Abstract
Twenty-seven strains of avian infectious bronchitis virus (IBV) were isolated from dead or diseased chickens at different chicken farms in South China during 2004–2008, of which the S1 gene was sequenced. Phylogenetic analysis of the S1 gene sequences of the isolated 27 strains together with 29 strains published in Genbank revealed that all IBV strains except for one isolated and one published were clustered into six distinct genotypes I-VI. 26 isolated strains belong to genotypes I, II, and III, forming a big phylogenetic branch without new predominant strains, whereas all five vaccine strains belong to genotype V that is evolutionarily distant from genotypes I, II, and III. The study of the protease cleavage motif within the S1 protein found 12 different cleavage motifs, of which 3 motifs are shared by both isolated and published strains, 2 motifs unique to isolated strains, and 7 motifs unique to published strains, further bolstering the notion of no new predominant strains. Alignment analysis of the S1 amino acid sequences indicated that the amino acid substitutions, insertions, and deletions are polymorphic and diverse, showing no sign of predominant genetic changes among the isolated strains. Taken together, there was no predominant new strain circulating in South China during 2004–2008. Nonetheless, circulating IBV strains have been continuously evolving with genetic compositions distant from vaccine strains; this explains why there have been constant but infrequent outbreaks in commercial flocks in South China during 2004–2008. Furthermore, in order to safe guard against the sudden emergence of new predominant strains, continuing surveillance of IBV strains circulating in the field is of extreme importance.
Keywords: Cleavage recognition sites, Infectious bronchitis virus, Sequence analysis, Phylogenetic analysis, Spike glycoprotein
1. Introduction
Avian infectious bronchitis virus (IBV), a member of the Coronaviridae (order Nidovirales, genus Coronavirus) family, is a highly infectious and contagious pathogen of domestic fowl, replicating primarily in the respiratory tract but also in some epithelial cells of the gut, kidney, and oviduct (Cavanagh, 2001, Cavanagh and Naqi, 2003, Cook et al., 2001). IBV genome consists of a single-stranded sense RNA encoding four structural proteins, envelope (E) glycoprotein, integral membrane (M) glycoprotein, phosphorylated nucleocapsid (N) protein, and spike (S) glycoprotein (Lai and Cavanagh, 1997, Stern and Sefton, 1982). S glycoprotein is posttranslationally cleaved into amino-terminal S1 (535 amino acids; 90-kDa) and carboxyl-terminal S2 (627 amino acids; 84-kDa) subunits by cellular proteases (Cavanagh et al., 1986b); S1 forms a receptor-binding site whereas S2 anchors S1 to the viral membrane. The protease cleavage site is usually associated with one or more pairs of basic amino acids (e.g., Arg-Arg-Ser-Arg-Arg) (Cavanagh et al., 1992).
S1 protein involves in infectivity, contains virus-neutralizing epitopes, serotype-specific sequences, and hemagglutinin activity (Cavanagh, 1983, Cavanagh and Davis, 1986, Cavanagh et al., 1986a, Karaca et al., 1992, Koch et al., 1990), and plays a major role in the tissue tropism and the induction of protective immunity (Cavanagh et al., 1988, Cavanagh et al., 1992). As the most variable protein, S1 protein sequences from different strains vary significantly, usually by between 2 and 25% at the amino acid level (Lai and Cavanagh, 1997). Various serotypes are believed to be generated by nucleotide insertions, deletions, point mutations, and/or RNA recombinations in S1 subunit (Jia et al., 1995, Kusters et al., 1987, Wang et al., 1993, Wang et al., 1994). The mutations in the N-terminal amino acids of S1 subunit directly lead to emergence of new serotypes and change of tissue tropism (Casais et al., 2003, Cavanagh et al., 1986a, Stern and Sefton, 1982). Certain studies indicated that IBV immunity was serotype-specific, small differences in S1 contributing to poor cross-protection (Cavanagh and Davis, 1986, Cavanagh et al., 1997, Gelb et al., 1997, Wang et al., 1994) and vaccine failures (Jia et al., 1996), while some groups reported that cross-protection could be provided by appropriate vaccine programmes against genetically or antigenically unrelated IBVs (Gelb et al., 2005, Cook et al., 1999). S1 gene sequence was reported as a good predictor of challenge of immunity in chickens (Ladman et al., 2006), the genetic analyses of IBV strains have been mainly focused on the S1 gene (Cavanagh et al., 1986b, Cavanagh et al., 1997, McFarlane and Verma, 2008).
Since IBV strains were first isolated and identified in China in 1982, infectious bronchitis (IB) caused by IBV has been an economically important disease to the poultry industry. Vaccines based on Massachusetts (Mass) serotype strains such as H120 and H52 have been used for many years worldwide (Hofstad, 1981). Moreover, various live or inactivated vaccines based on Mass serotype strains have been used for control of IBV infections in China. Vaccines have been generally effective, but new strains continue to emerge and cause clinical disease and production problems in vaccinated flocks (Bijlenga et al., 2004, Gelb et al., 2005, Nix et al., 2000). For example, based on neutralization test and the immune protection test, A2 strain is closely related to 4/91 serotype, spreading over Europe since its first isolation in UK in 1991 (Capua et al., 1999, Cavanagh et al., 2005, Cook et al., 1996, Gough et al., 1992, Liu et al., 2009b, Meulemans et al., 2001, Parsons et al., 1992, Xu et al., 2007, Zhao and Qin, 2002). QXIBV, a new IBV variant reported in China in 2004 (Liu and Kong, 2004), had become widespread in several countries in Europe (Terregino et al., 2008, Worthington et al., 2008). In recent years, nephropathogenic IBV strains had been circulating in China (Bing et al., 2007, Liu and Kong, 2004, Liu et al., 2006). However, the natures of circulating IBV strains in South China were not clear.
This study took on the task of deciphering the natures of the IBV strains circulating in commercial flocks in South China by carrying out a long-term surveillance program. During the program, 27 IBV strains were isolated in South China from clinical outbreaks occurred in the period of 2004–2008. The isolated IBV strains were molecularly characterized by sequencing the whole S1 genes. Sequence alignment and phylogenetic analysis of the isolated IBV strains together with other published IBV strains provided useful information as to the nature of the circulating IBV strains; and the implications of this study in the strategies for future protection of IBV infections are also discussed.
2. Materials and methods
2.1. Virus
Circulating field IBV strains were isolated from dead or diseased broilers at different chicken farms located in Guangdong, Guangxi, Sichuan, Hainan, Chongqing, and Hubei provinces during 2004–2008 (Table 1 ). Documented clinical signs included typical respiratory and nephropathogenic IB symptoms and pathological changes. Viruses were propagated by more than three passages in the allantoic cavities of 10-day-old specific pathogen free (SPF) chicken embryos. The allantoic fluid was collected after 48 h post-inoculation, frozen at −80 °C for RNA extraction, and viruses were identified as IBV by the observation of curled and dwarfed embryos and RT-PCR for the N protein gene.
