Abstract
A one step reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay was developed for detection of viral hemorrhagic septicaemia virus (VHS). A set of six primers were designed, based on the G-protein sequence of the VHS virus serotypes (He, F1, 23.75, Klapmolle and Rindsholm). The assay was optimised to amplify VHS RNA by incubation at 63 °C for only 1 h, and required only a simple water bath or heating block to provide a constant temperature of 63 °C. RT-LAMP amplification products were detected by visual inspection using SYBR Green I stain and had a ladder-like appearance when electrophoresed on an agarose gel. The detection limit of the RT-LAMP assay was found to be similar to the commonly used RT-PCR method: both methods detected VHS RNA at a dilution of 106. The assay was evaluated using clinical samples and the results indicated the suitability and simplicity of the test as a rapid, field diagnostic tool for VHS virus.
Keywords: RT-LAMP, VHS, Diagnosis, SYBR Green I, RT-PCR
1. Introduction
Animal Rhabdoviruses, belonging to the family Rhabdoviridae, infect a broad range of hosts throughout the animal kingdom, including insects, mammals and fish. Aquatic Rhabdoviruses cause significant diseases in fish reared as part of the worldwide salmonid farming industry (Rocha et al., 2004). Among fish Rhabdoviruses, viral hemorrhagic septicaemia virus (VHS), of the genus Novirhabdovirus, often causes pronounced cumulative mortality of salmonids, especially rainbow trout Oncorhynchus mykiss (Miller et al., 1998). The virus is enzootic in much of continental Europe (Wolf, 1988) and has been found in North America (King et al., 2001, Snow and Smail, 1999). VHS is a notifiable disease, included in List II of the European Union Directive 93/54 (1993), and is known by several names including Egtved disease and infectious kidney-swelling and liver degeneration.
The virus’ aetiology was established by Jensen (1965), and it is known to infect primarily rainbow trout, Oncorhynchus mykiss, brown trout, Salmo trutta, and to a lesser extent northern pike, Esox lucius (Jorgensen, 1980, Meier and Jorgensen, 1980), grayling, Thymallus thymallus, and whitefish, Coregonus sp. (Wizigmann et al., 1980, Ahne and Thomsen, 1985, Meier et al., 1986). The virus has also been isolated from free-living marine fish in the northern Pacific and Atlantic Oceans, the North Sea and the Baltic Sea (Meyers and Winton, 1995, Mortensen et al., 1999). Virus isolates from wild marine fish are serologically indistinguishable from normal fresh water isolates (Jorgensen and Olesen, 1987). VHS virus is transmitted by direct contact with either infected fish or contaminated water (Vestergard Jorgensen, 1973). Survivors from infection become carriers and shed the virus with urine and sexual fluids (Neukirch, 1985). Although the virus can be present on the surface of eggs, it is readily dissipated and there is no true vertical transmission (Vestergard Jorgensen, 1970). VHS infection in susceptible fish species is often lethal, due to impairment of osmotic balance; clinical signs include oedema and red haemorrhagic spots around the eyes and on the gills, and blood clots in body fat, viscera and muscle (Wolf, 1988).
Rapid diagnosis of VHS is necessary to prevent further spread of the disease (Miller et al., 1998). Serological tests have been used to diagnose VHS virus, and include the virus neutralisation test (Jorgensen, 1969, Jorgensen, 1972), a fluorescent antibody technique (Jorgensen, 1969, Enzmann, 1981) and enzyme linked immunosorbent assay (ELISA) (Olesen and Jorgensen, 1991). Yet serological tests along with other traditional means of diagnosing viral infection, including epizootiological data, direct light or electron microscopic observation and isolation of the virus in cell culture, are often time-consuming and expensive (Amos, 1985, Sanz and Coll, 1992). Recently, molecular biological techniques have been developed to improve diagnosis of fish pathogens by rapid and sensitive detection of pathogen RNA and DNA.
