Abstract
Attaching and effacing Escherichia coli are involved in diarrhea in 2 to 8-week old calves. The virulence factors of these bacteria include: (i) the secretion of proteins (i.e. EspB) involved in microvilli effacement, (ii) the production of the intimin, a 94 kDa outer membrane protein encoded by the eaeA gene and involved in the intimate attachment of bacteria to epithelial cell and (iii) the production of verotoxins: VT1 and/or VT2. We investigated the presence and the pathotype of these strains in several calf populations by colony hybridization or by genetic amplification. Using the colony hybridization method we showed first that only 5% of calves who died from diarrhea presented EaeA+ E. coli strains and secondly that 19% of healthy calves showed an asymptomatic carriage. However, using colony hybridization and genetic amplification, we identified EaeA+ strains in 91% of calves living in farms with recurrent diarrhea problems. In 66% of the calves, there was a correlation between the presence of AEEC and diarrhea. At the pathotype level, most of the EaeA+ isolates were negative for VT probes. In VT+ bacteria, the majority were VT1+. The number of VT positive bacteria was significantly higher in calves who died from diarrhea than in healthy or sick calves. This underlined the aggravating role of verotoxins in the disease. Moreover, only 25% of the bovine AEEC were positive with the EaeB probe. Surprisingly, the proportion of EaeB+ strains was significantly higher in healthy calves than in other populations.
Keywords: Cattle-Bacteria, Escherchia coli, Diarrhea
1. Introduction
Neonatal calf diarrhea involves the interaction of one or more pathogenic microorganisms (bacterial, viral, protozoal agents), the immune status of the calf, and one or more environmental factors (Butler and Clarke, 1994). Enterotoxigenic Escherichia coli (ETEC), rota and corona viruses and Cryptosporidium sp. together account for 75–95% of all cases of neonatal calf diarrhea (Acres, 1985; Tzipori, 1985).
More recently, attaching and effacing E. coli (AEEC) (Moon et al., 1983) have been implicated in diarrhea and dysentery mostly in 2 to 8-week old calves (Chanter et al., 1984; Mainil et al., 1993). “Attaching and effacing” was the term first used by Moon et al. (1983)to describe an intestinal lesion produced by E. coli: `attaching' indicates the intimate attachment of bacteria to the enterocyte; `effacing' signifies the localized effacement of brush border microvilli. Such a lesion appears when enteropathogenic E. coli (EPEC) or enterohaemorrhagic E. coli (EHEC) infect their host.
The pathogenesis of AEEC has been studied mainly on human EPEC strain E2348/69 (Nataro et al., 1985). It is a three-step model (Knutton, 1994). First, the bacteria adhere to the epithelial cell via the production of fimbriae. Second, the bacteria secrete proteins (i.e. EspB encoded by the eaeB gene) which are responsible for the transmission of a signal to the epithelial cell (Kenny and Finlay, 1995). These cells respond by an increase in phosphorylations and in intracellular Ca2+ level. The Ca2+ ions activate villin which severs actin filaments leading to the destruction of microvilli. Third, the bacteria adhere intimately to epithelial cells. This intimate attachment is correlated both with the expression by the bacteria of a 94 kDa outer membrane protein called intimin and with the accumulation of cytoskeletal compounds such as actin in the cell beneath the bound bacteria. The intimin is encoded by the eaeA gene. This gene is present in human and in bovine AEEC as demonstrated by colony hybridization studies using the EaeA probe (Jerse et al., 1990; Mainil et al., 1993). Moreover, the intimin has been demonstrated to be a virulence factor in vivo (Donnenberg et al., 1993a).
The name EHEC has been applied to verotoxigenic E. coli (VTEC) implicated in hemorrhagic colitis in humans (Levine, 1987). Verotoxins are E. coli cytotoxins which are lethal for cultured Vero cells (Konowalchuck et al., 1977). These toxins share a number of properties with Shiga toxin produced by Shigella dysenteriae and are therefore also called Shiga-like toxins (O'Brien and La Veck, 1983). Verotoxins consist of two groups, with VT1 (SLT-I) constituting one group and VT2 (SLT-II) and antigenically related toxins constituting a second group (Newland et al., 1988). The VT1 and VT2 encoding operons were sequenced (Jackson et al., 1987a, Jackson et al., 1987b). EPEC and EHEC strains have been described in several animal species including cattle (Chanter et al., 1984). A previous study indicated that most bovine AEEC were actually EHEC-like strains (Mainil et al., 1993).