Table 1.
IBV strains isolated from flocks in different provinces of South China.
| IBV isolates | Provincea | Years of isolation | Major clinical signs | Spike glycoprotein cleavage recognition motifs | Accession number |
|---|---|---|---|---|---|
| CQ04-1 | Chongqing | 2004 | Nephritis | RRFRR1 | GQ265952 |
| CQ04-2 | Chongqing | 2004 | Nephritis | RRSKR3 | GQ265953 |
| DY04 | Sichuan | 2004 | Nephritis | RRFRR1 | GQ265950 |
| NN04 | Guangxi | 2004 | Nephritis | RRSRR4 | GQ265951 |
| DY05 | Sichuan | 2005 | Nephritis | HRRRR2 | GQ265928 |
| GM05 | Guangdong | 2005 | Nephritis | RRFRR1 | GQ265931 |
| LZ05 | Guangdong | 2005 | Nephritis | RRFRR1 | GQ265943 |
| HN06-1 | Hainan | 2006 | Nephritis | RRFRR1 | GQ265938 |
| HN06-2 | Hainan | 2006 | Nephritis | RRFRR1 | GQ265939 |
| HY06 | Guangdong | 2006 | Nephritis | RRSKR3 | GQ265941 |
| SS06-1 | Guangdong | 2006 | Respiratory | RRSKR3 | GQ265935 |
| SS06-2 | Guangdong | 2006 | Respiratory | RRFRR1 | GQ265937 |
| DY07 | Sichuan | 2007 | Respiratory | HRRRR2 | GQ265927 |
| HY07 | Guangdong | 2007 | Nephritis | RRSKR3 | GQ265942 |
| LZ07 | Guangdon | 2007 | Respiratory | RRFRR1 | GQ265944 |
| NN07 | Guangxi | 2007 | Nephritis | HRRRR2 | GQ265946 |
| SS07 | Guangdong | 2007 | Respiratory | RRFRR1 | GQ265936 |
| TC07-1 | Guangdong | 2007 | Nephritis | HRRRR2 | GQ265947 |
| TC07-2 | Guangdong | 2007 | Respiratory | HRRKR5 | GQ265948 |
| ZX07 | Guangdong | 2007 | Nephritis | HRRRR2 | GQ265949 |
| FS08-1 | Guangdong | 2008 | Respiratory | RRFRR1 | GQ265929 |
| FS08-2 | Guangdong | 2008 | Respiratory | RRFRR1 | GQ265930 |
| GL08-1 | Guangxi | 2008 | Respiratory | HRRRR2 | GQ265932 |
| GL08-2 | Guangxi | 2008 | Respiratory | HRRRR2 | GQ265933 |
| HB08 | Hubei | 2008 | Respiratory | HRRRR2 | GQ265934 |
| HN08 | Hainan | 2008 | Nephritis | RRFRR1 | GQ265940 |
| XX08 | Guangdong | 2008 | Nephritis | RRFRR1 | GQ265945 |
Province where the viruses were isolated.
RRFRR: Arg-Arg-Phe-Arg-Arg (13).
HRRRR: His-Arg-Arg-Arg-Arg (8).
RRSKR: Arg-Arg-Ser-Lye-Arg (4).
RRSRR: Arg-Arg-Ser-Arg-Arg (1).
HRRKR: His-Arg-Arg-Lys-Arg (1).
2.2. Primers for S1 gene
A pair of primers for amplifying entire S1 gene were designed using Primer Premier 5.0 software based on alignment of GenBank sequences of several known IBV strains from China. The sense primer: 5′-TTG AAA ACT GAA CAA AAG ACC G-3′, and the anti-sense primer: 5′-TAC AAA ACC TGC CAT AAC TAA CAT-3′. The anticipated amplification segment is about 1760 bp encompassing the entire S1 gene including the protease cleavage motif.
2.3. RNA extraction, RT-PCR and PCR
Viral RNA was extracted using TRIzol reagent (TaKaRa, Japan) according to the manufacturer's instructions. The first-strand cDNA was synthesized using an PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa, Japan). According to the manufacturer's instructions, reverse transcription reaction mixture was set up as follows: 1 μl of Random 6 mers (50 μM), 1 μl of 10 mmol/L dNTPmix, 2 μl of RNA extract, 6 μl RNase free dH2O. The reaction mixture was incubated at 65 °C for 5 min, and then quickly frozen on ice. After cooling, reagents were added in the same tube as follows: 4 μl 5× PrimeScript Buffer, 0.5 μl of 40 U/μl RNase inhibitor, 1 μl of 200 U/μl PrimeScript Rtase, 4.5 μl RNase free dH2O. The total volume of reverse transcription reaction mixture was 20 μl; the reaction was carried out at 30 °C for 10 min, 42 °C for 30 min, 70 °C for 15 min. About 1 μl of the cDNA was used for amplification in PCR reactions. The PCR reactions (total volume of 50 μl) contained 5 μl of 10× PCR buffer, 4 μl of 2.5 mmol/l dNTP, 2 μl of 10 μmol/l of each of the two primers, and 0.25 μl of 2 U/μl Taq DNA polymerase. The PCR conditions for amplification were 94 °C for 3 min, 30 cycles of 94 °C for 30 s, 52 °C for 30 s, and 72 °C for 2 min, followed by 72 °C for 10 min. The products were analyzed on 1.0% agarose gel.
2.4. DNA cloning
PCR products of each RT-PCR were purified using Axy Prep™ DNA Gel Extraction Kit (AXYGEN). Purified PCR products ligated with a TA cloning vector, pMD18-T (TaKaRa, Japan), were transformed into DH5a E. coli competent cells. Cells carrying recombinant plasmid were screened on Luria-bertani (LB) agar plates containing Ampicillin (50 μg/ml). Positive clones were initially screened from three different PCRs through blue and white colonies; then the inserts were amplified by PCR which had the same conditions as that for the above-mentioned PCR amplification. Positive colonies were verified by EcoRI (TaKaRa, Japan) restriction enzyme digestion of the amplified inserts. The verified recombinant plasmid DNAs were used for sequencing.
2.5. Gene sequencing and analysis
The purified recombinant plasmids were sequenced by the dideoxy chain termination procedure using the automated ABI Prism 3730, Genetic Analyzer. Translation of the nucleotide sequence and sequence alignments was performed by the DNAStar software (DNASTAR Inc., USA).
Phylogenetic analyses were performed with the neighbor-joining method using MEGA version 4. The bootstrap values were determined from 1000 replicates of the original data.
2.6. IBV strains published in Genbank
Twenty-nine IBV strains published in Genbank were selected for the performance of phylogenetic and alignment analyses, including Beaudette, M41, Gray, Holte, Ark99, 4/91, 7/93, H120, H52, Ma5, W93, QXIBV, ZJ971, LX4, A2, SC021202, SAIBK, KQ6, CK/CH/LGD/04III, CK/CH/LGD/04II, CK/CH/LLN/06I, CK/CH/LGX/06I, SDZB0804, DE/072/92, Italy-02, K069-01, JP9578, 3468/07, and T07/02 (Table 2 ).
