be pointed out that the virus was detected only in feces from diarrheic calves. A survey on a larger number of animals, longitudinal cohort study or animal challenge study would be necessary to determine the clinical significance of BToV for calf diarrhea.
Bovine coronavirus is historically believed to be a major bovine enteric pathogen causing calf diarrhea, corroborated by pathologic studies (Boileau and Kapil, 2010). However, such a role has been challenged as some epidemiological studies could not demonstrate a statistically significant association between BCoV infection and calf diarrhea (Bartels et al., 2010, Bjorkman et al., 2003, Uhde et al., 2008). A recent cohort study on Dutch cattle farms even suggested potential opportunistic nature of BCoV infection with previous history of diarrhea (Bartels et al., 2010). In our study, BCoV was found to be significantly associated with calf diarrhea although its association strength with calf diarrhea was relatively weak (OR = 2.7) as compared to pathogens historically known to be causes of calf diarrhea such as BRV, C. parvum, E. coli K99+ and Salmonella. As reported by other investigators, the virus was also detected in some of the fecal samples (12.2%) from healthy calves. While this was initially suspected to be due to fecal shedding of a vaccine virus (Theil and McCloskey, 1995, Thiel et al., 2011), BRV-A was not detected concurrently in those BCoV-positive samples, noting that commercial vaccines for calf scouring contain both BCoV and BRV-A in a live form. Co-infection or other factors may contribute to diarrhea in association with BCoV infection as levels of BCoV in feces from diarrheic calves were significantly higher than those in feces from healthy calves. Such a quantitative difference may be a useful criterion in determining the clinical significance of BCoV detection during diagnostic investigation.
Bovine enterovirus is commonly present in gastrointestinal track in cattle and highly prevalent in high-density cattle farms (Jimenez-Clavero et al., 2005, Ley et al., 2002). The virus is also known to be stable in the environment (Jimenez-Clavero et al., 2005). Most of BEV infections are subclinical, although gastroenteritis and reproductive disease associated with BEV infection have been reported (Blas-Machado et al., 2007, Blas-Machado et al., 2011). In our study, detection of BEV did not demonstrate a statistically significant association with calf diarrhea (OR = 0.113). In fact BEV was more commonly detected in feces from healthy calves, which supports asymptomatic infection of BEV in bovine gastrointestinal track (Jimenez-Clavero et al., 2005, Ley et al., 2002).
Accurate and rapid diagnosis of pathogens of bovine enteric disease is important for quick and appropriate interventions in the field to mitigate losses (McGuirk, 2008). The detection frequency of BRV-A, BCoV and C. parvum was increased by 1.5–4.5 times after implementing the BEP, PCR-based testing in ISU-VDL. Such an increase in incidence and/or prevalence is more likely attributed to higher sensitivity and specificity of BEP than conventional tests and accurately reflects actual epidemiology of these pathogens in the field, while there was neither apparent increase of sample submissions nor change in personnel or fees associated with calf diarrhea testing at the lab were made during the study period. Interestingly, the detection frequency of C. parvum in diarrhea cases increased by 4.5 times (i.e., from 8.6% to 38.3%) after implementation of BEP, raising awareness of the epidemiological and clinical significance of C. parvum in the field. This observation is an example of the bias of test sensitivity on interpretation of infection prevalence or disease prevalence, which, in turn, can misguide veterinary practitioners or producers on disease intervention or animal management on farm. Continuous and frequent evaluation of the performance of diagnostic tests in context of impact on the animal (infection versus disease) is highly desired to minimize misclassification of data (David et al., 2005).
Our study was not an age-matched case–control study, which could introduce a bias into frequency of certain pathogen detection. Besides, use of diagnostic submissions for the study could also bias the study outcome as sick animals may have handled differently before samples were taken from the animals. Nonetheless, age distribution between cases and controls was similar. Many other factors, such as sex, breed, sampling season and farm management, were similar between case and control groups. Use of a PCR-based panel for all 11 targeted agents may have reduced potential bias due to section of different tests for different pathogens. Therefore, observed detection frequency and association strength between certain pathogens and calf diarrhea would be decent assessment.