The genetic diagnosis of AEEC can be performed either by hybridization using virulence gene-specific probes for eaeA, vt1 and vt2 genes (Mainil et al., 1993) or by specific amplification of virulence genes (China et al., 1996). We report here the detection of AEEC in calf populations in Belgium using these genetic tools.
2. Materials and methods
2.1. Animals populations and sampling
A total of 695 calves were included in this survey: 295, 2 to 10-week old calves which had died from diarrhea problems; 311, 4 to 6-week old healthy calves from five different farms without a serious diarrheal problem; and 89 newborn to 3-month old calves from seven farms with severe diarrhea problems. The E. coli were isolated from the intestinal contents of dead calves or from feces of the other calves. In the first two populations, samples were taken only once. In the third population, fecal samples were collected twice a week for up to 12 weeks after birth, whereas the other calves were sampled once. Fecal and intestinal samples were handled for recovery of E. coli and for PCR.
2.2. Escherichia coli recovery
For live calves, 100 mg of feces were diluted in Brain Heart Infusion (Atlas, 1993, BHI, Gibco, Paisley, Scotland). Ten-fold dilutions were plated on Gassner Agar (Merck 1282, Darmstadt, Germany) using a spiral plater (spiral system, Led Techno, Eksel, Belgium). The plates were incubated overnight at 37°C. Five lactose-positive colonies were picked up and stored at −70°C in 10% glycerol in 96 wells microplates (Greiner, Frickenhausen, Germany) until further characterization by colony hybridization and by serotyping. For dead calves, a sample from the intestinal content was suspended in saline. One drop was plated on Gassner Medium. The plates were incubated overnight at 37°C and three to five lactose positive colonies were picked up and kept at −70°C in 10% glycerol until further characterization. The Gassner plates from the third calf population were subsequently covered with a Whatman 541 paper filter (Maidstone, UK) that was treated to perform colony hybridization for quantification of the relative number of AEEC among E. coli population.
2.3. Colony hybridization
The protocol was adapted from Mainil et al. (1993). The colonies were planted out from the microplates on Luria Bertani (LB) Agar (Atlas, 1993) using a 96 teeth replicator. The plates were incubated overnight at 37°C. The colonies were blotted on a filter paper (Whatman 541, Maidstone, UK). The colonies were treated as follows: 3 min in SDS 10%, 15 min in NaOH 0.5 M NaCl 1.5 M and 2×5 min in Tris–HCl 0.5 M NaCl 1.5 M pH 7.5. The probes (Table 1 ) were labeled with α-P32-dCTP by random priming (DNA Labelling beads, Ready To Go, Pharmacia, Uppsala, Sweden). The filters were incubated with the probe for at least 6 h in hybridization solution (powdered skimmed milk 1%, SSC 6X, SDS 1%, salmon sperm DNA 20 μg/ml). The filters were washed three times for 10 min with SDS 0.1% SSC 2X and autoradiographied for at least 6 h at −70°C with a X-Ray film (Fuji, Tokyo, Japan).
Table 1.
Probes used for colony hybridization in this study
| Gene | Plasmid | Vector | Strain of origin | Restriction enzymes | Size (bp) | References |
| vt1 | pJN37-19 | pUC19 | 933 | BamHI | 1142 | Newland et al., 1988 |
| vt2 | pNN111-19 | pUC19 | 933 | PstI | 842 | Newland et al., 1988 |
| eaeB | pMSD3 | pACYC184 | E2348/69 | ClaI + NruI | 511 | Donnenberg et al., 1993b |
| eaeA | pCDV434 | pUC19 | E2348/69 | SalI + KpnI | 1050 | Jerse et al., 1990 |
2.4. Polymerase chain reaction
The PCR reaction was performed on the fecal samples of the third calf population. Five milliliter of LB medium were inoculated with 100 μl of feces. The culture was incubated overnight at 37°C under shaking. The PCR on eaeA gene was performed on the culture using the method previously described (China et al., 1996). The PCR on STIa (or STaP) gene was performed as described above using the following primers: B84 (TTTCTGTATTATCTTTCCCCTCTT) and B85 (AGCACAGGCAGGATTACAACAA). This PCR reaction generates a 179 bp amplicon which is specific to the STIa (So and McCarthy, 1980) gene and fails to amplify the STIb (or STaH) gene (Moseley et al., 1983).