Table 2.
IBV strains published in Genbank.
| IBV strains (country of origin) | Years of isolation | Serotype/Genotype/Pathogenicity type/organs used for virus isolation | Spike glycoprotein cleavage recognition motifs | Accession number |
|---|---|---|---|---|
| Beaudette (USA) | 1937 | Mass serotype | RRFRR1 | NC_001451 |
| M41 (USA) | 1956 | Mass serotype | RRFRR1 | DQ834384 |
| Gray (USA) | 1962 | Gray serotype | RRSRR4 | L14069 |
| Hotle (USA) | 1962 | Hotle serotype | RRSRR4 | L18988 |
| Ark99 (USA) | 1973 | Arkansas serotype | HRSRR6 | M99482 |
| 4/91 (UK) | 1992 | 793/B serotype | RRSRR4 | AF093794 |
| 7/93 (UK) | 1993 | 793/B serotype | RRSRR4 | Z83979 |
| H120 | Vaccine strain | Mass serotype | RRFRR1 | M21970 |
| H52 | Vaccine strain | Mass serotype | RRFRR1 | AF352315 |
| Ma5 | Vaccine strain | Mass serotype | RRFRR1 | AY561713 |
| W93 (China) | Vaccine strain | Nephropathogenicity | RRFRR1 | AY427818 |
| QXIBV (China) | 1997 | Proventriculus | HRRRR2 | AF193423 |
| ZJ971 (China) | 1997 | Proventriculus | RRFRR1 | AF352313 |
| LX4 (China) | 1999 | Nephropathogenicity | HRRRR2 | AY189157 |
| A2 (China) | 1996 | 4/91 serotype | HRRRR2 | AY043312 |
| SC021202 (China) | 2002 | Nephropathogenicity | RRHRR7 | AY237817 |
| SAIBK (China) | Nda | Nephropathogenicity | RRFRR1 | DQ288927 |
| KQ6 (China) | 2003 | Respiratory | RRFRR1 | AY641576 |
| CK/CH/LGD/04III (China) | 2004 | Kidney | RRLRR8 | DQ167135 |
| CK/CH/LGD/04II (China) | 2004 | Kidney | RRFRR1 | DQ167134 |
| CK/CH/LLN/06I (China) | 2006 | Tracheaa | HRPRR9 | EF213566 |
| CK/CH/LGX/06I (China) | 2006 | Respiratory | RRFRR1 | EF213580 |
| SDZB0804 (China) | 2008 | Kidneya | HRRRR2 | FJ210647 |
| DE/072/92 (USA) | 1992 | DE072 serotype | RRIRR10 | AIU77298 |
| Italy-02 (Italy) | 1999 | Respiratory | HRFRR11 | AJ457137 |
| K069-01 (Korea) | 2001 | Korean Genotype III | RRFRR1 | AY257061 |
| JP9758 (Japan) | 1995 | Nephropathogenicity | RRFKR12 | AY296746 |
| 3468/07 (Taiwan) | 2007 | Nda | RRSRR4 | EU822336 |
| T07/02 (Taiwan) | 2002 | TW II | RRFRR1 | AY606322 |
Nd, not documented.
RRFRR: Arg-Arg-Phe-Arg-Arg (13).
HRRRR: His-Arg-Arg-Arg-Arg (4).
RRSRR: Arg-Arg-Ser-Arg-Arg (5).
HRSRR: His-Arg-Ser-Arg-Arg (1).
RRHRR: Arg-Arg-His-Arg-Arg (1).
RRLRR: Arg-Arg-Leu-Arg-Arg (1).
HRPRR: His-Arg-Pro-Arg-Arg (1).
RRIRR: Arg-Arg-Ile-Arg-Arg (1).
HRFRR: His-Arg-Phe-Arg-Arg(1).
RRFKR: Arg-Arg-Phe-Lys-Arg(1).
3. Results
3.1. Twenty-seven IBV strains isolated during 2004–2008 in South China
From dead or diseased chickens suspected of IBV infection, 27 circulating field IBV strains were isolated from different chicken farms located in Guangdong, Guangxi, Sichuan, Hainan, Chongqing and Hubei provinces of South China during 2004–2008; the main features of the isolated IBV strains are summarized in Table 1. Eleven strains were from chickens showing typical respiratory clinical signs such as difficulty in breathing, nasal discharge, sneezing, tracheal rales, watery eyes, lethargy, and mucosal thickening with serous or catarrhaeal exudates in the nasal passages, sinuses, and trachea. Sixteen strains were from chickens showing typical nephritis symptoms such as enlarged and pale kidneys, frequently with urate deposits in the tubules, severe dehydration and weight loss. All 27 isolated strains were verified by the observation of curled and dwarfed embryos and RT-PCR for the N gene of IBV.
3.2. Wide range of homologies among S1 nucleotide and deduced amino acid sequences
S1 gene sequences of the 27 isolated IBV strains were delineated and submitted to the GenBank database (Table 1). Sequencing results showed that S1 genes contain mutations, insertions and/or deletions, resulting in different lengths of nucleotides and consequently different numbers of amino acids being encoded. As shown in Table 3 , S1 gene of the isolated strains contain 1617, 1620, 1626, 1632, 1638 nucleotides, and 539, 540, 542, 544, 546 amino acids, respectively. The similarity of the nucleotide and deduced amino acid sequences of S1 genes (data not shown) among the 27 isolated strains were 65.4–99.9% and 57.2–99.8%, respectively, indicating low homology and high variation among these strains. When the S1 genes of the 29 published IBV strains (Table 2) were included, the similarity of the nucleotide and amino acid sequence among the 56 isolates were 60.9–99.9% and 45.4–99.8%, respectively.
Table 3.
Different lengths of nucleotides and deduced amino acids of S1 glycoprotein gene of the 27 isolated IBV strains.
