Highlights
-
•
A novel IBV variant CK/CH/2010/JT-1 was identified.
-
•
A novel genotypic cluster IBV has emerged in China.
-
•
Isolate CK/CH/2010/JT-1 originated from homologous RNA recombination.
-
•
Isolate CK/CH/2010/JT-1 is highly virulent.
Keywords: Infectious bronchitis virus, Genome, Recombinant, Virulence
Abstract
The emergence of new infectious bronchitis virus (IBV) variants is often disastrous in the poultry industry. In this study, an IBV, CK/CH/2010/JT-1, was isolated from an H120- and 4/91-IBV-vaccinated flock in China. Antisera against vaccine strains H120 and 4/91 could not provide effective protection against CK/CH/2010/JT-1 in virus neutralization assays. CK/CH/2010/JT-1 could cause 43.75% mortality with respiratory and severe renal lesions in inoculated chickens. Phylogenetic analysis of the S1 gene showed that CK/CH/2010/JT-1 and 31 other isolates could be grouped as a new genotypic cluster. Recombination analysis revealed that three recombination events could be found in the genome of CK/CH/2010/JT-1 at positions 24709-365, 17160-19811 and 21136-21770. Whole-genome sequence analysis showed that CK/CH/2010/JT-1 originated from multiple template switches among QX-like, CK/CH/LSC/99I-, tl/CH/LDT3/03- and 4/91-type IBVs. All of these data demonstrated that CK/CH/2010/JT-1 is a new recombinant genotype IBV with high virulence. Our findings suggest that the surveillance of new genotype strains of IBV is very important for developing more effective anti-IBV strategies.
1. Introduction
Infectious bronchitis (IB) is a highly contagious disease caused by infectious bronchitis virus (IBV), which belongs to the family Coronaviridae and the genus Coronavirus. IBV infection can cause clinical pathological signs mainly in the respiratory tract, kidney and reproductive tract of chickens, resulting in huge economic losses in affected flocks (Cook et al., 2012). Little or no cross-protection occurs between different serotypes of IBV, and the increasing number of new serotypes of IBV is a major challenge for the prevention and control of IB (Cavanagh, 2007).
Although the IBV vaccine plays a vital role in controlling IB, it is still an epidemic across the world. IBV variants have been continuously emerging in China (Liu et al., 2014, Xue et al., 2012, Zhao et al., 2014). The nature of the large, single-RNA genome of IBV makes the virus variable through recombination or mutation. Recombination can cause the emergence and evolution of different IBV genotypes and different species of coronaviruses (Jackwood et al., 2010). More and more recombination events have been reported in IBV, and they are distributed throughout the entire genome (Brooks et al., 2004, Kuo et al., 2013, Thor et al., 2011). Recently, a new recombinant cluster of nephropathogenic IBVs able to cause death, respiratory signs and nephritis in infected chickens emerged in Korea (Lim et al., 2011, Lim et al., 2015). Therefore, studies on the function of recombination in the antigenicity and pathogenicity of IBV are very important, potentially allowing IBV evolution to be predicted and better strategies for IBV control to be developed.
In this study, an IBV, CK/CH/2010/JT-1, was isolated from a vaccinated flock in China, and its complete genomic sequence was determined. Genomic sequence analysis and pathogenicity studies revealed that CK/CH/2010/JT-1 is a virulent recombinant strain that belongs to a novel IB genotype.
2. Materials and methods
2.1. Virus isolation
In this study, we identified and isolated an IBV (CK/CH/2014/JT-1) from a broiler flock vaccinated with IBV H120 and 4/91, in which the birds showed severe respiratory symptoms and some of them died. Gross examination showed lesions in the different organs, especially in trachea and kidney. The morbidity and mortality in this flock were as high as approximate 50% and 10% respectively. Trachea, lung and kidney specimens of five 25-day-old chickens were collected from the broiler flock. Sample suspensions (10% w/v) of tissues prepared in sterile phosphate-buffered saline (PBS) were used for virus isolation (Liu et al., 2014). Ten passes were performed, and characteristic embryo changes, such as dwarfing, stunting, curling or death of the embryos, were observed between 2 and 7 days post-inoculation (dpi).
2.2. Cross-virus neutralization tests
Cross virus neutralization tests as described in the OIE manual (http://www.oie.int/international-standard-setting/terrestrial-manual/access-online/) were performed using anti-sera against Mass serotype vaccine strain H120 and 4/91 serotype vaccine strain 4/91 to determine their antigenic relationship. In brief, the viral titres of the Mass-type M41 and 4/91-type CK/CH/2014/TM98 strains were determined by inoculation of 10-fold dilutions into groups of five 10-day-old embryonated specific-pathogen-free (SPF) chicken eggs. The β method of cross-virus neutralization testing was performed using constant (102 50% embryo infectious doses [EID50s]) viral titres and 2-fold diluted serum against H120 and 4/91 in SPF chickens embryos. The EID50 and the end-point of each serum sample were calculated using the methods of Reed and Muench.