2.5. Other pathogens
When lactose-negative colonies were present on Gassner agar plates, they were tested by agglutination with an anti-salmonella polyvalent serum OMA (Sanofi Diagnostic, Pasteur, France). The positive colonies were further identified by API sugar sets (BioMérieux, Brussels, Belgium). The search for Cryptosporidium sp. was performed by dimethylsulphoxide-Ziehl-Neelsen staining (Pohjola et al., 1984). Due to the number of samples (1164), a systematic research of viruses was not performed.
2.6. Statistics
The frequencies were compared by the Fischer's exact test using the Graphpad Instat 2.01 Software.
3. Results
3.1. Prevalence and typing of AEEC in calves dead from diarrhea
Thirty-two isolates out of 970 tested (3.3%) were positive with the EaeA probe by the colony hybridization assay (Eae+ isolates). The Eae+ isolates originated from 15 calves out of 295 (5%). The 32 Eae+ isolates were tested with gene probes specific for three other potential virulence factors: VT1, VT2 and EspB. Sixteen of them (50%) were positive only with the EaeA probe, and were considered as EPEC, and the other sixteen (50%) were positive in addition with one or both VT probes, and were considered as EHEC. Of the EHEC isolates, 14 (87.5%) were VT1+ and two (12.5%) were VT1+ VT2+. Moreover, seven EaeA+ isolates (21%) were also EaeB+: two were EPEC and five were EHEC.
3.2. Prevalence and typing of AEEC in healthy calves
Two hundred fifty-five isolates out of 1322 tested (19%) were positive with the EaeA probe by the colony hybridization assay. The Eae+ isolates originated from 75 calves out of 311 (24%). One hundred and seventy-one of the EaeA+ isolates (67%) were EPEC and 84 (33%) were EHEC. Of the EHEC isolates, 75 (89%) were VT1+ and nine (11%) were VT2+. Moreover, 135 of the EaeA+ isolates (53%) were also positive with the EaeB probe: 102 were EPEC and 33 were EHEC.
3.3. Incidence and typing of AEEC in diarrheic calves
The proportion of probe-positive isolates is significantly higher in healthy calves than in calves dead from diarrhea. However, these healthy calves were sampled only once and it is possible either that they had diarrhea before the sampling and remained carriers or that the calves would have had diarrhea just after the sampling. To overcome this drawback, an epidemiological study was carried out consisting of a twice-a-week sampling of the same calves for up to 12 weeks after their birth. The seven chosen farms showed the same profile: recurrent diarrhea problems in spite of vaccination (against rotavirus, coronavirus and ETEC) and antibiotherapy. The presence of EPEC and EHEC was checked by colony hybridization and by PCR on fecal samples.
EaeA+ isolates were detected by colony hybridization in each farm (Fig. 1 ), but the percentage of positive fecal samples ranged from 11% to 63% according to the farm. On the basis of both the presence/absence of EaeA+ isolates and the presence/absence of diarrhea, the calves were classified into four groups: calves presenting diarrhea and AEEC, calves presenting AEEC but no diarrhea, calves presenting diarrhea but no AEEC, and calves presenting neither AEEC nor diarrhea (Fig. 2 ): 66% of calves had a correlation between the presence of AEEC and diarrhea although 25% calves showed an asymptomatic carriage. Feces from calves of 1 to 12 weeks of age were positive (Fig. 3 ). However, 90% of the positive samples originated from 2 to 8-week old calves and the calculated mean age was 5 weeks.
Fig. 1.