| Length (nt/aa) | Strains |
|---|---|
| 1617/539 | DY05, LZ07, ZX07 |
| 1620/540 | DY07, GM05, HB08, LZ05, NN07, SS06-2, SS07, TC07-1 |
| 1626/542 | CQ04-1, CQ04-2, DY04, FS08-1, FS08-2, GL08-1, GL08-2, HN06-1, HN08, HN06-2, NN04, XX08 |
| 1632/544 | HY06, HY07, SS06-1 |
| 1638/546 | TC07-2 |
3.3. 26 of the 27 isolated IBV strains grouped into three genotypes by phylogenetic analysis
Phylogenetic analysis was performed on S1 gene nucleotide sequences consisting of the 27 isolated and 29 published IBV strains. All IBV strains except for the isolated TC07-2 strain and the reference DE/072/92 strain were clustered into six distinct genetic groups or genotypes (I–VI) (Fig. 1 ). Genotype I was comprised of 10 isolated IBV strains and 5 published IBV strains from China, designated as A2-like group for the inclusion of A2 strain; genotype II of 2 isolated and 4 published IBV strains, designated as 4/91 group for the inclusion of 4/91 strain; genotype III of 14 isolated and 5 published strains; genotype IV of 2 published IBV strains; genotype V of 8 published IBV strains; and genotype VI of 4 published IBV strains. There are four noteworthy features of the constructed phylogenetic tree. First, all isolated IBV strains except for TC07-2 strain belong to genotypes I, II and III, forming a large evolutionary branch. Furthermore, the isolated IBV strains grouped in genotype I were most from 2007(5/10), and most of the isolated IBV strains from 2008(6/7) were grouped into genotype III, showing that there were no predominant new strains circulating in the fields of South China during 2004–2008. Second, 2 published IBV strains from Taiwan formed a unique genotype (i.e., genotype IV), implying that the evolution process in Taiwan is an isolated one. Third, all 5 known vaccine strains (i.e., H52, H120, M41, Ma5 and W93) were clustered into genotype V that is evolutionarily distant from genotypes I, II and III; this may explain why the isolated IBV strains survived the vaccination by the vaccine strains. Finally, 3 classical American and 1 Japanese strains formed genotype VI, and the isolated TC07-2 and published DE/072/92 strains showed the greatest evolutionary distances to all six major genotypes, but their significances are not clear. The results strongly suggest that different genotypes of IBVs were cocirculating in chicken flocks in South China.
Fig. 1.
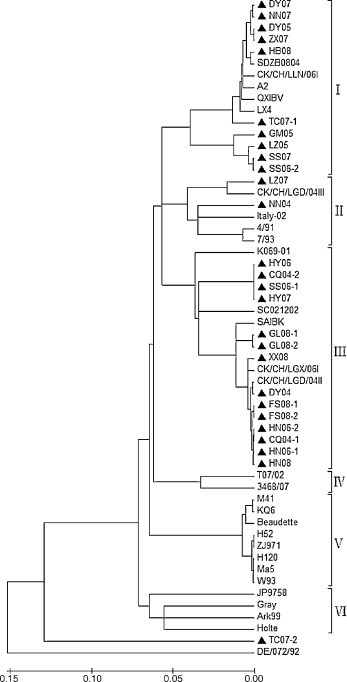
Phylogenetic tree constructed by neighbor-joining method (Mega 4.0). S1 nucleotide sequences of the 27 isolated and 29 published IBV strains were included for the phylogenetic analysis, where the 27 isolated IBV strains are marked with ante-black triangle.
3.4. Polymorphic protease cleavage recognition site motifs in S proteins
The spike glycoprotein of IBV is translated as a precursor protein (S0) and then cleaved into two subunits S1 and S2 by the protease during viral maturation (Cavanagh et al., 1986b). A motif at the cleavage site of the IBV S protein was generally comprised of RRXRR/S (X for any amino acid). Cleavage site motifs of the 27 isolated and 29 published IBV strains are listed in Table 1, Table 2, respectively. The 27 isolated strains contain 5 cleavage site motifs while the 29 published strains have 10 cleavage site motifs. The cleavage site motifs have the following four noteworthy features. First, in light of a total of 12 cleavage site motifs identified, the protease cleavage site motifs in S proteins are polymorphic. Second, 3 cleavage site motifs (RRFRR, HRRRR and RRSRR) are shared by both the isolated and published strains, where the shared motifs represented the majority of the strains (22/27 for isolated strains; 22/29 for published strains), and the majority of motifs have the recognized motif sequence RRXRR/S (14/27 for isolated strains; 21/29 for published strains). Third, 2 new motifs (RRSKR and HRRKR) emerged in the isolated strains, and in the meanwhile 7 motifs (HRSRR, RRHRR, RRLRR, HRPRR, RRIRR, HRFRR and RRFKR) identified in the published strains were not found in the isolated strains, indicating that the cleavage site motifs were undergoing a continuous evolution process. Finally, individual strains with high similarities in the phylogenetic tree usually share the same motifs, e.g., the lower branch of genotype I (RRFRR), the top branch of genotype III (RRSKR), the bottom branch of genotype III (RRFRR), and the genotype V (RRFRR). However, individual strains within one genotype have multiple motifs, e.g., genotype I having three motifs (RRFRR, HRRRR, and HRPRR), genotype II having 4 motifs (RRFRR, RRSRR, RRLRR, and HRFRR), and genotype III having 4 motifs (RRFRR, HRRRR, RRSKR, and RRHRR); indicating that the contribution of the cleavage site motif to one strain's genotype is limited.
3.5. Alignment analysis of nucleotide and deduced among amino acid sequence of S1 protein
Alignment analysis of S1 complete nucleotide and deduced amino acid sequences of all 27 isolated and 29 published IBV strains were performed. Strains in genotypes I–VI showed amino acid similarities of 84.6–99.8%, 78.9–94.8%, 82.6–99.8%, 84.9%, 94.4–99.6% and 77.2–80.7%, respectively. The S1 proteins of the 27 isolated strains had amino acid sequence similarities of 46.4–99.8%, and aa similarities of 58.7–79.5% to the Mass-type vaccine strain H120. The alignment analysis of the deduced amino acid sequences revealed three high variable regions located between amino acid residues 53–76, 94–141, and 253–292 (numbering using S1 sequence of Mass 41 as reference); the featured deletions, insertions and mutations were summarized in Table 4 . In line with the phylogenetic analysis discussed earlier, 27 isolated strains share no similarities in the listed amino acid positions with vaccine strains (e.g., H120) in genotype V. Interestingly, TC07-2 strain had a 4 amino acids insertion (Gln-Lys-Glu-Pro) at the region of 283–284, and this insertion was reported for the first time from this study (Table 4); the significance of this insertion in vial pathogenicity is not clear yet. Moreover, BLAST searches revealed that TC07-2 did not show close similarity (<80%) to any S1 gene sequences available.
Table 4.
Sequence alignment of amino acid residues of the S1 glycoprotein of IBV strains with the H120 vaccine strain.
