Highlights
-
•
BVDV NS5B-specific nanobodies were identified.
-
•
NS5B-specific nanobody Nb1 suppresses BVDV infection and replication.
-
•
Nb1 interacts with NS5B protein during BVDV infection.
Keywords: BVDV, Intrabody, Nanobody, NS5B, Antiviral effect
Abstract
Bovine viral diarrhea virus (BVDV) infection causes significant economic losses to the cattle industry worldwide and still represents a huge pressure on agricultural production. Thus, the development of novel anti-BVDV strategies are urgently needed. The nonstructural protein 5 (NS5B) of BVDV is essential for viral replication. Further, the camel single-domain antibody (nanobody) represents a promising antiviral approach with the advantages of small size, stable structure, high specificity and solubility, and the recognition of specific epitopes. However, no NS5B-specific nanobodies against BVDV have been reported. In this study, NS5B-specific nanobodies were isolated from a phage display library of variable domains of Camellidae heavy chain-only antibodies (VHHs). Further, an MDBK cell line stably expressing Nb1 was established to explore antiviral activity. Results showed that Nb1 could markedly suppress BVDV replication and interact with the BVDV NS5B protein. This suggests that nanobodies have potential for the development of novel antiviral drugs against BVDV infection.
1. Introduction
Bovine viral diarrhea (BVD), which is caused by BVD virus (BVDV) infection, is one of the most important viral diseases of cattle, causing enormous economic losses to the livestock industry worldwide (Suda et al., 2018). At the same time, it is also one of the main pollutants of cattle-derived biological products (Yarnall and Thrusfield, 2017). Due to the complexity of disease pathogenesis, there is no effective means to control or treat BVDV infection (Quinet et al., 2016). Therefore, it is imperative to develop an efficient antiviral strategy to combat BVDV infection in the cattle industry.
BVDV is a positive-strand RNA virus that belongs to the Pestivirus genus of the Flaviviridae family. The RNA genome of BVDV is approximately 12.5 kb, consisting of a single large open reading frame with a UTR on both the 5′ and 3′ ends and encodes a polymerized protein that is then processed by host and viral proteases into the capsid protein, three envelope glycoproteins (Erns, E1, E2) and seven or eight non-structural proteins (NSPs) comprising Npro, P7, NS2/3, NS4A, NS4B, NS5A, and NS5B (Collett et al., 1991; Tautz et al., 1997). The first four proteins are structural, whereas the rest are nonstructural and function in viral assembly, replication, and host immune evasion. NS5B, which is located at the carboxyl terminus of the polyprotein, is highly conserved among pestiviruses, and has been confirmed to possess RNA-dependent RNA polymerase (RdRp) activity, and is responsible for transcription and replication of the viral genome (Zhong et al., 1998). Due to these unique characteristics, NS5B is an ideal target for the development of antiviral drugs against BVDV.
Single-domain antibodies, also known as nanobodies (Nbs), are derived from the heavy chain antibody variable region (VHH) in camelids and considered as the smallest available intact antigen-binding fragments (Hamers-Casterman et al., 1993; Muyldermans et al., 2009). Nanobodies have desirable properties such as small volume (15 kDa), good antigen binding performance, strong tissue penetration and high stability (Zou et al., 2015); these attractive features make them beneficial for immunoassays and therapeutic applications. In addition, for the majority of nanobodies, their intrinsic stability is sufficient for proper folding and intracellular function (Rothbauer et al., 2006). For example, ALX-0171, a trivalent nanobody that binds fusion proteins on the surface of RSV, thereby inhibiting viral replication, is a novel, inhaled biotherapeutic; it is in development for the treatment of RSV infections in infants and is currently in a clinical phase III trial (Detalle et al., 2016; Larios Mora et al., 2018). In addition to some nanobodies as drugs for clinical applications, some studies have referred to the antiviral effects of specific nanobodies in vitro. For example, a novel nanobody Nb25 was found to effectively block duck HAV replication by targeting a conserved B cell epitope of this virus (Xue et al., 2019). Another nanobody NbMS10, which targets the MERS-CoV receptor-binding region significantly abrogates virus replication via its neutralization activity (Zhao et al., 2018). Further, Intracellularly-expressed Nb6 was found to potently suppress porcine reproductive and respiratory syndrome replication (Liu et al., 2015). Nevertheless, there have been no reports on the use of nanobodies for antiviral therapy against BVDV to date.
In this study, a Bactrian camel was immunized with soluble NS5B recombinant protein and a VHH library was constructed. Then, eight NS5B specific nanobodies were isolated by phage display and MDBK cell lines stably expressing these nanobodies were established using lentiviral packaging technology. in vitro results indicated that one of these nanobodies, Nb1 could strongly suppress BVDV replication. This is the first report of an anti-BVDV nanobody against NS5B, which could lead to the development of further novel anti-BVDV strategies.
2. Materials and methods
2.1. Ethics statement
The animal studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Northwest Agriculture and Forestry University (NWAFU). The animal protocols were approved by the IACUC of the (NWAFU) (20150017/08).