A reverse transcriptase–polymerase chain reaction (RT-PCR) has been developed to detect VHS (Bruchhof et al., 1995, Einer-Jensen et al., 1995, Miller et al., 1998, Strommen and Stone, 1998, Guillou et al., 1999, Williams et al., 1999). However, diagnosis of VHS using this PCR test requires expensive equipment and reagents, rendering it unfavourable for use on a large-scale basis, or on-site. A novel, rapid and sensitive technique called loop-mediated isothermal amplification (LAMP) has been developed by Notomi et al. (2000) and is capable of amplifying DNA or RNA under isothermal conditions with high specificity and efficiency. LAMP is based on the principle of autocycling strand displacement DNA synthesis. The reaction is performed by a DNA polymerase with high strand displacement activity and a set of two specific inner primers and two outer primers (Notomi et al., 2000). A reverse transcriptase coupled LAMP assay has been used to detect West Nile virus (Parida et al., 2004) and for rapid detection of SARS (severe acute respiratory syndrome) coronavirus (Thai et al., 2004).
The aim of the present study was to develop a one-step, single-tube, accelerated RT-LAMP reaction for rapid detection of different serotypes of VHS virus. The specificity and sensitivity of the method were assessed, and its applicability as a diagnostic test was evaluated.
2. Materials and methods
2.1. Viruses
VHS virus serotypes (He, Klapmolle, Rindsholm, and 23.75) were kindly provided by Dr. Fichtner, Federal Research Institute for Animal Health, Insel Riems, Germany. The VHS virus serotype (F1) and infectious hematopoietic necrosis virus (IHNV) were provided by Dr. Widemann, Department of Virology, Central Institute of Animal Health, Bavaria, Germany.
2.2. RNA extraction
Fish tissues suspected of being infected with VHS (spleen, kidney and brain) were preserved in RNA later. These were then processed by grinding thoroughly in liquid nitrogen with a mortar and pestle. Twenty milligram of tissue powder was then placed into a RNase-free, liquid nitrogen-cooled 2 ml microcentrifuge tube with 600 μl lysis buffer. The lysate was transferred directly into a QIAshredder spin column (QIAGEN GmbH, Hilden, Germany) and centrifuged at maximum speed (18,000 × g) for 2 min. One volume (600 μl) of 70% ethanol was added to the supernatant and extraction completed as per the manufacture's instructions. RNA was eluted in RNase-free water (70 °C) and stored at −80 °C.
Genomic viral RNA was extracted from 140 μl of infected culture supernatant using the QIAamp viral RNA kit (QIAGEN GmbH, Hilden, Germany) according to the manufacture's instructions. After elution of the RNA in elution buffer, it was stored at −80 °C until required.
2.3. LAMP primers
Six LAMP primers were designed, based on the G-protein sequence of the VHS virus serotypes: F1, Klapmolle, Rindsholm, He and 23.75 (GenBank accession number AF345857, AF345858, AF345859, U28798 and U28799, respectively). The forward inner primer, FIP, consisted of complementary sequence of F1 (24 nt), a TTTT linker and the sense sequence of the F2 (19 nt): 5′-GATCCACCGATACTGTTTTTGGGGTTTTCCCGTTCTTC CCTGAACCC-3′. The backward inner primer, BIP, contained a sense sequence of B1 (21 nt), a TTTT linker and the complementary sequence of the B2 (18 nt): 5′-ARGGGG TYTGCACARCCTCGC TTTT CGACKYGGGRCAAKGGGC-3′. The outer primers consisted of the sense sequence of F3 (19) 5′-GGSAAGCAAGGAYCACGAG-3′; and complementary sequence of B3 (21 nt) 5′-CAGGTGTCCYTCTAGTGTTTC-3′.
To accelerate amplification, two loop primers were used: a loop forward primer (loop-F, 21 nt): 5′-GTTATGTCCTTATGGACATTG-3′; and a loop backward primer (loop-B; 19 nt): 5′-GTCAAACTCATTGGCAGGG-3′. The location of the primers within the gene fragment is shown in (Fig. 1 ).
Fig. 1.

Nucleotide sequence of the G-protein fragment of VHS virus (serotype F1 as example) (GenBank accession number AF345857) used to construct the inner and outer primers (indicated in bold and italic). The inner primers FIP and BIP comprise the complementary sequences to F1 and sense sequence of F2, and the sense sequence of B1 and complementary sequence of B2, respectively.