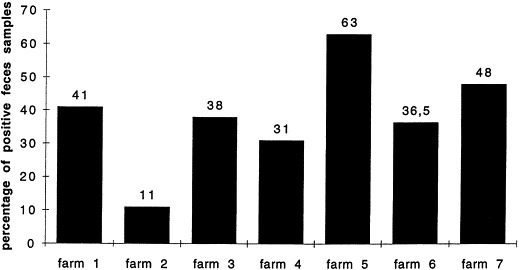
Percentage of EaeA+ feces samples per farm. The presence of EaeA positive strains in the feces samples has been determined by PCR and colony hybridization for each tested farm.
Fig. 2.
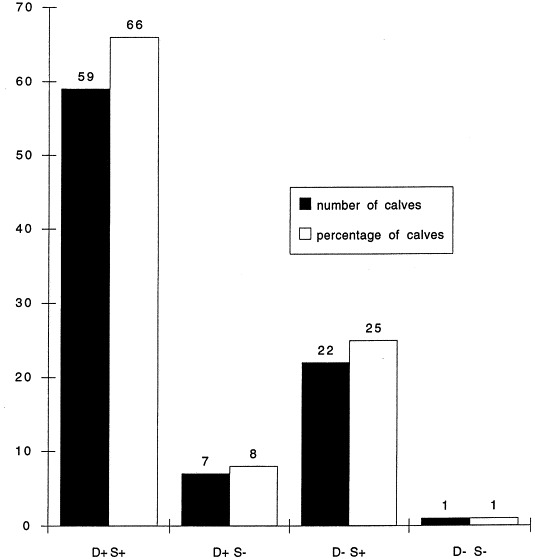
Number (■) or percentage (□) of calves in respect to the presence or absence of diarrhea (D+ or D−) and to the presence or the absence of EaeA+ strains (S+ or S−). These positive strains were determined by PCR or colony hybridization.
Fig. 3.
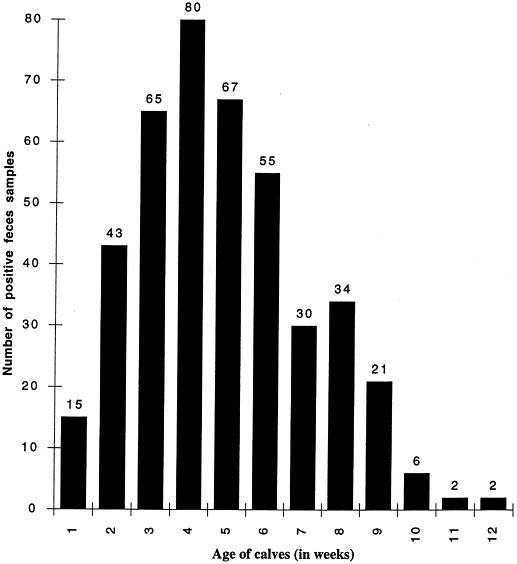
Number of EaeA+ feces samples in function of the calves age in weeks. The presence of EaeA positive strains in the feces samples has been determined by PCR and colony hybridization.
The relative percentage of AEEC among the fecal E. coli populations was estimated by colony hybridization of the filters made from 50 original Gassner agar plates after overnight growth. The number of E. coli per ml of feces ranged from 103 to 5×107 and the relative percentage of EaeA+ colonies ranged from 10% to 90% with a mean of 60±32.5%.
All samples were negative for the presence of Salmonella. The first two samples of all calves were tested by PCR for the presence of ETEC: four calves out of 89 (4.5%) tested positive. In two of the seven farms, cryptosporidia were observed in fecal smears. In the first one (Farm 2), there was a correlation between diarrhea and the presence of cryptosporidia, since 70% of the calves with diarrhea were positive. In the second farm (Farm 3), the situation was less clear since 65% of the calves with diarrhea were negative for cryptosporidia.
Four hundred and ninety isolates out of 5416 tested (9%) were positive with the EaeA probe by the colony hybridization assay. The Eae+ isolates were detected in 429 of the 1164 fecal samples (37%) originating from 79 calves out of 89 (89%). Three hundred and sixty-three of the EaeA+ isolates (74%) were EPEC and 127 (26%) were EHEC. Of the EHEC isolates, 114 (90%) were VT1+, seven (5%) were VT2+, and six (5%) were VT1+VT2+. Moreover, 42 of the EaeA+ isolates (8.5%) were also positive with the EaeB probe: 11 were EPEC and 31 were EHEC (Table 2 ).