In conclusion, co-infection of multiple pathogens is common in calf diarrhea cases although clinical significance/role of each pathogen in diarrhea may vary and remains to be further studied for some pathogens. C. parvum and BRV-A appear to be the primary enteric pathogens significantly contributing to calf diarrhea under conditions presented in the study. Frequent detection of bovine caliciviruses, such as BNoV and Nebovirus, in feces from diarrheic calves raises the need to pay attention to these viruses with respect to the management of enteric disease on farm. Use of a PCR-based testing panel (e.g., multiplex real-time PCRs) covering a wide range of known and potential pathogens with defined sensitivity and specificity is strongly recommended for monitoring/surveillance of populations for diseases, particularly when dealing with multifactorial diseases such as calf diarrhea or bovine respiratory disease complex. Such a screening test for multiple pathogens would be useful for not only studying the host-agent ecology, disease expression and dynamics in a population but also developing an effective intervention strategy for disease control or prevention. In addition, further characterization of pathogens with high rate of mutation on the on-going basis may be necessary to keep a vaccine-based intervention strategy effective.
Acknowledgments
The authors would like to thank Jessica Boor and Jacqueline Thomas for their excellent assistance in sample collection from VDL submissions. The authors are grateful to Drs. Annette O’Connor and Grant Dewell for their critical review of the manuscript. The study was supported in part by funding from Calf Scouring Fund, USDA Cooperative State Research, Education, and Extension Service (Award No. 2007-35102-18115), the National Research Foundation on behalf of the Korean Ministry of Education, Science and Technology (Award No. 2010-0024447) and VDL R&D Fund.
References
- Acha S.J., Kuhn I., Jonsson P., Mbazima G., Katouli M., Mollby R. Studies on calf diarrhoea in Mozambique: prevalence of bacterial pathogens. Acta Vet. Scand. 2004;45:27–36. doi: 10.1186/1751-0147-45-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini S., Brodard I., Jaussi A., Wollschlaeger N., Frey J., Miserez R., Abril C. Real-time multiplex PCR assays for reliable detection of Clostridium perfringens toxin genes in animal isolates. Vet. Microbiol. 2008;127:179–185. doi: 10.1016/j.vetmic.2007.07.024. [DOI] [PubMed] [Google Scholar]
- Bartels C.J., Holzhauer M., Jorritsma R., Swart W.A., Lam T.J. Prevalence, prediction and risk factors of enteropathogens in normal and non-normal faeces of young Dutch dairy calves. Prev. Vet. Med. 2010;93:162–169. doi: 10.1016/j.prevetmed.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman C., Svensson C., Christensson B., de Verdier K. Cryptosporidium parvum and Giardia intestinalis in calf diarrhoea in Sweden. Acta Vet. Scand. 2003;44:145–152. doi: 10.1186/1751-0147-44-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blas-Machado U., Saliki J.T., Boileau M.J., Goens S.D., Caseltine S.L., Duffy J.C., Welsh R.D. Fatal ulcerative and hemorrhagic typhlocolitis in a pregnant heifer associated with natural bovine enterovirus type-1 infection. Vet. Pathol. 2007;44:110–115. doi: 10.1354/vp.44-1-110. [DOI] [PubMed] [Google Scholar]
- Blas-Machado U., Saliki J.T., Sanchez S., Brown C.C., Zhang J., Keys D., Woolums A., Harvey S.B. Pathogenesis of a bovine enterovirus-1 isolate in experimentally infected calves. Vet. Pathol. 2011;48:1075–1084. doi: 10.1177/0300985810395728. [DOI] [PubMed] [Google Scholar]