The reverse transcriptase-polymerase chain reaction used specific primers VG1 and VGR, which amplified a 1524 bp segment, and semi-nested primers VD5 and VD3, which amplified a 443 bp segment of VHS cDNA (Bruchhof et al., 1995): VG1: 5′-ATGGAATGGAACACTTTTTTC-3′, VGR: 5′-TCAGACCGTCTGACTTCTGGA-3′; VD5: 5′-TCCCGCTATCAGTCACCAG-3′; VD3: 5′-TGTGATCATGGGTCCTGGTG-3′.
2.4. RT-PCR-based detection
RT-PCR was performed using a TITANIUM™ one-step RT-PCR kit (BD Clontech, Heidelberg, Germany). For a 50 μl reaction volume, 1 μg of RNA was mixed with 10× one-step buffer, dNTPs mix, recombinant RNase inhibitor, thermostabilising reagent, GC-melt, Oligo (dT) primer, 45 μM of each VG1 and VGR VHS specific primers, and RT-TITANIUM™ Taq enzyme mix. The reaction conditions comprised: incubation at 50 °C for 1 h, then denaturation at 94 °C for 5 min; followed by 39 cycles of 94 °C for 30 s; 52 °C for 40 s and 72 °C for 40 s. There was a terminal extension step of 72 °C for 5 min.
Following RT-PCR, 3 μl of the product was added to 47 μl of semi-nested PCR reaction mixture: comprising 1.1× ReddyMix PCR Master mix (ABgene, Hamburg, Germany: 75 mM Tris–HCl (pH 8.8), 20 mM (NH4)2SO4, 1.5 mM MgCl2, 0.01% Tween 20, 0.2 mM each of dATP, dCTP, dGTP, dTTP; 1.25 U Taq DNA polymerase and red dye for electrophoresis), and 20 pmol of each VHS specific primers VD5 and VD3. The reaction entailed initial denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 52 °C for 40 s and 72 °C 40 s. There was a terminal extension step of 72 °C for 5 min.
2.5. RT-LAMP assay
The LAMP reaction was performed in a 25 μl reaction volume containing: 20 mM Tris–HCl (pH 8.8), 10 mM KCl, 6.5 mM MgSO4, 10 mM (NH4)2−SO4, 0.1% Triton x-100, 1.6 M betaine, deoxynucleotide triphosphates 2.8 mM each, 1.6 μM each FIP and BIP, 0.8 μM each loop-F and loop-B, 0.2 μM each F3 and B3 primers, 8 U Bst DNA polymerase (New England BioLabs, GmbH, Frankfurt, Germany), 1.0 U Avian Myeloblastosis Virus (AMV) reverse transcriptase (Invitrogen, Groningen, The Netherlands) and 2 μl RNA template. RNA template was omitted from one reaction as a negative control. The mixture was incubated at 63 °C for 60 min and then heated at 80 °C for 2 min to terminate the reaction.
2.6. Analysis of amplification products
RT-PCR and RT-LAMP reaction products were analysed by gel electrophoresis with 1.5% agarose in Tris acetate–EDTA buffer, TAE (0.04 M Tris acetate, 1 mM EDTA), stained with ethidium bromide and visualised on a UV transilluminator. A 100 bp DNA molecular weight standard ladder (Cambrex Bio Science, Inc., Rockland, ME, USA) was used. Success of the RT-LAMP reaction was also gauged by visual inspection: 1 μl of 1:10 diluted SYBR Green I Nucleic acid gel stain, 10,000× concentration in DEMSO (Cambrex Bio Science, Rockland, Inc., ME, USA) was added to the reaction tube and any colour change noted.
2.7. RT-LAMP specificity
The ability of the assay to amplify only VHS RNA was appraised by testing it with RNA from infectious hematopoietic necrosis virus a related Rhabdoviruses. RNA from non-infected fish was used as a negative control to determine any non-specific amplification.
2.8. RT-LAMP sensitivity
Ten-fold serial dilutions of the used VHS serotypes RNA were tested by both RT-LAMP and RT-PCR to determine the detection limits of each assay.
2.9. Applicability of the RT-LAMP assay
The suitability of the assay to diagnose VHS was evaluated by comparing detection results of RT-LAMP assay on 26 experimentally infected rainbow trout with VHS serotype F1 (Mattes, 2004) against RT-PCR results on the same samples.