2.3. RT-PCR amplification and sequencing
Viral RNA was extracted from 200 μl of allantoic fluid from the inoculated eggs using TRIzol reagent (Introvigen, Grand Island, USA). First-strand cDNA was synthesized using random hexamers (Introvigen, Grand Island, USA). The complete genomic sequence of strain CK/CH/2010/JT-1 was amplified with 26 pairs of primers, including the 5′- and 3′-terminal segments. Primer sequences and locations were designed corresponding to the genomic sequence of strains ck/CH/IBTZ/2012 and ck/CH/LJL/110302 (GenBank accession numbers KF663559 and KC136209, respectively). The polymerase chain reaction (PCR) conditions for amplification included 95 °C for 5 min; 30 cycles of 94 °C for 1 min, 55 °C for 1 min for P1-P16 or 52 °C for 1 min for P17-P23, 72 °C for 2 min; followed by 72 °C for 10 min. The 5′- and 3′- ends of the viral genome were amplified using 5′- and 3′-random amplification of cDNA ends (RACE) Kits (TaKaRa, Dalian, China) according to the operating instructions. The amplified products were analysed by 1.0% agarose gel electrophoresis, then purified with a Gel Extraction Kit (Qiagen, Hilden, Germany) and ligated into the pGEM-T Easy vector (Promega, Madison, USA). Each fragment of the viral genome was sequenced at least three times, and the consensus sequence was determined.
2.4. Sequence and phylogenetic analysis
The complete genome and gene sequences of isolate CK/CH/2010/JT-1 and of IBV reference strains obtained from GenBank were aligned and analysed using the ClustalW multiple alignment method in the MegAlign program of DNASTAR software (version 7.1; DNAstar, Madison, USA). A phylogenetic tree was constructed from the nucleotide sequences using MEGA4.0 software (www.megasoftware.net) with UPGMA or neighbour-joining statistical methods. The phylogeny test method chosen was bootstrap with 1000 bootstrap replicates. Evolutionary distances were computed by the pairwise distance method using the maximum composite likelihood model. BLASTN analysis was performed by services available on http://blast.ncbi.nlm.nih.gov/Blast.cgi
The aligned nucleotide sequences of the complete genomes were analysed with the Recombination Detection Program (RDP4, Version 4.36) to detect potential within-gene recombination events. Seven detection methods in RDP v.4.36, including RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan and 3Seq, were used to confirm the recombination events. Only transferred gene fragments where at least 5 detection methods p-value ≤ 1 × 10−14 were accepted. Recombination events and recombination breakpoints were further confirmed by simplot and bootscan analysis using the SimPlot program (version 3.5.1.). Nucleotide identity was performed by the Kimura (2-parameter) method with a transition–transversion ratio of 2, and the window width and step size were 200 and 20 bp, respectively.
2.5. GenBank accession numbers
The genomic sequence of IBV strain CK/CH/2010/JT-1 was submitted to the GenBank database and assigned the accession number KU361187. The GenBank accession numbers of strains M41 and CK/CH/2014/TM98 are DQ834384 and KU361199, respectively.
2.6. Pathogenicity studies
Twenty-one 3-day-old SPF white leghorn chickens were randomly divided into two groups. Sixteen birds in group 1 were inoculated intranasally with 105 EID50s of IBV strain CK/CH/2010/JT-1 in 0.2 ml PBS. Five birds in group 2 were inoculated with 0.2 ml PBS as non-infected controls. The birds were housed in isolators. All birds were observed daily for signs of disease (e.g., dishevelled feathers, depression, respiratory signs or diarrhoea) and death for 28 dpi. Gross pathologic changes in tissues such as trachea, lung and kidney were observed. Serum samples collected from chickens that survived were checked for IBV antibodies using a commercial enzyme-linked immunosorbent assay (ELISA) kit (IDEXX Laboratories, Westbrook, ME, USA). The endpoint titres were calculated according to the manufacturer’s instructions, and titres of more than 396 were considered positive for IBV antibody. All experiments complied with the institutional animal care guidelines and were approved by the University of Yangzhou Animal Care Committee.
A histopathological assay and haematoxylin–eosin (HE) staining of the tissues were performed as previously described (Zhang et al., 2013). Briefly, the tracheae, lungs, and kidneys of the sick chickens were fixed in 10% neutral formalin, dehydrated in alcohol, and embedded in paraffin. They were then stained with HE and screened via light microscopy.