- Boileau M.J., Kapil S. Bovine coronavirus associated syndromes. Vet. Clin. North Am. Food Anim. Pract. 2010;26:123–146. doi: 10.1016/j.cvfa.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.I., Han J.I., Sun D., Park S.I., Cooper V., Schwartz K., Yoon K. Detection and molecular characterization of bovine norovirus among bovine diarrhea cases in US Midwest. Proceedings Conference of Research Workers in Animal Disease; Chicago, IL; 2011. p. 054. [Google Scholar]
- Cho Y.I., Kim W.I., Liu S., Kinyon J.M., Yoon K.J. Development of a panel of multiplex real-time polymerase chain reaction assays for simultaneous detection of major agents causing calf diarrhea in feces. J. Vet. Diagn. Invest. 2010;22:509–517. doi: 10.1177/104063871002200403. [DOI] [PubMed] [Google Scholar]
- David K.G., Sullivan K.M., Barker N.D. Springer Science + Business Media; LLC, 223 Spring Street, New York, NY 10013, USA: 2005. ActiveEpi companion Textbook; pp. 225–230. [Google Scholar]
- de la Fuente R., Garcia A., Ruiz-Santa-Quiteria J.A., Luzon M., Cid D., Garcia S., Orden J.A., Gomez-Bautista M. Proportional morbidity rates of enteropathogens among diarrheic dairy calves in central Spain. Prev. Vet. Med. 1998;36:145–152. doi: 10.1016/S0167-5877(98)00077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente R., Luzon M., Ruiz-Santa-Quiteria J.A., Garcia A., Cid D., Orden J.A., Garcia S., Sanz R., Gomez-Bautista M. Cryptosporidium and concurrent infections with other major enterophatogens in 1 to 30-day-old diarrheic dairy calves in central Spain. Vet. Parasitol. 1999;80:179–185. doi: 10.1016/S0304-4017(98)00218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bartolo I., Ponterio E., Monini M., Ruggeri F.M. A pilot survey of bovine norovirus in northern Italy. Vet. Rec. 2011;169:73. doi: 10.1136/vr.d2625. [DOI] [PubMed] [Google Scholar]
- Duckmanton L., Carman S., Nagy E., Petric M. Detection of bovine torovirus in fecal specimens of calves with diarrhea from Ontario farms. J. Clin. Microbiol. 1998;36:1266–1270. doi: 10.1128/jcm.36.5.1266-1270.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarezi M.C., Cardoso T.C., Dutra I.S. Genotyping of Clostridium perfringens isolated from calves with neonatal diarrhea. Anaerobe. 2008;14:328–331. doi: 10.1016/j.anaerobe.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Garcia A., Ruiz-Santa-Quiteria J.A., Orden J.A., Cid D., Sanz R., Gomez-Bautista M., de la Fuente R. Rotavirus and concurrent infections with other enteropathogens in neonatal diarrheic dairy calves in Spain. Comp. Immunol. Microbiol. Infect. Dis. 2000;23:175–183. doi: 10.1016/S0147-9571(99)00071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurjar A.A., Hegde N.V., Love B.C., Jayarao B.M. Real-time multiplex PCR assay for rapid detection and toxintyping of Clostridium perfringens toxin producing strains in feces of dairy cattle. Mol. Cell. Probes. 2008;22:90–95. doi: 10.1016/j.mcp.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Guy R.A., Payment P., Krull U.J., Horgen P.A. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 2003;69:5178–5185. doi: 10.1128/AEM.69.9.5178-5185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschek B., Klein D., Benetka V., Herrera C., Sommerfeld-Stur I., Vilcek S., Moestl K., Baumgartner W. Detection of bovine torovirus in neonatal calf diarrhoea in Lower Austria and Styria (Austria) J. Vet. Med. B Infect. Dis. Vet. Public Health. 2006;53:160–165. doi: 10.1111/j.1439-0450.2006.00936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoet A.E., Nielsen P.R., Hasoksuz M., Thomas C., Wittum T.E., Saif L.J. Detection of bovine torovirus and other enteric pathogens in feces from diarrhea cases in cattle. J. Vet. Diagn. Invest. 2003;15:205–212. doi: 10.1177/104063870301500301. [DOI] [PubMed] [Google Scholar]