3. Results
3.1. Optimisation of the RT-LAMP reaction
Different primer concentrations, primer combinations, amplification temperatures, and amounts of reverse transcriptase were tested to determine the optimal RT-LAMP conditions. Optimal amplification of VHS RNA was achieved by incubation of FIP, BIP, F3, B3 primers, 8 U of Bst DNA polymerase and 1 U of AMV reverse transcriptase enzyme with the target RNA at 63 °C for 1 h. The amplification product appeared as a ladder-like pattern (many bands of different molecular weights) on the gel (Fig. 2 ). There was no amplification of the negative control. The assay was able to amplify all five serotypes of VHS RNA (Fig. 2). Two additional primers, loop-F and loop-B, were subsequently added to enhance and accelerate the reaction down to 30 min (Fig. 3 ). There was no difference in amplification pattern using the additional primers.
Fig. 2.
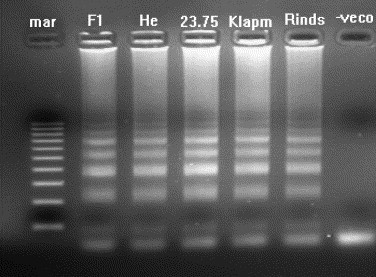
Agarose gel showing RT-LAMP amplification products of VHS virus RNA which appears as multiple bands of different molecular weights. Lanes: mar = 100 bp DNA molecular weight standard; F1, LAMP products from RNA of VHS serotype F1; He, VHS serotype He; 23.75, VHS serotype 23.75; Klapm, VHS serotype Klapmolle; Rinds, VHS serotype Rindsholm; −veco, negative control.
Fig. 3.
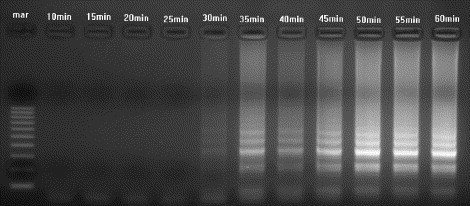
Agarose gel showing the effect of varying the amplification time in the RT-LAMP reaction. Amplification using primers FIP, BIP, F3, B3, loop-F and loop-B was carried out at 63 °C for the times indicated. Mar = 100 bp DNA molecular weight standard. VHS RNA was detected as early as 30 min.
Colour changes were noted on visual inspection of RT-LAMP reaction tubes after addition of diluted SYBR Green I: positive samples turned green, while negative samples and no template control reactions remained orange (Fig. 4 ). These observations agreed with gel electrophoresis results.
Fig. 4.

Appearance of RT-LAMP products visualised with SYBR Green I stain. (1) reaction positive for VHS RNA showing green colouration; (2) positive VHS LAMP reaction with RNA dilution 103; (3) positive VHS LAMP reaction with RNA dilution 106; (4) negative reaction showing orange colour; (5) no template negative control showing orange colour.
3.2. Specificity of the RT-LAMP assay
The specificity of the VHS-LAMP primers and conditions to VHS virus was confirmed by its aptitude to amplify only RNA from VHS virus serotypes, with no amplification of IHNV or non-infected fish (Fig. 5 ).
Fig. 5.
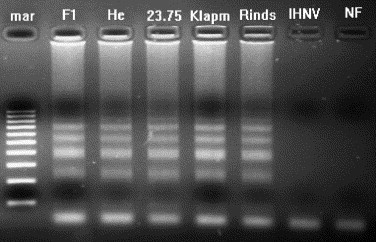
Agarose gel highlighting the specificity of the RT-LAMP primers to VHS virus RNA. The reaction was carried out at 63 °C for 1 h using the 6 primer set. Lanes: F1, He, 23.75, Klapm; and Rinds, amplification products of VHS virus RNA serotypes (F1, He, 23.75, Klapmolle and Rindsholm); IHNV, no amplification product was detected for IHN virus RNA; NF, no amplification product was detected using RNA from non-infected fish; mar = 100 bp molecular weight marker.
3.3. Sensitivity of the RT-LAMP assay
Amplification of 10-fold serial dilutions of VHS RNA by RT-PCR and RT-LAMP revealed that both tests could detect viral RNA to a dilution of 1 in 106 (Fig. 6, Fig. 7 ). The same result was obtained with the all tested VHS serotypes.
Fig. 6.