3. Results
3.1. Sera against mass- and 4/91-type IBV could not completely neutralize isolate CK/CH/2010/JT-1
The last dilution of each serum against H120 and 4/91, which protected 50% of the embryos against 102 EID50s of strains M41 and CK/CH/2014/TM98, was tested at 1:9.85 and 1:25.46, respectively. However, serum against H120 could not neutralize 102 EID50s of CK/CH/2010/JT-1, while the end-point of serum against 4/91 that could neutralize 102 EID50s of CK/CH/2010/JT-1 was only 1:2.14. These results indicate that the CK/CH/2010/JT-1 isolate was antigenically distinct from the IBV Mass and 4/91 serotypes.
3.2. CK/CH/2010/JT-1 is a novel genotype of IBV
By the third passage, typical symptoms, such as curling, dwarfing and stunting, were observed in the eggs. Inoculated egg death appeared at the 4th passage, and the inoculated eggs had all died by the 6th-10th passages. The virus was identified as IBV by RT-PCR and named CK/CH/2010/JT-1. The complete genome sequence of CK/CH/2010/JT-1 was obtained by assembling 25 overlapping sequences, with a 27670-nucleotide genome. The complete genome encodes several different genes, and components of the related genes are ordered 5′ UTR -1a-1b-S-3a,b,c(E)-M-4b-5a,b-N-UTR3′. Sequence analysis showed that CK/CH/2010/JT-1 shares the highest complete genome sequence identity with GX-YL9 (96.3%) and GX-YL5 (96.1%) but only 44.3%-89.2% S1 nucleic acid identity with 59 reference strains (Fig. 1 ). BLASTN analysis showed that the S1 gene of CK/CH/2010/JT-1 shares 90%-99% similarity with 78 isolates from China in recent years. Phylogenetic analysis of the S1 gene of IBVs revealed that CK/CH/2010/JT-1 and 31 other isolates described in BLASTN analysis are grouped as a new cluster (Fig. 1), which is separated from the previously identified serotypes and genotypes.
Fig. 1.
Phylogenetic tree of the S1 gene from infectious bronchitis viruses (IBVs).
A phylogenetic tree was constructed with the UPGMA method using MEGA version 5.05. Bootstrap values were determined from 1000 replicates of the original data. The CK/CH/2010/JT-1 strain is marked with a black solid triangle, and strains described in BLASTN analysis are marked with black hollow triangles.
3.3. Isolate CK/CH/2010/JT-1 originated from recombination
To examine the sequence characteristics of isolate CK/CH/2010/JT-1, potential recombination events were analysed. Three recombination events were identified in the genome of CK/CH/2010/JT-1. The assessment using RDP4 software revealed four recombination events in the CK/CH/2010/JT-1 genome (Table 1 ). The parent strains CQ04-1, SAIBK and GX-YL5 were grouped into genotype CK/CH/LSC/99I, while LX4 and ck/CH/LDL/091022 were QX-like genotypes, and 4/91 and partridge/GD/S14/2003 were genotypes 4/91 and tl/CH/LDT3/03 (Fig. 1). Three recombinant events were further confirmed by Simplot (Fig. 2 A) and Bootscan analysis (Fig. 2B). Recombinant gene fragments were considered recombinants if any crossover event appeared between two putative parental strains. Obvious recombination signals were found in simplot and bootscan analyses of recombinant events 1, 3 and 4. The similarities and breakpoints also agreed with the RDP software. However, the recombinant gene fragments shared low homology with the parental sequences, similarity analysis could not further confirm recombination in event 2. Furthermore, whole non-structural protein (nsp) 15; genes 5a, 5b and N; the 3′UTR and parts of the 5′UTR; nsp 14 and nsp 16; and genes S and M were involved in the recombination events.
Table 1.
Information on recombination events detected in CK/CH/2010/JT-1.
| Recombination eventa | Breakpoints |
Genesb | Minor parentc (similarity) | Major parentd (similarity) | Detection methods (p-value) | |
|---|---|---|---|---|---|---|
| Beginning | Ending | |||||
| 1 | 24709 | 365 | M, 5a, 5b, N, 3′UTR, 5′UTR | CQ04-1 (98.2%) | ck/CH/LDL/091022 (97.5%) | RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan, 3Seq (6.466 × 10−38, 6.262 × 10−38, 2.011 × 10−36, 5.316 × 10−23, 8.846 × 10−20, 4.244 × 10−23 and 5.036 × 10−35) |
| 2 | 2036 | 5249 | 1a | SAIBK (94.0%) | LX4 (91.8%) | RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan, 3Seq (2.132 × 10−60, 1.100 × 10−79, 4.597 × 10−98, 4.725 × 10−19, 3.018 × 10−24, 1.473 × 10−41 and 3.952 × 10−60) |
| 3 | 17160 | 19811 | 1b | ck/CH/LDL/091022 (98.9%) | GX-YL5 (98.2%) | RDP, GENECONV, BootScan, Chimaera, SiScan, 3Seq (6.520 × 10−20, 3.033 × 10−18, 1.124 × 10−18, 4.699 × 10−15, 5.056 × 10−17 and 1.476 × 10−21) |
| 4 | 21136 | 21770 | S1 | 4/91 (96.4%) | partridge/GD/S14/2003 (93.1%) | RDP, GENECONV, MaxChi, Chimaera, 3Seq (1.056 × 10−38, 2.420 × 10−39, 2.134 × 10−19, 3.918 × 10−21, and 1.623 × 10−54) |
Only transferred gene fragments where at least 5 detection methods p-value ≤ 1 × 10−14 are included in the table.