| Strains | 52–55 | 71–76 | 106–107 | 117–118 | 140 | 185 | 253 | 281 | 284–285 | 317 | Genotype |
|---|---|---|---|---|---|---|---|---|---|---|---|
| H120 | ISSE | HGG--------RVV | DT | --HV | -. | S. | Q | N | ----PS | E | V |
| DY07 | STNY | KDV……..YNQ | EI | SVAG | D | Q | ….SG | A | I | ||
| NN07 | STNY | KDV……..YNQ | EI | SVAG | . | . | D | Q | ….SG | A | I |
| DY05 | STNY | KDV……..YNQ | EI | SGAG | . | . | D | Q | ….SG | A | I |
| ZX07 | STNY | KDV……..YNQ | EI | SGAG | . | . | D | Q | ….SG | A | I |
| HB08 | STNY | KDV……..YNQ | EI | SGTG | . | . | D | Q | ….SG | A | I |
| SDZB0804 | STNY | KDV……..YNQ | EI | SGTG | . | . | D | Q | ….SG | A | I |
| CK/CH/LLN/06I | STNY | KDV……..YNQ | EI | SGAG | . | . | D | Q | …..IG | A | I |
| A2 | STNH | KDV……..YNQ | EI | SGTG | . | . | E | Q | …..SG | A | I |
| QXIBV | STNY | KDV……..YNQ | EI | SGSG | . | . | E | Q | ….SG | A | I |
| LX4 | S-NY | KDV……..YNQ | EI | SGSG | . | . | E | Q | ….SG | A | I |
| TC07-1 | STNY | KDV……..YNQ | EI | SGRG | . | . | E | Q | ….SG | A | I |
| GM05 | STNY | KDV……..YNQ | EI | IGTG | . | . | E | Q | ….SG | A | I |
| LZ05 | STNY | KDV……..YNQ | EI | SGIG | . | . | E | Q | L…VG | H | I |
| SS07 | STNY | KDV……..YNQ | EI | SGTG | . | . | E | Q | L…VG | H | I |
| SS06-2 | STNY | KDV……..YNQ | EI | SGTG | . | . | E | Q | L…VG | H | I |
| LZ07 | STNY | KDV……..YNQ | EI | SGTG | . | . | G | S | ….SG | P | II |
| CK/CH/LGD-04III | STNK | FNYSNGN.DVGYNN | .F | N--- | . | G | D | S | ….SG | P | II |
| NN04 | STNK | FNYTNGN.DVGYNN | .F | N--- | . | G | D | S | ….SG | P | II |
| Italy-02 | V.T. | SWS……..KNF | .F | SGTG | S | G | D | P | S----G | P | II |
| 4/91 | VFNG | YES……..YNI | .F | SQQG | F | G | D | S | S….G | P | II |
| 7/93 | VFNE | YES……..HNI | HV | SQPG | V | G | D | S | S….G | P | II |
| K069-01 | STNY | KDV……..YNQ | EI | RGSG | . | . | . | S | ….SG | . | III |
| HY06 | FTNK | FNYSNGN-DIGYNN | .F | N--- | . | G | K | A | ….TG | Q | III |
| CQ04-2 | FTNK | FNYSNGN-DIGYNN | .F | N--- | . | G | K | A | ….TG | Q | III |
| SS06-1 | FTNK | FNYSNGN-DIGYNN | .F | N--- | . | G | K | A | ….TG | Q | III |
| HY07 | FTNK | FNYSNGN-DIGYNN | .F | N--- | . | G | K | A | ….TG | Q | III |
| SC021202 | STNR | FNYTNGN-DVGYNN | .V | N--- | . | G | E | S | ….QG | Q | III |
| SAIBK | STNK | FNYTNGN-DVGYNN | .F | N--- | . | G | E | S | ….QG | . | III |
| GL08-1 | YTNK | FNYTNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| GL08-2 | STNK | FNYTNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| XX08 | STNR | FNYTNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| CK/CH/LGX/06I | STNK | FNYTNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| CK/CH/LGD/04II | STNK | FNYTNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| DY04 | STNK | FNYNNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| FS08-1 | STNK | FNYTNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| FS08-2 | STNK | FNYTNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| HN06-2 | STNK | FNYTNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| CQ04-1 | STNK | FNYTNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| HN06-1 | STNK | FNYTNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| HN08 | STNK | FNYTNGN-DVGYNN | .F | N--- | . | G | K | P | ….QG | Q | III |
| T07/02 | VTTR | Q……….Y.F | EV | STQG | . | . | . | S | ….PG | . | VI |
| 3468/07 | V..Q | S.D……..T.F | .F | SGSG | . | . | . | Q | ….SG | . | VI |
| M41 | …. | ………….. | .. | ..YD | . | . | . | . | …… | . | V |
| KQ6 | …. | ………….. | .. | ..YD | . | . | . | . | …… | . | V |
| Beaudette | …. | ………….. | .. | …G | . | . | . | . | …… | . | V |
| H52 | …. | ………….. | .. | …. | . | . | . | . | …… | . | V |
| ZJ971 | …. | D…………. | .. | …. | . | . | . | . | …… | . | V |
| Ma5 | …. | ………….. | .. | …G | . | . | . | . | …… | . | V |
| W93 | …. | ..S……….. | .. | ..QG | . | . | . | . | …… | . | V |
| JP9758 | V.Q. | .WS……..KNF | NI | SGNN | D | G | E | P | T…VG | . | IV |
| Gray | V.E. | FWS……..KNF | HF | VGAG | H | G | D | P | N…-G | A | IV |
| Ark99 | V… | GYS……..KNF | SY | SGSN | H | A | D | P | T…-G | . | IV |
| Holte | VFQ. | GWS……..KNF | .I | HDG- | D | A | D | D | T…-G | . | IV |
| TC07-2 | FFN. | VHS……..LN. | TI | NRQG | N | G | E | Q | QKEPSP | S | |
| DE/072/92 | …K | G……….VTI | SF | TG-H | Q | A | N | E | G…-- | . |
Dot (.) indicates amino acid identical to that of H120 strain.
Dash (-) indicates amino acid deletion in comparison with H120 strain.
4. Discussion
This paper is a periodic report on our ongoing surveillance program with dual objectives, identifying the IBV strains that have escaped immune defenses conferred by vaccination and have caused persistent but infrequent outbreaks in commercial chicken farms, and preparing for the possible challenges of epidemic or pandemic IBV outbreaks in case of the emergence of a new predominant IBV strain in the field. Our survey included only broilers because most of chicken flocks in poultry industry in South China were broilers. The inclusion of only broilers in our survey did not mena that IBV was not associated with disease problems in laying and breeding chickens in South China; instead, we believe that our survey of broilers was indicative for all chickens including laying and breeding chickens. Our data showed that all 27 isolated IBV strains from South China during 2004–2008 were evolutionarily distant from the vaccine strains used for current vaccinations, explaining at least partially why vaccination failed in occasions where new IBV strains emerged and caused persistent but infrequent outbreaks. Our data also demonstrated that no new predominant IBV strains were being emerged during 2004–2008, but the continuing evolution of the escaped IBV strains makes the emergence of a new predominant strain as a matter of time, mandating of our efforts in carrying out continuing surveilance of emerging new IBV strains in order to prevent possible epidemic or pandemic outbreaks of IBV infections in commercial flocks.
In China, IB was first reported in 1972, and IBVs were found nearly all over the country in the following years. IBV infection is commonly followed by secondary bacterial infection, such as with Escherichia coli (Smith et al., 1985, Vandekerchove et al., 2004), resulting in complicated morbidity and increased mortality. Live-attenuated and inactivated vaccines have been widely used, resulting in effective control of IB (Cook, 2001). However, the outbreaks of IBV infection has been persistent but infrequent in vaccinated flocks in recent years (Liu and Kong, 2004, Liu et al., 2006, Liu et al., 2009a, Bing et al., 2007, Xu et al., 2007), causing clinical disease and production problems in vaccinated flocks (Nix et al., 2000, Farsang et al., 2002). The isolation of IBV strains from vaccinated flocks in China in recent years (Liu and Kong, 2004, Liu et al., 2006, Liu et al., 2009a, Bing et al., 2007, Xu et al., 2007) demonstrated that the evolution of IBV strains in the vaccinated flocks might eventually break down the immune defenses conferred by current vaccines, urging the investment of our efforts in tracking the IBV strains circulating in the commercial flocks.