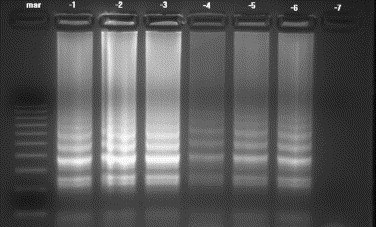
Agarose gel showing RT-LAMP reactions using 10-fold serial dilutions of template RNA. The assay detected VHS RNA at a dilution one in 106. Lanes: −1 = template concentration 10−1; −2 = 10−2; −3 = 10−3; −4 = 10−4; −5 = 10−5; −6 = 10−6; −7 = 10−7; mar = 100 bp DNA molecular weight standard.
Fig. 7.
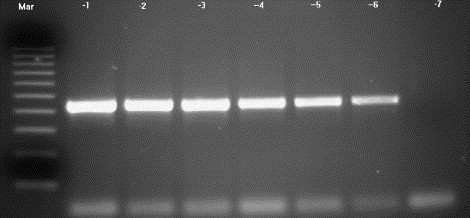
Agarose gel showing VHS RT-PCR using 10-fold serial dilutions of template RNA. The RT-PCR produced a 443 bp amplification product and detected VHS RNA at a dilution of 1 in 106. Lanes: −1 = template concentration 10−1; −2 = 10−2; −3 = 10−3; −4 = 10−4; −5 = 10−5; −6 = 10−6; −7 = 10−7; mar = 100 bp DNA molecular weight standard.
3.4. Applicability of the RT-LAMP assay
RT-LAMP detected VHS RNA from 26 infected samples, which also tested positive by the RT-PCR test. This result indicates that the developed RT-LAMP assay can feasibly be used as a diagnostic test of VHS infection.
4. Discussion
Viral infections pose a serious threat to the aquaculture industry and are responsible for significant financial losses (Caipang et al., 2004). It is clearly important to rapidly identify and differentiate the causative agents of fish diseases to prevent further disease transmission or outbreaks (Bruchhof et al., 1995). VHS is one of several Rhabdoviruses that affects the aquaculture industry, and causes disease with high mortality among salmonids (Miller et al., 1998). Difficulties associated with isolation of viruses from clinical specimens has lead to development of rapid and reliable virus detection assays (Parida et al., 2004) including many RT-PCR assays developed for diagnosis of VHS (Bruchhof et al., 1995, Einer-Jensen et al., 1995, Miller et al., 1998, Strommen and Stone, 1998, Guillou et al., 1999, Williams et al., 1999). Despite the simplicity of the RT-PCR reaction, its requirements for a high precision thermacycler and elaborate methods for detecting the amplified products have restricted its wide use as a routine diagnostic tool (Thai et al., 2004). To overcome these issues, Notomi et al. (2000) developed a novel nucleic acid amplification assay: loop-mediated isothermal amplification. The LAMP reaction has been used successfully to detect fish viral infections (Caipang et al., 2004, Gunimaladevi et al., 2004, Gunimaladevi et al., 2005), and has been applied to the detection of VHS in the present study.
The RT-LAMP technique is rapid, straightforward and includes a simple visual method for detecting positive results. The reaction is executed in a single tube and only requires a simple water bath or heating block to provide a constant temperature of 63 °C for 1 h. The LAMP reaction relies on autocycling strand displacement DNA synthesis and utilises a set of four specially designated inner and outer primers (Notomi et al., 2000) which are used during the initial steps of the reaction. During the subsequent cycling reactions, only the inner primers are used for strand displacement DNA synthesis (Parida et al., 2004). Each inner primer contains two distinct sequences corresponding to the sense and antisense sequences of the target DNA. The primers form stem-loop structures which initiate self-priming DNA synthesis, and serve as the starting material for the subsequent LAMP cycling reaction (Kuboki et al., 2003). To further reduce the reaction time, two additional loop primers were used to hybridise to the stem-loops formed by the outer primers. The loops hybridised by the inner primers subsequently prime strand displacement DNA synthesis (Nagamine et al., 2002).