“Genes” indicates the coding sequences contained within the fragment introduced by recombination.
The “minor parent” is the sequence closely related to that from which sequences in the proposed recombinant region may have been derived.
The “major parent” is the sequence closely related to that from which the greater part of the recombinant’s sequence may have been derived.
Fig. 2.
Simplot (A) and Bootscan (B) analyses of the complete genomic sequence of strain CK/CH/2010/JT-1.
Reference strains partridge/GD/S14/2003 (red), 4/91 (yellow), ck/CH/LDL/091022 (blue) and GX-YL5 (brown) were used as putative parental strains. The y-axis gives the percentage of identity or permuted trees respectively, and the x-axis gives the location of the query sequence. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
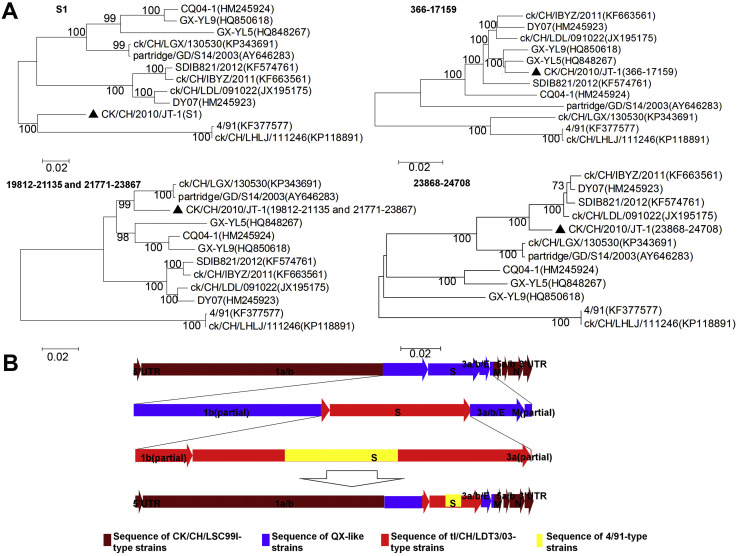
To further track the origins of the other parts of isolate CK/CH/2010/JT-1′s genome, we cut the complete genome sequence into different fragments according to recombination analysis and conducted a pairwise comparison of the fragment sequences of isolate CK/CH/2010/JT-1 with four QX-like, three CK/CH/LSC/99I-type, two tl/CH/LDT3/03-type and two 4/91-type strains. Genomic positions at 366-17159 of CK/CH/2010/JT-1 appeared very similar to CK/CH/LSC/99I-type strains GX-YL5 and GX-YL9, while genomic positions at 19812-21135 and 21171-23867 fell into the same group with tl/CH/LDT3/03-type IBVs. However, the CK/CH/2010/JT-1 isolate fell into the group of QX-like strains from location 23868-24708 in the phylogenetic tree (Fig. 3 A).
Fig. 3.
Evolution analysis of strain CK/CH/2010/JT-1.
(A) Phylogenetic trees of the S1 gene and genomic positions 366-17159, 19812-21135, 21771-23867, and 23868-24708 of ck/CH/2010/JT-1 by the neighbour-joining method using MEGA version 5.05.
(B) Recombination process of strain CK/CH/2010/JT-1′s genome sequence.
CK/CH/LSC/99I-type (brown), QX-like (blue), tl/CH/LDT3/03-type (red) and 4/91-type (yellow) strains. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
These results strongly suggested that CK/CH/2010/JT-1 originated from homologous RNA recombination events from multiple template switches among QX-like, CK/CH/LSC/99I-, tl/CH/LDT3/03- and 4/91-type IBVs (Fig. 3B), and the genome approximate positions 1-17159 and 24709-27670, 17160-19811 and 23868-24708, 19812-21135 and 21171-23867, and 21136-21770 possibly arose from CK/CH/LSC/99I-type, QX-like, tl/CH/LDT3/03-type and 4/91-type IBVs, respectively.