- Hoet A.E., Smiley J., Thomas C., Nielsen P.R., Wittum T.E., Saif L.J. Association of enteric shedding of bovine torovirus (Breda virus) and other enteropathogens with diarrhea in veal calves. Am. J. Vet. Res. 2003;64:485–490. doi: 10.2460/ajvr.2003.64.485. [DOI] [PubMed] [Google Scholar]
- Hosmillo M.D., Jeong Y.J., Kim H.J., Collantes T.M., Alfajaro M.M., Park J.G., Kim H.H., Kwon H.J., Park S.J., Kang M.I., Park S.I., Cho K.O. Development of universal SYBR Green real-time RT-PCR for the rapid detection and quantitation of bovine and porcine toroviruses. J. Virol. Methods. 2010;168:212–217. doi: 10.1016/j.jviromet.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo M.M., Kirkland P.D., Mohler V.L., Perkins N.R., Gunn A.A., House J.K. Prevalence of major enteric pathogens in Australian dairy calves with diarrhoea. Aust. Vet. J. 2011;89:167–173. doi: 10.1111/j.1751-0813.2011.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Clavero M.A., Escribano-Romero E., Mansilla C., Gomez N., Cordoba L., Roblas N., Ponz F., Ley V., Saiz J.C. Survey of bovine enterovirus in biological and environmental samples by a highly sensitive real-time reverse transcription-PCR. Appl. Environ. Microbiol. 2005;71:3536–3543. doi: 10.1128/AEM.71.7.3536-3543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachim A., Eckert E., Petry F., Bialek R., Daugschies A. Comparison of viability assays for Cryptosporidium parvum oocysts after disinfection. Vet. Parasitol. 2003;111:47–57. doi: 10.1016/s0304-4017(02)00329-1. [DOI] [PubMed] [Google Scholar]
- Jor E., Myrmel M., Jonassen C.M. SYBR Green based real-time RT-PCR assay for detection and genotype prediction of bovine noroviruses and assessment of clinical significance in Norway. J. Virol. Methods. 2010;169:1–7. doi: 10.1016/j.jviromet.2010.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon J., Guenau E., Asdrubal P., Pothier P., Ambert-Balay K. Possible novel nebovirus genotype in cattle, France. Emerg. Infect. Dis. 2011;17:1120–1123. doi: 10.3201/eid1706.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelling C.L., Steffen D.J., Cooper V.L., Higuchi D.S., Eskridge K.M. Effect of infection with bovine viral diarrhea virus alone, bovine rotavirus alone, or concurrent infection with both on enteric disease in gnotobiotic neonatal calves. Am. J. Vet. Res. 2002;63:1179–1186. doi: 10.2460/ajvr.2002.63.1179. [DOI] [PubMed] [Google Scholar]
- Kirisawa R., Takeyama A., Koiwa M., Iwai H. Detection of bovine torovirus in fecal specimens of calves with diarrhea in Japan. J. Vet. Med. Sci. 2007;69:471–476. doi: 10.1292/jvms.69.471. [DOI] [PubMed] [Google Scholar]
- Larson R.L., Tyler J.W. Reducing calf losses in beef herds. Vet. Clin. North Am. Food Anim. Pract. 2005;21:569–584. doi: 10.1016/j.cvfa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Ley V., Higgins J., Fayer R. Bovine enteroviruses as indicators of fecal contamination. Appl. Environ. Microbiol. 2002;68:3455–3461. doi: 10.1128/AEM.68.7.3455-3461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlum C.E., Haugerud S., Shivers J.L., Rossow K.D., Goyal S.M., Collins J.E., Faaberg K.S. Detection of bovine viral diarrhea virus by TaqMan reverse transcription polymerase chain reaction. J. Vet. Diagn. Invest. 2002;14:120–125. doi: 10.1177/104063870201400205. [DOI] [PubMed] [Google Scholar]
- Martella V., Banyai K., Matthijnssens J., Buonavoglia C., Ciarlet M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010;140:246–255. doi: 10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- McDonough S.P., Stull C.L., Osburn B.I. Enteric pathogens in intensively reared veal calves. Am. J. Vet. Res. 1994;55:1516–1520. [PubMed] [Google Scholar]