Due to the variation in the glycoprotein gene sequence of the VHS serotypes, degenerative primers were designed from an alignment of the G-protein sequences of VHS serotypes He, F1, 23.75, Klapmolle and Rindsholm. These primers were then aligned against G-protein sequences of IHNV serotypes, WRAC, SRCV and RB, to check that they would not bind; which was confirmed by testing in a reaction with IHNV RNA (Fig. 5). The LAMP primers are suitable for amplification of at least five VHS serotypes (Fig. 2).
The specificity of the reaction was extremely high because it uses six primers that recognise eight distinct regions on the target DNA (Notomi et al., 2000, Nagamine et al., 2002). The amplified products appeared as a ladder-like pattern on the gel due to the formation of a mixture of stem-loop DNA products with varying stem lengths, and cauliflower-like structures of multiple loops formed by annealing between alternately inverted repeats of the target sequence in the same strand (Thai et al., 2004). The assay used SYBR Green I DNA staining as a rapid, specific method for detection of amplification products. Visual detection is possible due to the high specificity and amplification efficiency of LAMP (Iwamoto et al., 2003), and to the high binding affinity of SYBR Green I to DNA (Karlsen et al., 1995). Visual inspection is a superior detection method as there is no need for gel electrophoresis and staining with ethidium bromide; only 1 μl of diluted SYBR Green I was required to visualise the reaction products: a distinct green colour indicated a positive result while orange indicated negative. All samples which tested positive by visual inspection were positive also when analysed by electrophoresis; there were no samples which were negative by visual examination but which tested positive by electrophoresis.
The RT-LAMP assay detected VHS RNA at a dilution of 1 in 106, which was equivalent to the limit of RT-PCR. The amplification efficiency of the LAMP method was extremely high under isothermal conditions at the optimal temperature for the polymerase; any inhibition reactions at the latter stages of amplification (Kalinina et al., 1997) were less likely to occur compared with PCR (Mori et al., 2001). The suitability of the RT-LAMP assay for detection of VHS RNA in infected samples was successfully validated.
In conclusion, the developed RT-LAMP assay is extremely rapid, cost effective, sensitive and specific for detection of VHS RNA. The test requires only a water bath and is completed in 1 h compared with 4–5 h for RT-PCR. Considerably less time is required to obtain a result using SYBR Green I stain, compared with traditional gel electrophoresis. The assay is suitable to be used under field conditions for the rapid diagnosis of VHS, which would allow expedited control and hygiene measures to be implemented to prevent spread of infection.
Acknowledgements
We would like to express our grateful thanks to Dr. D. Fichtner and Dr. Bergmann, Federal Research Institute for Animal Health, Insel Riems, Germany and Dr. Widemann, Department of Virology, Central Institute of Animal Health, Bavaria, Germany for providing us with the VHS and IHN viruses. We also thank Dr. Marianne Mattes for providing samples of the experimentally infected rainbow trout.
References
- Ahne W., Thomsen I. Occurrence of viral hemorrhagic septicaemia virus in wild whitefish Coregonus sp. Zentralbl. Vet. Reihe. B. 1985;32:73–75. doi: 10.1111/j.1439-0450.1985.tb01940.x. [DOI] [PubMed] [Google Scholar]
- Amos K.H. Fish Health Section. third ed. American Fisheries Society; Corvallis: 1985. Procedures for the detection and identification of certain fish pathogens; p. 114. [Google Scholar]
- Bruchhof B., Marquardt O., Enzmann P.J. Differential diagnosis of fish pathogenic Rhabdoviruses by reverse transcriptase-dependent polymerase chain reaction. J. Virol. Methods. 1995;55:111–119. doi: 10.1016/0166-0934(95)00051-u. [DOI] [PubMed] [Google Scholar]