3.4. Isolate CK/CH/2010/JT-1 is highly virulent
Animal experiments for CK/CH/2010/JT-1 in chicken were performed to evaluate the pathogenicity of isolate CK/CH/2010/JT-1. At 3 dpi, birds in group 1 inoculated with CK/CH/2010/JT-1 appeared depressed with ruffled feathers and huddled together. At 4 dpi, the birds in group 1 developed respiratory symptoms, most of the birds breathing with head shaking and a few breathing with open mouth and lifted up neck. Notably, infected birds began to die at 4 dpi, which continued until 13 dpi, with the death rate reaching 43.75% (Fig. 4 A). No deaths or clinical signs were observed in group 2.
Fig. 4.
Chickens experimentally infected with IBV isolate CK/CH/2010/JT-1.
(A) Survival of chickens after inoculation with IBV strain CK/CH/2010/JT-1.
(B) Antibody responses to IBV in sera at 28 days post-inoculation.
Among the seven dead birds in group 1, slight haemorrhage with serous catarrhal exudates could be seen in the trachea (Fig. 5 B). Typical kidney lesions were found in all dead chickens, the affected kidneys from the dead chickens showing predominant gross lesions that were pale, mottled, and swollen, with renal tubules and ureters that were distended with excess urate (Fig. 5D). Clinical signs in the surviving birds tended to disappear gradually, and the rest of the pathological changes were present by 28 dpi. No gross lesions were observed in any bird in group 2. Antibody responses in birds that survived at 28 dpi were measured. All of the chickens showed positive reactions (Fig. 4B), and the mean titre induced by the CK/CH/2010/JT-1 strain was 702.38 at 28 dpi. These data clearly demonstrated that strain CK/CH/2010/JT-1 isolated here was a virulent IBV.
Fig. 5.
Gross lesions in trachea and kidney tissues from chickens experimentally infected with IBV strain CK/CH/2010/JT-1.
A, normal trachea control; B, tracheas of dead chickens at 8 dpi showed haemorrhage with serous catarrhal exudates; C, normal kidney control; D, kidneys of dead chickens at 8 dpi showed swollen tubules and ureters distended with excess urate.
Microscopic examination of the tracheal tissues revealed extensive degeneration and necrosis of the ciliated epithelial cells, sometimes with pseudoacinar structures resulting from dropout of dead cells (Fig. 6 B). Lung lesions included haemorrhage, congestion, and lymphocytic infiltration in the alveolar lumen (Fig. 6D). Severe renal lesions were characterized by degeneration and necrosis of renal tubular epithelial cells, lymphocytic infiltration in the interstitium, and exfoliated renal tubular epithelial cells, with erythrocytes being frequently observed (Fig. 6F).
Fig. 6.
Histopathologic analysis (haematoxylin-eosin stain) of chickens infected with IBV strain CK/CH/2010/JT-1.
A, normal trachea tissue control; B, trachea tissue infected with CK/CH/2010/JT-1 displays extensive dropout, degeneration, and necrosis of ciliated epithelial cells and lymphocytic infiltration (black arrow). Scale bar = 50 μm; C, normal lung tissue control; D, lung tissue infected with CK/CH/2010/JT-1 displays haemorrhage and congestion (black arrow). Scale bar = 200 μm; E, normal kidney control; F, kidney tissue infected with CK/CH/2010/JT-1 displays severe renal lesions, including degeneration, necrosis of renal tubular epithelial cells, and exfoliated renal tubular epithelial cells (black arrow). Scale bar = 50 μm.
4. Discussion
Recently, various live-attenuated and inactivated vaccines have been widely and extensively used to prevent IB disease in chicken farms in China. However, the IB vaccine program has not been very effective, as new IBV serotypes, genotypes and variants are sometimes isolated from vaccinated chickens (Feng et al., 2015, Han et al., 2011). This study identified an IBV, CK/CH/2010/JT-1, and 31 other isolates grouped into a novel genotypic cluster different from previously identified genotypes. Although 23 of the 31 isolates were reported in other five papers, we found that among the 23 isolates, 13 isolates were clustered into a genotype with reference strains A2 and QXIBV (Luo et al., 2012), 9 isolates were grouped with reference strains 4/91 and UK 7/93 (Feng et al., 2014, Ji et al., 2011, Li et al., 2010), and one isolate showed far evolutionary distances with reference tl/CH/LDT3/03 strains, but grouped into tl/CH/LDT3/03-type (Sun et al., 2011). In our study, the CK/CH/2010/JT-1 and 31 strains showed a low S1 nucleic acid identity with reference strains. Phylogenetic analysis of the S1 gene showed these strains could be grouped as a new genotypic cluster which was different from the previously identified genotypes IBV in China. Some vaccines, such as Mass-type H120 and 4/91-type 4/91 vaccine strains, could not provide effective protection against CK/CH/2010/JT-1 infection. In addition, the CK/CH/2010/JT-1-like isolates form a new genotypic cluster, which is similar to a new IBV cluster in Korea (Lim et al., 2011).