In this study 27 IBV strains were successfully isolated from dead or diseased chickens from different chicken farms in South China during 2004–2008, of which S1 genes were sequenced. Phylogenetic analysis indicated that the 27 isolated IBV strains were concentrated in genotypes I, II and III, and had a large evolution distance to genotype V which is comprised primarily of vaccine strains (Fig. 1). Phylogenetic analysis also showed that the 27 isolated IBV strains had diverse genetic origins for the isolated viruses were dispersed in three genotypes (genotype I being comprised of 3 strain from 2005, 5 from 2007, 1 from 2006 and 1 from 2008; genotype II of 1 strain from 2004 and 1 strain from 2007; and genotype III of 3 strains from 2004, 4 from 2006, 1 from 2007, and 6 from 2008). It is plausible to presume that if one strain becomes prodominant, more and more isolated strains shall be concentrated into one group as the time progresses. However, the phylogenetic analysis failed to show such characteristics. Therefore, it is safe to conclude that there were no predominant strains circulating in commercial chicken farms in South China during 2004–2008.
S protein has a protease cleavage site motif that is essential for cleaving it into S1 and S2 subunits during viral maturation (Cavanagh et al., 1986b). There was report on study of the relationship between the cleavage site motifs and host cell range, serotype, geographic origin, and pathogenicity (Jackwood et al., 2001). The analysis of the cleavage site motifs in the 27 isolated IBV strains revealed two new cleavage site motifs, showing continuing evolution of IBV strains in the fields. The most dominant motif (RRFRR) in the 27 isolated IBV strains is shared by the majority of published IBV strains. Interestingly, the motif (HRRRR) is only present in IBV strains isolated in China, implying the continuity of evolution; its significance in viral pathogenicity and vaccine preparation is not clear yet. In addition, the cleavage site motifs do not correlate with the genotypes for one genotype has multiple motifs. This is not a total surprise since it was reported that the cleavage site motifs had no relationship with pathogenicity and tissue tropism (Jackwood et al., 2001). Furthermore, respiratory and nephropathogenic strains shared the cleavage site motifs, indicating that the cleavage site motif was not a determinant to pathotypes.
The genetic diversity of the 27 isolated strains was manifested by the low similarities of S1 genes. S1 genes of the 27 isolated strains had average amino acid sequence similarities of 79.6% (varied from 0.2 to 54.6%), and amino acid similarities of 58.7–79.5% to the Mass-type vaccine strain H120, which has been widely used for vaccine production in China for decades, indicating that point mutations, deletions, and insertions of S1 protein were frequent in IBVs in South China under vaccination. Previous study showed that the first 300 amino acids in the N-terminal of the S1 protein of IBV had a high frequency of variations, called highly variable region (HVR) (Kusters et al., 1989). Analysis of the S1 amino acid sequences in this study indicated that the three hypervariable regions (HVRs) in S1 protein in this study were similar to previous studies (Cavanagh et al., 1988, Moore et al., 1997, Liu et al., 2006, Schikora et al., 2003).
The reisolation of vaccine virus was an important issue for our survey program. This was addressed in two ways: genetic analysis and phylogenetic tree clustering. For S1 gene, no field isolate showed more than 84% (data not shown) similarity of nucleotide sequence to vaccine strains (genotype V), and no field isolate was clustered to vaccine strains in phylogenetic tree. Therefore, we could reasonably rule out that any field isolate was a reisolation of vaccine virus. Nonetheless, the fact that NN04 and LZ07 were grouped with 4/91 in Group II in Fig. 1 was intriguing because it was possible they might be originated from the leak of 4/91 strain from experiments done in China even though no vaccine based on 4/91 strain was licensed in China. This possibility was highly unlikely because their S1 amino acid sequences were very different. As shown in Table 4, amino acids from 52 to 55 and from 71 to 76 were VFNG and YES……..YNI for 4/91, STNK and FNYTNGN.DVGYNN for NN04, and STNY and KDV……..YNQ for LZ07.
QXIBV was first isolated in China in 1996 (reported in Chinese), associated with proventriculitis. In 2004, this virus was reported associated predominantly with various forms of renal pathology in China (Liu and Kong, 2004). The QX-like IBV strains more recently have been widely detected in many European countries, becoming a dominant genotype in some European countries, and are causing severe losses to both the layer and broiler industry (Worthington et al., 2004, Worthington and Jones, 2006). In addition, QX-like strains have been reported associated with false layers, swollen kidneys. In the present study, 6 field isolates were clustered to QXIBV in a branch (Fig. 1), of which, 4 show typical nephritis symptoms and 2 show typical respiratory clinical signs. In Worthington et al. (2008), the European QX-like IBV strains have been detected in flocks of broilers, broiler breeders and commercial layers. Of these flocks, 86% had respiratory signs, 22% had wet litter or enteric problems, 14% had increased mortality, 2% had swollen kidneys and 2% had arthritis. In 60% of the layers there was a reduction of egg production and quality. In the phylogenetic tree analysis with QX-like IBV strains from both Europe and China (data not shown), it is of interest that the QX-like IBV strains disclosed in Worthington et al. (2008) were clustered into a different branch than the one formed by the isolates from China, indicating that the QX-like IBV strains have undergone divergent evolution paths in Europe and China.
In conclusion, the present study has demonstrated that the circulating IBV strains in commercial flocks in South China had were genetically diverse and underwent continuing evolution; the emergence of new IBV strains will continue to cause persistent but infrequent outbreaks of IBV infections in commercial flocks. Furthermore, the present study challenges the feasibility of inclusion of new variant strains for vaccine preparation for better protection against IBV infection because it is unpredictable of which new variant strains will emerge. Finally, the present study revealed that there were no new predominant IBV strains circulating in the fields in South China yet, but it is reasonable to expect that such predominant IBV strains will eventually emerge under the increased immune pressures due to increased vaccination of commercial flocks. Therefore, this study manifests the importance of continuing surveillance of new IBV strains in order to better prepare for next epidemic or pandemic outbreaks of IBV infections in commercial flocks.
Acknowledgements
This work was supported by the grants from State Public Industry Scientific Research Programs (nyhyzx07-038, 2007GYJ019) and Major Programs of Science Technology Strategic Plan (2007A020400006) of Guangdong, People's Republic of China. We thank Dr. George D. LIU for kindly revising the manuscript.