- Caipang C.M.A., Haraguchi I., Ohira T., Hirono I., Aoki T. Rapid detection of a fish iridovirus using loop-mediated isothermal amplification (LAMP) J. Virol. Methods. 2004;121:155–161. doi: 10.1016/j.jviromet.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Einer-Jensen K., Olesen N.J., Lorenzen N., Jorgensen P.E.V. Use of polymerase chain reaction (PCR) to differentiate serologically similar VHS virus isolates from Europe and America. Vet. Res. 1995;26:464–469. [PubMed] [Google Scholar]
- Enzmann P.J. Rapid identification of VHVS from trout by immunofluorescence. In: Hennessen W., editor. vol. 49. S. Karger; Basel, München, Paris, London, New York, Sydney: 1981. pp. 57–62. (Fish Biologics, Serodiagnostics and Vaccines. Developments in Biological Standardization). [Google Scholar]
- Guillou J.P., Merle G., Henault S., Hattenberger A.M. Detection of viral hemorrhagic septicaemia virus (VHS) in rainbow trout (Oncorhynchus mykiss) by reverse transcription followed by polymerase chain reaction. Diagnostic validation. Vet. Res. 1999;30:49–60. [PubMed] [Google Scholar]
- Gunimaladevi I., Kono T., Venugopal M.N., Sakai M. Detection of koi herpesvirus in common carp, Cyprinus carpio L., by loop-mediated isothermal amplification. J. Fish Dis. 2004;27:583–589. doi: 10.1111/j.1365-2761.2004.00578.x. [DOI] [PubMed] [Google Scholar]
- Gunimaladevi I., Kono T., LaPatra S.E., Sakai M. A loop mediated isothermal amplification (LAMP) method for detection of infectious hematopoietic necrosis virus (IHNV) in rainbow trout (Oncorhynchus mykiss) Arch. Virol. 2005;150:899–909. doi: 10.1007/s00705-004-0468-7. [DOI] [PubMed] [Google Scholar]
- Iwamoto T., Sonobe T., Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 2003;41:2616–2622. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.H. Research on the virus of Egtved disease. Ann. N.Y. Acad. Sci. 1965;126:422–426. doi: 10.1111/j.1749-6632.1965.tb14292.x. [DOI] [PubMed] [Google Scholar]
- Jorgensen P.E.V. Serological identification of Egtved virus. Bull. Off. Int. Epizootiol. 1969;69:985. [PubMed] [Google Scholar]
- Jorgensen P.E.V. Egtved virus: antigenic variation in 76 virus isolates examined in neutralization tests and by means of the fluorescent antibody technique. In: Mawdesley-Thomas L.E., editor. Diseases of Fish. Academic Press; London: 1972. p. 333. [Google Scholar]
- Jorgensen P.E.V. Egtved virus: the susceptibility of brown trout and rainbow trout to eight virus isolates and the significance of the findings for the VHS control. In: Ahne W., editor. Fish Diseases. Third COPRAQ Session. Springer-Verlag; Berlin, Heidelberg, New York: 1980. p. 37. [Google Scholar]
- Jorgensen P.E.V., Olesen N.J. Cod ulcus syndrome rabdovirus is indistinguishable from the Egtved (VHS) virus. Bull. Eur. Assoc. Fish Pathol. 1987;7:73–74. [Google Scholar]
- Kalinina O., Lebedeva I., Brown J., Silver J. Nanoliter scale PCR with TaqMan detection. Nucleic Acids Res. 1997;25:1999–2004. doi: 10.1093/nar/25.10.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsen F., Steen H., Nesland J. SYBR Green I DANN staining increases the detection sensitivity of viruses by polymerase chain reaction. J. Virol. Methods. 1995;55:153–156. doi: 10.1016/0166-0934(95)00053-w. [DOI] [PubMed] [Google Scholar]
- King J.A., Snow M., Smail D.A., Raynaerd R.S. Distribution of viral hemorrhagic septicaemia virus in wild fish species of the North Sea, north east Atlantic Ocean and Irish Sea. Dis. Aquat. Organ. 2001;47:81–86. doi: 10.3354/dao047081. [DOI] [PubMed] [Google Scholar]
- Kuboki N., Inoue N., Sakurai T., Di Cello F., Grab D.J., Suzuki H., Sugimoto C., Igarashi I. Loop-mediated isothermal amplification for detection of African trypanosomes. J. Clin. Microbiol. 2003;41:5517–5524. doi: 10.1128/JCM.41.12.5517-5524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes, M., 2004. Untersuchungen zur Empfänglichkeit zweier Regenbogenforellen-stämme gegenüber Tetracapsuloides bryosalmonae, Yersinia ruckeri und dem Viralen Hämorrhagischen Septikämie-Virus. Ph.D. Thesis. University of Munich, pp. 144 (in German language).