- McGuirk S.M. Disease management of dairy calves and heifers. Vet. Clin. North Am. Food Anim. Pract. 2008;24:139–153. doi: 10.1016/j.cvfa.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijovski J.Z., Poljsak-Prijatelj M., Steyer A., Barlic-Maganja D., Koren S. Detection and molecular characterisation of noroviruses and sapoviruses in asymptomatic swine and cattle in Slovenian farms. Infect. Genet. Evol. 2010;10:413–420. doi: 10.1016/j.meegid.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Milnes A.S., Binns S.H., Oliver S.L., Bridger J.C. Retrospective study of noroviruses in samples of diarrhoea from cattle, using the Veterinary Laboratories Agency's Farmfile database. Vet. Rec. 2007;160:326–330. doi: 10.1136/vr.160.10.326. [DOI] [PubMed] [Google Scholar]
- Moore M.M., Feist M.D. Real-time PCR method for Salmonella spp. targeting the stn gene. J. Appl. Microbiol. 2007;102:516–530. doi: 10.1111/j.1365-2672.2006.03079.x. [DOI] [PubMed] [Google Scholar]
- Nunez A., McNeilly F., Perea A., Sanchez-Cordon P.J., Huerta B., Allan G., Carrasco L. Coinfection by Cryptosporidium parvum and porcine circovirus type 2 in weaned pigs. J. Vet. Med. B Infect. Dis. Vet. Public Health. 2003;50:255–258. doi: 10.1046/j.1439-0450.2003.00664.x. [DOI] [PubMed] [Google Scholar]
- Olesen B., Neimann J., Bottiger B., Ethelberg S., Schiellerup P., Jensen C., Helms M., Scheutz F., Olsen K.E., Krogfelt K., Petersen E., Molbak K., Gerner-Smidt P. Etiology of diarrhea in young children in Denmark: a case–control study. J. Clin. Microbiol. 2005;43:3636–3641. doi: 10.1128/JCM.43.8.3636-3641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto P.H., Clarke I.N., Lambden P.R., Salim O., Reetz J., Liebler-Tenorio E.M. Infection of calves with bovine norovirus GIII. 1 Strain Jena virus: an experimental model to study the pathogenesis of norovirus infection. J. Virol. 2011;85:12013–12021. doi: 10.1128/JVI.05342-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.I., Jeong C., Kim H.H., Park S.H., Park S.J., Hyun B.H., Yang D.K., Kim S.K., Kang M.I., Cho K.O. Molecular epidemiology of bovine noroviruses in South Korea. Vet. Microbiol. 2007;124:125–133. doi: 10.1016/j.vetmic.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.I., Jeong C., Park S.J., Kim H.H., Jeong Y.J., Hyun B.H., Chun Y.H., Kang M.I., Cho K.O. Molecular detection and characterization of unclassified bovine enteric caliciviruses in South Korea. Vet. Microbiol. 2008;130:371–379. doi: 10.1016/j.vetmic.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Oh E.H., Park S.I., Kim H.H., Jeong Y.J., Lim G.K., Hyun B.H., Cho K.O. Molecular epidemiology of bovine toroviruses circulating in South Korea. Vet. Microbiol. 2008;126:364–371. doi: 10.1016/j.vetmic.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putignani L., Menichella D. Global distribution, public health and clinical impact of the protozoan pathogen cryptosporidium. Interdiscip. Perspect. Infect. Dis. 2010:1–39. doi: 10.1155/2010/753512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., Pankovics P., Egyed L. Detection of genotype 1 and 2 bovine noroviruses in Hungary. Vet. Rec. 2009;165:537–538. doi: 10.1136/vr.165.18.537. [DOI] [PubMed] [Google Scholar]
- Reynolds D.J., Morgan J.H., Chanter N., Jones P.W., Bridger J.C., Debney T.G., Bunch K.J. Microbiology of calf diarrhoea in southern Britain. Vet. Rec. 1986;119:34–39. doi: 10.1136/vr.119.2.34. [DOI] [PubMed] [Google Scholar]