Coronaviruses are identified in a wide variety of animals and divided into three distinct groups based on genotypic and serological characterization (Brian and Baric, 2005, Lai and Cavanagh, 1997). Coronaviruses have a high frequency of recombination due to the unique mechanism of viral replication. IBV is a member of the genus Gamma Coronavirus. Novel IBV variants could emerge mainly via two approaches: recombination and mutation (Jackwood et al., 2012, Toro et al., 2012). In this paper, we found the CK/CH/2010/JT-1-like strains underwent three recombination events. The recombination fragments were located in approximate positions 17160-24708, 19812-23876 and 21136-21770. The genome approximate positions 1-17159 and 24709-27670, 17160-19811 and 23868-24708, 19812-21135 and 21171-23867, and 21136-21770 of the CK/CH/2010/JT-1-like IBV possibly arose from CK/CH/LSC/99I-type, QX-like, tl/CH/LDT3/03-type and 4/91-type IBVs respectively (Fig. 3B). However the chronological order of the recombination events needs further investigation. Recently, nine genotypes of IBV have been circulating in China. The predominant genotype is QX-like IBV, which has spread widely in Asia, Europe and Africa since it was first reported in 1996 in China (Cook et al., 2012, Knoetze et al., 2014, Toffan et al., 2011). The second dominant genotype detected in China is the CK/CH/LSC/99I-type (Han et al., 2011). The co-circulation of QX-like, CK/CH/LSC/99I, vaccine, tl/CH/LDT3/03 and 4/91 genotype strains in the same chicken farm provided the possibility of new recombinants like CK/CH/2010/JT-1. Previous studies found that the recombination hot spots tended to be located in the 1a (nsp2 and nsp3), 1b (nsp16), S1, envelope, membrane genes and the 3′UTR (Jackwood et al., 2012). Most of the recombination breakpoints identified in isolate CK/CH/2010/JT-1 are located in these genes. To our knowledge, this recombinant virus is different from those in other reports (Liu et al., 2014, Wu et al., 2016, Xu et al., 2016, Zhao et al., 2014). All these data indicate that the new isolate is recombined with homologous RNA recombination events from multiple template switches among QX-like, CK/CH/LSC/99I-, tl/CH/LDT3/03- and 4/91-type IBV strains.
A new genotype virus could acquire new biological characteristics, including virulence from gene recombination. The IBV spike and replicase genes are known to be determinants of pathogenicity (Armesto et al., 2009, Wickramasinghe et al., 2011). The spike gene also plays a key role in IBV tissue tropism (Wickramasinghe et al., 2011). The Mass-type M41 and 4/91-type 793/B could cause the respiratory signs without death in infected chickens (Benyeda et al., 2009). Recently, the Chinese QX-like IBV virulent strains were found to cause 10%-50% mortality in infected chickens (Feng et al., 2015, Liu et al., 2009, Shi et al., 2011, Sun et al., 2011), while the virulent CK/CH/LSC/99I-type strains YN and ck/CH/LDL/07I and the tl/CH/LDT3/03-type strain tl/CH/LDT3/03 caused 65%, 30%, and 80% mortality with severe kidney lesions in infected chickens, respectively (Feng et al., 2012, Feng et al., 2015, Liu et al., 2005). The isolate CK/CH/2010/JT-1 could cause respiratory and severe renal disease and resulted in deaths of 43.75% experimental infections of 3-day-old SPF chickens. These results indicate the CK/CH/2010/JT-1 isolate is highly virulent. The replicase gene of isolate CK/CH/2010/JT-1 was derived from QX-like, CK/CH/LSC/99I- and tl/CH/LDT3/03-type strains, and the spike gene possibly originated from 4/91- and tl/CH/LDT3/03-type strains. All of these features yield 43.75% mortality with respiratory and severe renal disease in CK/CH/2010/JT-1 infected chickens. The altered 4/91 fragment in the spike gene may not affect the tissue tropism of isolate CK/CH/2010/JT-1; the tissue tropism of the virus may have more than one pathogenicity factor, an issue to be studied in the future.
5. Conclusion
In this study, an IBV, named CK/CH/2010/JT-1, was identified from an IBV-vaccinated flock in China. CK/CH/2010/JT-1-like viruses are grouped as a new genotypic cluster different from previously identified genotypes. Genome sequence analysis suggested that isolate CK/CH/2010/JT-1 originated from QX-like, CK/CH/LSC/99I, tl/CH/LDT3/03 and 4/91 genotypes. CK/CH/2010/JT-1 of IBV is very virulent in chickens. Such novel recombinants should receive more attention during viral surveillance for better development of anti-IBV strategies.