- Meier W., Jorgensen P.E.V. Isolation of VHS virus from pike fry (Esox lucius) with hemorrhagic symptoms. In: Ahne W., editor. Fish Diseases. Third COPRAQ Session. Springer verlag; Berlin, Heidelberg, New York: 1980. p. 817. [Google Scholar]
- Meier W., Ahne W., Jorgensen P.E.V. Fish viruses: viral hemorrhagic septicaemia in whitefish (Coregonus sp.) J. Appl. Icthyol. 1986;4:181–186. [Google Scholar]
- Meyers T.R., Winton J.R. VHS-virus in North America. In: Faisal M., Hetrick F.M., editors. vol. 5. Pergamon, Elsevier Sciences; Oxford: 1995. pp. 3–24. (Annual Review of Fish Diseases). [Google Scholar]
- Miller T.A., Rapp J., Wastlhuber U., Hoffmann R.W., Enzmann P.J. Rapid and sensitive reverse transcriptase polymerase chain reaction based detection and differential diagnosis of fish pathogenic Rhabdoviruses in organ samples and cultured cells. Dis. Aquat. Organ. 1998;34:13–20. doi: 10.3354/dao034013. [DOI] [PubMed] [Google Scholar]
- Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Mortensen H.F., Heuer O.E., Lorenzen N., Otte L., Olesen N.J. Isolation of VHSV from wild marine fish species in the Baltic Sea, Kattegat, Shagerrak and the North Sea. Virus Res. 1999;63:95–106. doi: 10.1016/s0168-1702(99)00062-3. [DOI] [PubMed] [Google Scholar]
- Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Neukirch M. Uptake, multiplication and excretion of VHSV in rainbow trout. In: Ellis A.E., editor. Fish and Shellfish Pathology. Academic Press; London: 1985. pp. 295–300. [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen N.J., Jorgensen P.E.V. Rapid detection of viral hemorrhagic septicaemia virus in fish by ELISA. J. Appl. Ichth. 1991;7:183–186. [Google Scholar]
- Parida M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 2004;42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha A., Ruiz S., Tafalla C., Coll J.M. Conformation- and fusion-defective mutations in the hypothetical phospholipids-binding and fusion peptides of viral hemorrhagic septicaemia salmonid Rhabdovirus protein G. J. Virol. 2004;78:9115–9122. doi: 10.1128/JVI.78.17.9115-9122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz F., Coll J. Techniques for diagnosing viral diseases of salmonid fish. Dis. Aquat. Organ. 1992;13:211–223. [Google Scholar]
- Snow M., Smail D.A. Experimental susceptibility of turbot Scophthalmus maximus to viral hemorrhagic septicaemia virus isolated from cultivated turbot. Dis. Aquat. Organ. 1999;38:163–168. doi: 10.3354/dao038163. [DOI] [PubMed] [Google Scholar]
- Strommen H.K., Stone D.M. Detection of VHS virus in fish tissues by semi-nested polymerase chain reaction (PCR) In: Barnes A.C., Davidson G.A., Hiney M.P., McIntosh D., editors. Methodology in Fish Disease Research. Fisheries Research Services; Aberdeen: 1998. [Google Scholar]
- Thai H.T.C., Le M.Q., Vuong C.D., Parida M., Minekawa H., Tsugunori N., Hasebe F., Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of sever acute respiratory syndrome Coronavirus. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergard Jorgensen The survival of VHS virus associated with trout eggs. Riv. Ital. Pisci. Ittiop. 1970;5:13–14. [Google Scholar]
- Vestergard Jorgensen Artificial transmission of VHS of rainbow trout. Riv. Ital. Pisci. Ittiop. 1973;8:101–102. [Google Scholar]
- Williams K., Blake S., Sweeney A., Singer J.T., Nicholson B.L. Multiplex reverse transcriptase PCR assay for simultaneous detection of three fish viruses. J. Clin. Microbol. 1999;37:4139–4141. doi: 10.1128/jcm.37.12.4139-4141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wizigmann G., Baath C., Hoffmann R. Isolation of viral hemorrhagic septicemia virus from fry of rainbow trout, pike, and grayling. Zentralbl. Vet. Reihe B. 1980;27:79–81. [PubMed] [Google Scholar]
- Wolf K. Cornell University Press; Ithaca: 1988. Fish Viruses and Fish Viral Diseases. p. 476. [Google Scholar]