Table 2.
Detection of EaeA+ bacteria and determination of their pathotypes
| Healthy calves | Calves in farms with diarrhea problemsc | Calves dead of diarrhea | |
|---|---|---|---|
| Animal tested (n) | 311 | 89 | 295 |
| Age range (weeks) | 4 to 6 | 0 to 10 | 0 to 10 |
| Faeces sample tested (n) | 311 | 1164 | 295 |
| Faeces with EaeA+ bacteria (n) | 75a | 429b | 15a |
| E. coli isolates (n) | 1322 | 5416 | 970 |
| E. coli EaeA+ (n) | 255a | 490a | 32a |
| eaeA+ | 171a | 363a | 16a |
| eaeA+ VT1+ | 75a | 114a | 14a |
| eaeA+ VT2+ | 9a | 7a | Oa |
| eaeA+ VT1+ VT2+ | 0a | 6a | 2a |
By colony hybridization.
bBy colony hybridization and/or PCR.
cAmong the seven farms involved in the screening, the percentage of positive samples for EaeA+ bacteria ranged from 11% to 63%.
4. Discussion
Diarrhea represents an increasing and recurrent problem in young calves, from 1 week up to 12 weeks of age, especially in suckling beef calves, in farms where the neonatal diarrhea problems have regressed thanks mainly to vaccination and higher hygiene standards. Among the possible infectious agents, the `attaching and effacing' strains of E. coli (AEEC; Moon et al., 1983) have been associated with diarrhea and dysentery in calves up to 2 months of age (Butler and Clarke, 1994; Mainil et al., 1993). However no large scale epidemiological study has been carried out to verify this association mainly because of the difficulty in the diagnosis of the AEEC. We performed such a study in calves dead from diarrhea, in diarrheic calves, and in healthy calves, based on the detection, either by colony hybridization or by PCR, of the eaeA gene coding for the intimin protein, an adhesin which plays a central role in the pathogenesis of AEEC, which is essential for the production of AE lesions by bovine AEEC (China et al., 1997; Goffaux et al., 1997), and which is produced in vivo during infection in man (Donnenberg et al., 1993a).
AEEC are clearly present in large numbers in farms with recurrent problems of diarrhea with, however, variation from farm to farm. Moreover, in the literature, the rates of detection and isolation of AEEC are a function of the calf age (Fig. 3) with the calculated mean age at 5 weeks which is in perfect correlation with previous studies (Butler and Clarke, 1994). Although the proportion of AEEC strains seems to be related to the proportion of animal loss in the farms as indicated by the fact that in farm 5 presenting the highest incidence of AEEC, 35 calves died from diarrhea the year before this study. Nevertheless, the AEEC strains do not very often cause the death of the calves, as evidenced by the low percentage of positive isolates from dead calves. These two calf populations were however different so that no definite conclusion can be drawn from these data.
The role of AEEC strains in the development of diarrhea may also be questioned as asymptomatic carriage appears to be frequent in healthy calves both from farms with and without problems of diarrhea. However, the history of these healthy calves is unknown: they may have recovered from the disease at the time of sampling or succumbed thereafter. Moreover the proportion of calves with diarrhea and AEEC (66%) was significantly higher than the proportion of healthy calves with AEEC (25% in farms with diarrhea and 24% in farms without diarrhea).