Competing interest
The authors declared that they have no conflict of interest.
Acknowledgment
This study was made possible by funding from the Priority Academic Program Development of Jiangsu Province Higher Education Institutions (PAPD).
References
- Armesto M., Cavanagh D., Britton P. The replicase gene of avian coronavirus infectious bronchitis virus is a determinant of pathogenicity. PLoS One. 2009;4:e7384. doi: 10.1371/journal.pone.0007384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyeda Z., Mato T., Suveges T., Szabo E., Kardi V., Abonyi-Toth Z., Rusvai M., Palya V. Comparison of the pathogenicity of QX-like, M41 and 793/B infectious bronchitis strains from different pathological conditions. Avian Pathol. 2009;38:449–456. doi: 10.1080/03079450903349196. [DOI] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J.E., Rainer A.C., Parr R.L., Woolcock P., Hoerr F., Collisson E.W. Comparisons of envelope through 5B sequences of infectious bronchitis coronaviruses indicates recombination occurs in the envelope and membrane genes. Virus Res. 2004;100:191–198. doi: 10.1016/j.virusres.2003.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cook J.K., Jackwood M., Jones R.C. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41:239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- Feng J., Hu Y., Ma Z., Yu Q., Zhao J., Liu X., Zhang G. Virulent avian infectious bronchitis virus, People's Republic of China. Emerg. Infect. Dis. 2012;18:1994–2001. doi: 10.3201/eid1812.120552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng K., Xue Y., Wang F., Chen F., Shu D., Xie Q. Analysis of S1 gene of avian infectious bronchitis virus isolated in southern China during 2011–2012. Virus Genes. 2014;49:292–303. doi: 10.1007/s11262-014-1097-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng K., Xue Y., Wang J., Chen W., Chen F., Bi Y., Xie Q. Development and efficacy of a novel live-attenuated QX-like nephropathogenic infectious bronchitis virus vaccine in China. Vaccine. 2015;33:1113–1120. doi: 10.1016/j.vaccine.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Sun C., Yan B., Zhang X., Wang Y., Li C., Zhang Q., Ma Y., Shao Y., Liu Q., Kong X., Liu S. A 15-year analysis of molecular epidemiology of avian infectious bronchitis coronavirus in China. Infect. Genet. Evol. 2011;11:190–200. doi: 10.1016/j.meegid.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Boynton T.O., Hilt D.A., McKinley E.T., Kissinger J.C., Paterson A.H., Robertson J., Lemke C., McCall A.W., Williams S.M., Jackwood J.W., Byrd L.A. Emergence of a group 3 coronavirus through recombination. Virology. 2010;398:98–108. doi: 10.1016/j.virol.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Hall D., Handel A. Molecular evolution and emergence of avian gammacoronaviruses. Infect. Genet. Evol. 2012;12:1305–1311. doi: 10.1016/j.meegid.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Xie J., Chen F., Shu D., Zuo K., Xue C., Qin J., Li H., Bi Y., Ma J., Xie Q. Phylogenetic distribution and predominant genotype of the avian infectious bronchitis virus in China during 2008–2009. Virol. J. 2011;8:184. doi: 10.1186/1743-422X-8-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoetze A.D., Moodley N., Abolnik C. Two genotypes of infectious bronchitis virus are responsible for serological variation in KwaZulu-Natal poultry flocks prior to 2012. Onderstepoort J. Vet. Res. 2014;81:1–10. doi: 10.4102/ojvr.v81i1.769. [DOI] [PubMed] [Google Scholar]
- Kuo S.M., Kao H.W., Hou M.H., Wang C.H., Lin S.H., Su H.L. Evolution of infectious bronchitis virus in Taiwan: positively selected sites in the nucleocapsid protein and their effects on RNA-binding activity. Vet. Microbiol. 2013;162:408–418. doi: 10.1016/j.vetmic.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Xue C., Chen F., Qin J., Xie Q., Bi Y., Cao Y. Isolation and genetic analysis revealed no predominant new strains of avian infectious bronchitis virus circulating in South China during 2004–2008. Vet. Microbiol. 2010;143:145–154. doi: 10.1016/j.vetmic.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim T.H., Lee H.J., Lee D.H., Lee Y.N., Park J.K., Youn H.N., Kim M.S., Lee J.B., Park S.Y., Choi I.S., Song C.S. An emerging recombinant cluster of nephropathogenic strains of avian infectious bronchitis virus in Korea. Infect. Genet. Evol. 2011;11:678–685. doi: 10.1016/j.meegid.