- Saif L.J., Smith K.L. Enteric viral infections of calves and passive immunity. J. Dairy Sci. 1985;68:206–228. doi: 10.3168/jds.S0022-0302(85)80813-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks O.C., Atikovic E., Blackwood A.D., Lu J., Noble R.T., Domingo J.S., Seifring S., Sivaganesan M., Haugland R.A. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution. Appl. Environ. Microbiol. 2008;74:745–752. doi: 10.1128/AEM.01843-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snodgrass D.R., Terzolo H.R., Sherwood D., Campbell I., Menzies J.D., Synge B.A. Aetiology of diarrhoea in young calves. Vet. Rec. 1986;119:31–34. doi: 10.1136/vr.119.2.31. [DOI] [PubMed] [Google Scholar]
- Sting R. Detection of beta2 and major toxin genes by PCR in Clostridium perfringens field isolates of domestic animals suffering from enteritis or enterotoxaemia. Berl. Munch. Tierarztl. Wochenschr. 2009;122:341–347. [PubMed] [Google Scholar]
- Theil K.W., McCloskey C.M. Rotavirus shedding in feces of gnotobiotic calves orally inoculated with a commercial rotavirus–coronavirus vaccine. J. Vet. Diagn. Invest. 1995;7:427–432. doi: 10.1177/104063879500700401. [DOI] [PubMed] [Google Scholar]
- Thiel B., Sudbrink D., Haase C., Larson L.J., Schultz S., Kurth K., Schultz R. How long are viruses detected by PCR in blood and nasal swabs of calves vaccinated with infectious vaccines?. Proceedings Conference of Research Workers in Animal Disease; Chicago, IL; 2011. p. 077. [Google Scholar]
- Trotz-Williams L.A., Wayne Martin S., Leslie K.E., Duffield T., Nydam D.V., Peregrine A.S. Calf-level risk factors for neonatal diarrhea and shedding of Cryptosporidium parvum in Ontario dairy calves. Prev. Vet. Med. 2007;82:12–28. doi: 10.1016/j.prevetmed.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhde F.L., Kaufmann T., Sager H., Albini S., Zanoni R., Schelling E., Meylan M. Prevalence of four enteropathogens in the faeces of young diarrhoeic dairy calves in Switzerland. Vet. Rec. 2008;163:362–366. doi: 10.1136/vr.163.12.362. [DOI] [PubMed] [Google Scholar]
- United States Agriculture D.o. 2007. Dairy 2007. Part II: Changes in the U.S. Dairy Cattle Industry, 1991–2007; pp. 57–61. [Google Scholar]
- van der Poel W.H., van der Heide R., Verschoor F., Gelderblom H., Vinje J., Koopmans M.P. Epidemiology of Norwalk-like virus infections in cattle in The Netherlands. Vet. Microbiol. 2003;92:297–309. doi: 10.1016/s0378-1135(02)00421-2. [DOI] [PubMed] [Google Scholar]
- West D.M., Sprigings K.A., Cassar C., Wakeley P.R., Sawyer J., Davies R.H. Rapid detection of Escherichia coli virulence factor genes using multiplex real-time TaqMan PCR assays. Vet. Microbiol. 2007;122:323–331. doi: 10.1016/j.vetmic.2007.01.026. [DOI] [PubMed] [Google Scholar]
- Wilhelmi I., Roman E., Sanchez-Fauquier A. Viruses causing gastroenteritis. Clin. Microbiol. Infect. 2003;9:247–262. doi: 10.1046/j.1469-0691.2003.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise A.G., Monroe S.S., Hanson L.E., Grooms D.L., Sockett D., Maes R.K. Molecular characterization of noroviruses detected in diarrheic stools of Michigan and Wisconsin dairy calves: circulation of two distinct subgroups. Virus Res. 2004;100:165–177. doi: 10.1016/j.virusres.2003.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Williamson W.M., Hewitt J., Rivera-Aban M., Lin S., Ball A., Scholes P., Greening G.E. Sensitive multiplex real-time reverse transcription-PCR assay for the detection of human and animal noroviruses in clinical and environmental samples. Appl. Environ. Microbiol. 2007;73:5464–5470. doi: 10.1128/AEM.00572-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz H., Turan N., Altan E., Bostan K., Yilmaz A., Helps C.R., Cho K.O. First report on the phylogeny of bovine norovirus in Turkey. Arch. Virol. 2011;156:143–147. doi: 10.1007/s00705-010-0833-7. [DOI] [PubMed] [Google Scholar]