The pathogenesis of AEEC in young calves is probably more complex than the pathogenesis of K99+ enterotoxigenic E. coli in newborn calves and may well include:
(i) a quantitative aspect: the absolute and relative numbers of AEEC to the commensal enteric bacterial flora may be very important. The existence of a threshold under which no disease will occur is possible. The comparison of PCR and colony hybridization results showed that the number of positive samples was higher by PCR (429) than by colony-hybridization (142). Since PCR is a more sensitive technique, we can consider that the number of AEEC present in many positive sample was low but maybe sufficient to give diarrhea. Indeed, the lesions in enterohemorrhagic colitis could be located on a small portion of the intestine;
(ii) a qualitative aspect: AEEC strain population may be heterogeneous and only some of them with different pathotypes may cause diseases. Also the nature of the disease can be different according to the pathotype of the isolates, EPEC or EHEC. It is well known that cattle are carriers of EHEC strains which are pathogenic not only for people but also for newborn calves (i.e. O157:H7 strains); however VT-positive AEEC were significantly more frequent (p<0.05) in calves dead of diarrhea than in other populations emphasizing the fact that verotoxins could act as aggravating factors (Tesch and O'Brien, 1991). Determination of the serotype may help in confirming this hypothesis but was not performed on the AEEC isolated from live calves in this study. Otherwise AEEC isolates from dead calves in Belgium belong to O5, O18, O26, O111, and O118 serogroups (Mainil et al., 1993; data not shown), and the rate of isolation of O157 isolates is very low in Belgium at slaughterhouses (Mainil et al., unpublished data; G. Daube, personal communication). On another aspect, only 25% of the EaeA+ strains were EaeB+ and the proportion of EaeB+ AEEC was significantly higher (p<0.05) (53%) in healthy calves than in sick and healthy calves (8% and 21% respectively);
(iii) a synergistic aspect: AEEC may well act in synergy with other noninfectious parameters (drop in colostral protection, antibiotic treatment) and/or infectious agents: Salmonella was not detected during our surveys, but well cryptosporidia, although their presence was not clearly correlated with diarrhea, and virus which were not looked for. Synergistic association between rota- and coronaviruses is well documented (Radostits et al., 1994).
Acknowledgements
We thank Dorothy and Marie-Paule for their critical reading of this manuscript. This work was supported by grant n°5631A of IRSIA (Institut pour l'encouragement de la Recherche appliquée à l'Industrie et à l'Agriculture). We thank the farmers for their kind collaboration.
References
- Acres S.D. Enterotoxigenic E. coli infections in newborn calves: a review. J. Dairy Sci. 1985;68:229–256. doi: 10.3168/jds.S0022-0302(85)80814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlas, R.M., 1993. In: Parks, L.C. (Ed.), Handbook of Microbiological Media. CRC Press, Boca Raton, pp. 523
- Butler, D.G., Clarke, R.C., 1994. Diarrhea and dysentery in calves. In: Gyles, C.L. (Ed.), Escherichia coli in domestic animals and humans. CAB international, Oxon, pp. 91–116
- Chanter N, Morgan J.H, Bridger J.C, Hall G.A, Reynolds D.R. Dysentery in gnotobiotic calves caused by atypical Escherichia coli. Vet. Rec. 1984;114:71. doi: 10.1136/vr.114.3.71. [DOI] [PubMed] [Google Scholar]
- China, B., Pirson, V., Jacquemin, E., Pohl, P., Mainil, J.G., 1997. Pathotypes of bovine verotoxygenic Escherichia coli isolates producing attaching and effacing (AE) lesion in ligated intestinal loop assay in rabbits. In: Francis, D., Benfield, D. (Eds.), Mechanisms in the Pathogenesis of Enteric Diseases. Plenum Press, New York, NY, pp. 311–316
- China B, Pirson V, Mainil J. Typing of bovine attaching and effacing Escherichia coli by multiplex in vitro amplification of virulence-associated genes. Appl. Environ. Microbiol. 1996;62:3462–3465. doi: 10.1128/aem.62.9.3462-3465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg, M.S., Tzipori, S., McKee, M.L., O'Brien, A.D., Alroy, J., Kaper, J.B., 1993a. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in porcine model. J. Clin. Invest. 92, 1418–1424 [DOI] [PMC free article] [PubMed]