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim T.H., Youn H.N., Yuk S.S., Kwon J.H., Hong W.T., Gwon G.B., Lee J.A., Lee J.B., Lee S.W., Song C.S. Successful cross-protective efficacy induced by heat-adapted live attenuated nephropathogenic infectious bronchitis virus derived from a natural recombinant strain. Vaccine. 2015;33:7370–7374. doi: 10.1016/j.vaccine.2015.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Chen J., Chen J., Kong X., Shao Y., Han Z., Feng L., Cai X., Gu S., Liu M. Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas) J. Gen. Virol. 2005;86:719–725. doi: 10.1099/vir.0.80546-0. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhang X., Wang Y., Li C., Han Z., Shao Y., Li H., Kong X. Molecular characterization and pathogenicity of infectious bronchitis coronaviruses: complicated evolution and epidemiology in china caused by cocirculation of multiple types of infectious bronchitis coronaviruses. Intervirology. 2009;52:223–234. doi: 10.1159/000227134. [DOI] [PubMed] [Google Scholar]
- Liu S., Xu Q., Han Z., Liu X., Li H., Guo H., Sun N., Shao Y., Kong X. Origin and characteristics of the recombinant novel avian infectious bronchitis coronavirus isolate ck/CH/LJL/111054. Infect. Genet. Evol. 2014;23:189–195. doi: 10.1016/j.meegid.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Qin J., Chen F., Xie Q., Bi Y., Cao Y., Xue C. Phylogenetic analysis of the S1 glycoprotein gene of infectious bronchitis viruses isolated in China during 2009–2010. Virus Genes. 2012;44:19–23. doi: 10.1007/s11262-011-0657-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X.M., Zhao Y., Gao H.B., Jing Z., Wang M., Cui H.Y., Tong G.Z., Wang Y.F. Evaluation of recombinant fowlpox virus expressing infectious bronchitis virus S1 gene and chicken interferon-gamma gene for immune protection against heterologous strains. Vaccine. 2011;29:1576–1582. doi: 10.1016/j.vaccine.2010.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Han Z., Ma H., Zhang Q., Yan B., Shao Y., Xu J., Kong X., Liu S. Phylogenetic analysis of infectious bronchitis coronaviruses newly isolated in China, and pathogenicity and evaluation of protection induced by Massachusetts serotype H120 vaccine against QX-like strains. Avian Pathol. 2011;40:43–54. doi: 10.1080/03079457.2010.538037. [DOI] [PubMed] [Google Scholar]
- Thor S.W., Hilt D.A., Kissinger J.C., Paterson A.H., Jackwood M.W. Recombination in avian gamma-coronavirus infectious bronchitis virus. Viruses. 2011;3:1777–1799. doi: 10.3390/v3091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffan A., Monne I., Terregino C., Cattoli G., Hodobo C.T., Gadaga B., Makaya P.V., Mdlongwa E., Swiswa S. QX-like infectious bronchitis virus in Africa. Vet. Rec. 2011;169:589. doi: 10.1136/vr.d7636. [DOI] [PubMed] [Google Scholar]
- Toro H., van Santen V.L., Jackwood M.W. Genetic diversity and selection regulates evolution of infectious bronchitis virus. Avian Dis. 2012;56:449–455. doi: 10.1637/10072-020212-Review.1. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe I.N., de Vries R.P., Grone A., de Haan C.A., Verheije M.H. Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J. Virol. 2011;85:8903–8912. doi: 10.1128/JVI.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yang X., Xu P., Zhou L., Zhang Z., Wang H. Genome sequence and origin analyses of the recombinant novel IBV virulent isolate SAIBK2. Virus Genes. 2016;52:509–520. doi: 10.1007/s11262-016-1337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Liu X.Y., Zhao Y., Chen Y., Zhao J., Zhang G.Z. Characterization and analysis of an infectious bronchitis virus strain isolated from southern China in 2013. Virol. J. 2016;13:40. doi: 10.1186/s12985-016-0497-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Xie Q., Yan Z., Ji J., Chen F., Qin J., Sun B., Ma J., Bi Y. Complete genome sequence of a recombinant nephropathogenic infectious bronchitis virus strain in China. J. Virol. 2012;86:13812–13813. doi: 10.1128/JVI.02575-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Li Y., Lin J., Wan S., Gao H., Zhang L., Zheng J., Zhang P. Comparison of hematoxylin-eosin staining and methyl violet staining for displaying ghost cells. Eye Sci. 2013;28:140–143. [PubMed] [Google Scholar]
- Zhao Y., Liu X.Y., Cheng J.L., Wu Y.P., Zhang G.Z. Molecular characterization of an infectious bronchitis virus strain isolated from northern China in 2012. Arch. Virol. 2014;159:3457–3461. doi: 10.1007/s00705-014-2213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]