References
- Bijlenga G., Cook J.K., Gelb J., Jr., de Wit J.J. Development and use of the H strain of avian infectious bronchitis virus from the Netherlands as a vaccine: a review. Avian Pathol. 2004;33:550–557. doi: 10.1080/03079450400013154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing G.X., Liu X., Pu J., Liu Q.F., Wu Q.M., Liu J.H. Different genotypes of nephropathogenic infectious bronchitis viruses co-circulating in chicken population in China. Virus Genes. 2007;35(2):333–337. doi: 10.1007/s11262-007-0100-5. [DOI] [PubMed] [Google Scholar]
- Capua I., Minta Z., Karpinska E., Mawditt K., Britton P., Cavanagh D., Gough R.E. Co-circulation of four types of infectious bronchitis virus (793/B, 624/I, B1648 and Massachusetts) Avian Pathol. 1999;28:587–592. doi: 10.1080/03079459994380. [DOI] [PubMed] [Google Scholar]
- Casais R., Dove B., Cavanagh D., Britton P. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J. Virol. 2003;77:9084–9089. doi: 10.1128/JVI.77.16.9084-9089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus IBV: structural characterization of the spike protein. J. Gen. Virol. 1983;64:2577–2583. doi: 10.1099/0022-1317-64-12-2577. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. A nomenclature for avian coronavirus isolates and the question of species status. Avian Pathol. 2001;30:109–115. doi: 10.1080/03079450120044506. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986;67:1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Cook J.K.A., Li D., Kant A., Koch G. Location of the amino-acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992;21:33–43. doi: 10.1080/03079459208418816. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Darbyshire J.H., Peters R.W. Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or hemagglutination-inhibiting antibody, or induce chicken tracheal protection. J. Gen. Virol. 1986;67:1435–1442. doi: 10.1099/0022-1317-67-7-1435. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Mockett A.P. Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes. Virus Res. 1988;11:141–150. doi: 10.1016/0168-1702(88)90039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Pappin D., Binns M.M., Boursnell M., Brown T. Coronavirus IBV: partial amino terminal sequencing of spike polypeptide S2 identifies the sequence Arg-Arg-Phe-Arg-Arg at the cleavage site of the spike precursor propolypeptide of IBV strains Beaudette and M41. Virus Res. 1986;4:133–143. doi: 10.1016/0168-1702(86)90037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Ellis M.M., Cook J.K.A. Relationship between variation in the S1 spike protein of infectious bronchitis virus and the extent of cross-protection. Avian Pathol. 1997;26:63–74. doi: 10.1080/03079459708419194. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Naqi S. Infectious bronchitis. In: Saif Y.M., Barnes H.J., Glisson J.R., Fadly A.M., McDougald L.R., Swayne D.E., editors. Diseases of Poultry. 11th ed. Iowa State University Press; Ames, IA: 2003. pp. 101–119. [Google Scholar]
- Cavanagh D., Picault J.P., Gough R., Hess M., Mawditt K., Britton P. Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs. Avian Pathol. 2005;34:20–25. doi: 10.1080/03079450400025414. [DOI] [PubMed] [Google Scholar]
- Cook J.K. Current status of infectious bronchitis virus infections in chickens and prospects for control by vaccines. Proceedings of the Proceedings of the XIIth International Congress of the World Veterinary Poultry Association; Cairo-Egypt; 2001. pp. 30–39. [Google Scholar]
- Cook J.K., Chester J., Baxendale W., Greenwood N., Huggins M.B., Orbell S.J. Protection of chickens against renal damage caused by a nephropathogenic infectious bronchitis virus. Avian Pathol. 2001;30:423–426. doi: 10.1080/03079450120066421. [DOI] [PubMed] [Google Scholar]
- Cook J.K., Orbell S.J., Woods M.A., Huggins M.B. A survey of the presence of a new infectious bronchitis virus designated 4/91 (793B) Vet. Rec. 1996;138:178–180. doi: 10.1136/vr.138.8.178. [DOI] [PubMed] [Google Scholar]
- Cook J.K.A., Orbell S.J., Woods M.A., Huggins M.B. Breadth of protection of the respiratory tract provided by different live-attenuated infectious bronchitis vaccines against challenge with infectious bronchitis viruses of heterologous serotypes. Avian Pathol. 1999;28:477–485. doi: 10.1080/03079459994506. [DOI] [PubMed] [Google Scholar]
- Farsang A., Ros C., Renstrom L.H., Baule C., Soós T., Belák S. Molecular epizootiology of infectious bronchitis virus in Sweden indicating the involvement of a vaccine strain. Avian Pathol. 2002;31:229–236. doi: 10.1080/03079450220136530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb J., Jr., Keeler C.L., Jr., Nix W.A., Rosenberger J.K., Cloud S.S. Antigenic and S-1 genomic characterization of the Delaware variant serotype of infectious bronchitis virus. Avian Dis. 1997;41:661–669. [PubMed] [Google Scholar]
- Gelb J., Jr., Weisman Y., Ladman B.S., Meir R. S1 gene characteristics and efficacy of vaccination against infectious bronchitis virus field isolates from the United States and Israel (1996 to 2000) Avian Pathol. 2005;34:194–203. doi: 10.1080/03079450500096539. [DOI] [PubMed] [Google Scholar]
- Gough R.E., Randall C.J., Dagless M., Alexander D.J., Cox W.J., Pearson D. A ‘new’ strain of infectious bronchitis virus infecting domestic fowl in Great Britain. Vet. Rec. 1992;130:493–494. doi: 10.1136/vr.130.22.493. [DOI] [PubMed] [Google Scholar]
- Hofstad M.S. Cross-immunity in chickens using seven isolates of avian infectious bronchitis virus. Avian Dis. 1981;25:650–654. [PubMed] [Google Scholar]
- Karaca K., Naqi S., Gelb J., Jr. Production and characterization of monoclonal antibodies to three infectious bronchitis virus serotypes. Avian Dis. 1992;36:903–915. [PubMed] [Google Scholar]
- Koch G., Hartog L., Kant A., van Roozelaar D.J. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J. Gen. Virol. 1990;71:1929–1935. doi: 10.1099/0022-1317-71-9-1929. [DOI] [PubMed] [Google Scholar]
- Kusters J.G., Niesters H.G.M., Bleumink-Pluym N.M.C., Davelaar F.G., Horzinek M.C., van der Zeijist B.A.M. Molecular epidemiology of infectious bronchitis virus in The Netherlands. J. Gen. Virol. 1987;68:343–352. doi: 10.1099/0022-1317-68-2-343. [DOI] [PubMed] [Google Scholar]
- Kusters J.G., Niesters H.G., Lenstra J.A., Horzinek M.C., van der Zeijst B.A. Phylogeny of antigenic variants of avian coronavirus IBV. Virology. 1989;169(1):217–221. doi: 10.1016/0042-6822(89)90058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Hilt D.A., Callison S.A., Lee C.W., Plaza H., Wade E. Spike glycoprotein cleavage recognition site analysis of infectious bronchitis virus. Avian Dis. 2001;45:366–372. [PubMed] [Google Scholar]