- Donnenberg, M.S., Yu, J., Kaper, J.B., 1993b. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J. Bacteriol. 175, 4670–4680 [DOI] [PMC free article] [PubMed]
- Goffaux F, Mainil J, Pirson V, Charlier G, Pohl P, Jacquemin E, China B. Bovine attaching and effacing Escherichia coli possess a pathogenesis island related to the LEE of the human enteropathogenic Escherichia coli strain E2348/69. FEMS Microbil. Let. 1997;154:415–421. doi: 10.1111/j.1574-6968.1997.tb12676.x. [DOI] [PubMed] [Google Scholar]
- Jackson, M.P, Newland, J.W., Holmes, R.K., O'Brien, A.D. 1987a. Nucleotide sequence analysis of the structural genes for shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb. Pathog. 2, 147–153 [DOI] [PubMed]
- Jackson, M.P., Neill, R.J., O'Brien, A.D., Holmes, R.K., Newland, J.W., 1987b. Nucleotide sequence analysis and comparison of the structural gene for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli 933. FEMS Microbiol. Lett. 44, 109–114 [DOI] [PubMed]
- Jerse A.E, Yu J, Tall B.D, Kaper J.B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of ataching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny B, Finlay B.B. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc. Natl. Acad. Sci. USA. 1995;92:7991–7995. doi: 10.1073/pnas.92.17.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton, S., 1994. Attaching and effacing Escherichia coli. In: Gyles, C.L. (Ed.), Escherichia coli in Domestic Animals and Humans. CAB International, Oxon, pp. 567–91
- Konowalchuck J, Speirs J.L, Stavric S. Vero response to a cytotoxin of Escherichia coli. Infect. Immun. 1977;18:775–779. doi: 10.1128/iai.18.3.775-779.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M.M. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic and enteroadherent. J. Infect. Dis. 1987;155:377–389. doi: 10.1093/infdis/155.3.377. [DOI] [PubMed] [Google Scholar]
- Mainil J.G, Jacquemin E, Kaeckenbeeck A, Pohl P. Association between the effacing gene (eae) and the Shiga-like toxin-encoding genes in Escherichia coli isolates from cattle. Am. J. Vet. Res. 1993;54:1064–1068. [PubMed] [Google Scholar]
- Moon H.W, Whipp S.C, Argenzio R.A, Levine M.M, Gianella R.A. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 1983;53:1340–1351. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley S.L, Hardy J.M, Huq M.I, Echevarria P, Falkow S. Isolation and Nucleotide seuqence determiantion of a gene encoding a heat stable enterotoxin of Escherichia coli. Infect. Immun. 1983;39:1167–1174. doi: 10.1128/iai.39.3.1167-1174.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro J.P, Scaletsky I.C.A, Japer J.B, Levine M.M, Trabulsi L.R. Plasmid-mediated factors conferring diffuse and localized adherence of enteropathogenic Escherichia coli. Infect. Immun. 1985;48:378–383. doi: 10.1128/iai.48.2.378-383.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newland J.W, Neill R.G. DNA probe for shiga-like toxins I and II and for toxin-converting bacteriophages. J. Clin. Microbiol. 1988;26:1292–1297. doi: 10.1128/jcm.26.7.1292-1297.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien A.D, La Veck G.D. Purification and characterization of a Shigella dysenteriae 1-like toxin produced by Escherichia coli. Infect. Immun. 1983;40:675–683. doi: 10.1128/iai.40.2.675-683.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjola, S., Jokipii, L., Jokipii, A. M.M., 1984. Dimethylsulphoxide-Ziehl-Neelsen staining technique for detection of cryptosporidial oocysts. The Veterinary Record, 115, 442–443 [DOI] [PubMed]
- Radostits, O.M, Blood, D.C., Gay, G.C., 1994. Veterinary Medicine, 8th ed. Baillière Tindall, London, p. 1020
- So M, McCarthy B.J. Nucleotide sequence of transposon Tn1681 encoded a heat-stable toxin (ST) and its identification in enteroenterotoxigenic Escherichia coli strains. Proc. Nat. Acad. Sci. USA. 1980;77:4011–4015. doi: 10.1073/pnas.77.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesch V.L, O'Brien A.D. The pathogenic mechanisms of shiga and Shiga-like toxins. Mol. Microbiol. 1991;5:1817–1822. doi: 10.1111/j.1365-2958.1991.tb00805.x. [DOI] [PubMed] [Google Scholar]
- Tzipori, S., 1985. The relative importance of enteric pathogens affecting neonates of domestic animals. In: Cornelius, C.E., Simpson, C.F. (Eds.), Advances in Veterinary Science and Comparative Medicine, vol. 29. Academic Press, Toronto, pp. 103–206 [PubMed]