- Jia W., Karaca K., Parrish C.R., Naqi S.A. A novel variant of infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 1995;140:259–271. doi: 10.1007/BF01309861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W., Wang X., Parrish C., Naqi S.A. Analysis of the serotype-specific epitopes of avian infectious bronchitis virus strains Ark99 and Mass41. J. Virol. 1996;70:7255–7259. doi: 10.1128/jvi.70.10.7255-7259.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladman B.S., Loupos A.B., Gelb J., Jr. Infectious bronchitis virus S1 gene sequence comparison is a better predictor of challenge of immunity in chickens than serotyping by virus neutralization. Avian Pathol. 2006;35:127–133. doi: 10.1080/03079450600597865. [DOI] [PubMed] [Google Scholar]
- Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Kong X. A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and nonvaccinated flocks in China. Avian Pathol. 2004;33:321–327. doi: 10.1080/0307945042000220697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhang Q., Chen J., Han Z., Liu X., Feng L., Shao Y., Kong X., Rong J., Tong G. Genetic diversity of avian infectious bronchitis coronavirus strains isolated in China between 1995 and 2004. Arch. Virol. 2006;151:1133–1148. doi: 10.1007/s00705-005-0695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhang X., Wang Y., Li C., Liu Q., Han Z., Zhang Q., Kong X., Tong G. Evaluation of the protection conferred by commercial vaccines and attenuated heterologous isolates in China against the CK/CH/LDL/97I strain of infectious bronchitis coronavirus. Vet. J. 2009;179(1):130–136. doi: 10.1016/j.tvjl.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.L., Su J.L., Zhao J.X., Zhang G.Z. Complete genome sequence analysis of a predominant infectious bronchitis virus (IBV) strain in China. Virus Genes. 2009;38(1):56–65. doi: 10.1007/s11262-008-0282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane R., Verma R. Sequence analysis of the gene coding for the S1 glycoprotein of infectious bronchitis virus (IBV) strains from New Zealand. Virus Genes. 2008;37(3):351–357. doi: 10.1007/s11262-008-0273-6. [DOI] [PubMed] [Google Scholar]
- Meulemans G., Boschmans M., Decaesstecker M., Berg T.P.v.d., Denis P., Cavanagh D., van den Berg T.P. Epidemiology of infectious bronchitis virus in Belgian broilers: a retrospective study, 1986 to 1995. Avian Pathol. 2001;30:411–421. doi: 10.1080/03079450120066412. [DOI] [PubMed] [Google Scholar]
- Moore K.M., Jackwood M.W., Hilt D.A. Identification of amino acids involved in a serotype and neutralization specific epitope within the S1 subunit of avian infectious bronchitis virus. Arch. Virol. 1997;142:2249–2256. doi: 10.1007/s007050050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix W.A., Troeber D.S., Kingham B.F., Keeler C.L., Jr., Gelb J., Jr. Emergence of subtype strains of the Arkansas serotype of infectious bronchitis virus in Delmarva broiler chickens. Avian Dis. 2000;44:568–581. [PubMed] [Google Scholar]
- Parsons D., Ellis M.M., Cavanagh D., Cook J.K.A. Characterisation of an infectious bronchitis virus isolated from vaccinated broiler breeder flocks. Vet. Rec. 1992;131:408–411. doi: 10.1136/vr.131.18.408. [DOI] [PubMed] [Google Scholar]
- Schikora B.M., Shih L.M., Hietala S.K. Genetic diversity of avian infectious bronchitis virus California variants isolated between 1988 and 2001 based on the S1 subunit of the spike glycoprotein. Arch. Virol. 2003;148:115–136. doi: 10.1007/s00705-002-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.W., Cook J.K.A., Parsell Z.E. The experimental infection of chickens with mixtures of infectious bronchitis virus and Escherichia coli. J. Gen. Virol. 1985;66:777–786. doi: 10.1099/0022-1317-66-4-777. [DOI] [PubMed] [Google Scholar]
- Stern D.F., Sefton B.M. Coronavirus proteins: biogenesis of avian infectious bronchitis virus virion proteins. J. Virol. 1982;44:794–803. doi: 10.1128/jvi.44.3.794-803.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terregino C., Toffan A., Serena Beato M., De Nardi R., Vascellari M., Meini A., Ortali G., Mancin M., Capua I. Pathogenicity of a QX strain of infectious bronchitis virus in specific pathogen free and commercial broiler chickens, and evaluation of protection induced by a vaccination programme based on the Ma5 and 4/91 serotypes. Avian Pathol. 2008;37(5):487–493. doi: 10.1080/03079450802356938. [DOI] [PubMed] [Google Scholar]
- Vandekerchove D., De Herdt P., Laevens H., Butaye P., Meulemans G., Pasmans F. Significance of interactions between Escherichia coli and respiratory pathogens in layer hen flocks suffering from colibacillosis-associated mortality. Avian Pathol. 2004;33:298–302. doi: 10.1080/030794504200020399. [DOI] [PubMed] [Google Scholar]
- Wang L., Junker D., Collisson E.W. Evidence of natural recombination within the S1 gene of infectious bronchitis virus. Virology. 1993;192:710–716. doi: 10.1006/viro.1993.1093. [DOI] [PubMed] [Google Scholar]
- Wang L., Junker D., Hock L., Ebiary E., Collison E.W. Evolutionary implications of genetic variations in the S1 gene of infectious bronchitis virus. Virus Res. 1994;34:327–338. doi: 10.1016/0168-1702(94)90132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington K.J., Currie R.J.W., Jones R.C. A reverse transcriptase-polymerase chain reaction survey of infectious bronchitis virus genotypes in Western Europe from 2002 to 2006. Avian Pathol. 2008;37:247–257. doi: 10.1080/03079450801986529. [DOI] [PubMed] [Google Scholar]
- Worthington K.J., Jones R.C. An update in the European RT-PCR IBV survey and recent findings on a novel IBV genotype. Proceedings of Vth Symposium on Avian Corona and Pneumovirus Infections; Rauischholzhausen, Germany; 2006. pp. 176–188. [Google Scholar]
- Worthington K.J., Savage C., Naylor C.J., Wijmenga W., Jones R.C. An RT-PCR survey of infectious bronchitis virus genotypes in the UK and selected European countries between 2002 and 2004 and the results from a vaccine trial. Proceedings of the IV Symposium on Avian Corona and Pneumovirus Infections; Rauischholzhausen, Germany; 2004. pp. 101–133. [Google Scholar]
- Xu C., Zhao J., Hu X., Zhang G. Isolation and identification of four infectious bronchitis virus strains in China and analyses of their S1 glycoprotein gene. Vet. Microbiol. 2007;122(1–2):61–71. doi: 10.1016/j.vetmic.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Zhao J.X., Qin Z.M. Studies on the isolated strain of IBV which was similar to 4/91. Chin. J. Prevent. Vet. Med. 2002;24:360–363. (in Chinese, with English abstract) [Google Scholar]