2.2. Cells and viruses
HEK293 T cells and Madin Darby Bovine Kidney (MDBK) cells were both purchased from China Center for Type Culture Collection (CCTCC, Beijing, China) and were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies Corporation, New York, CA, USA) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic-antimycotic (Life Technologies Corporation, Shanghai, China) at 37 °C in a 5% CO2 atmosphere.
BVDV strain Oregon C24 V (CVCC no. AV69) was obtained from the CCTCC and was propagated in MDBK cells, briefly described as follows, MDBK cells in T75 flasks were inoculated with 0.1 MOI of BVDV in 7 ml DMEM when cells reached 80% confluence. After incubating them for 1 h at 37 °C, the medium was replaced with fresh 3% FBS + DMEM and cells were further cultured for 48–60 h until abundant visible cytopathic effects (CPEs) were observed. Cells and supernatants were harvested and freeze-thawed three times, which was followed by centrifugation at 2000 × g for 10 min at 4 °C. Supernatants were harvested and aliquoted in 1.5 ml EP tubes. Viruses were titrated in MDBK cells and stored at −80 °C until further use.
2.3. Expression and purification of BVDV NS5B recombinant protein
The gene encoding the NS5B segment was amplified by PCR from BVDV cDNA and cloned into the pET28a prokaryotic expression vector (Novagen, Darmstadt, Germany), creating the recombinant plasmid pET28a-NS5B. The primers used for PCR amplification are listed in Supplemental Table 1. Then the pET28a-NS5B plasmids were transformed into E. coli Transetta (DE3) (Transgene Biotech, Beijing, China) cells to express NS5B protein through induction with 1 mM IPTG for 18 h at 25 °C. The bacteria were collected and centrifuged, followed by resuspension in buffer A (20 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole, and 1 mM DTT). After sonication and centrifugation, the cleared cell lysate was loaded onto a Ni-NTA column (Qiagen, Hilden, Germany) and eluted using imidazole (250 mM), which was followed by loading onto a Superdex 200 gel filtration column (GE Healthcare Life Science, Pittsburgh, USA). Peak fractions containing the NS5B-His recombinant protein was pooled and analyzed by SDS-PAGE and Western blotting.
2.4. Bactrian camel immunization and library construction
A 4 years old male Alashan Bactrian camel was immunized subcutaneously at days 1, 14, 28, 42, 56, and 70 with 5 mg soluble NS5B recombinant protein combined with Freund’s complete adjuvant according to a previous report (Vincke et al., 2012), and serum antibody titers after the last immunization were evaluated via indirect enzyme-linked immunosorbent assay (iELISA), described briefly as follows: 96-well microplates coated with NS5B-His recombinant protein (200 ng/well) were incubated with serially diluted camel serum, followed by incubation with mouse anti-camel antiserum (1:2000, prepared in our lab) as the secondary antibody and then HRP-conjugated goat anti-mouse IgG (1:5000, Jackson ImmunoResearch Laboratories, West Grove, PA, USA) as the third antibody. Serum antibody titers were calculated using GraphPad Prism 5.0.
After the last immunization, peripheral blood mononuclear cells (PBMCs) were isolated from a 250 ml blood sample using Leucosep® tubes (Greiner Bio-One, Frickenhausen, Germany). Then, 5 μg total RNA was extracted and cDNA was synthesized by reverse transcription-PCR (RT-PCR) using a TransScript cDNA Synthesis SuperMix (TransGen Biotech, China). Subsequently, the VHH genes were amplified by nested PCR (∼400 bp) and ligated into the phagemid vector pCANTAB 5E (GE Healthcare Life Science, Pittsburgh, USA) to construct a bimodal camel heavy chain antibody variable region library.
2.5. Screening and identification of NS5B-specific nanobodies
Biopanning was performed as previously described (Vincke et al., 2012) with the following modifications. NS5B-specific nanobodies were screened by three rounds of panning using phage display technology. Then, the enrichment of specific phage particles was monitored using anti-M13/HRP conjugate ELISA together with phage titration. From this, 121 colonies were picked randomly and induced with 1 mM IPTG to express soluble VHHs with an E-Tag. All recombinant VHHs-E-Tag proteins were extracted from the periplasm and tested for their capacity to bind the NS5B protein using iELISA with an anti-E-Tag antibody (Genscript, Piscataway, NJ, USA). Finally, all positive clones identified were sequenced, and nanobodies were grouped according to their CDR3 sequence. In addition, nanobodies with better affinity were selected for subsequent experiments.
2.6. Establishment of MDBK cell lines stably expressing nanobodies
VHH genes were amplified with primers Nb-F and Nb-R using the pCANTAB 5E-VHH plasmid as a template and inserted into the pTRIP-CMV-Puro vector (prepared in our laboratory) at the XbaI and BamHI restriction sites to construct the pTRIP-CMV-NbsEGFP-Puro recombinant plasmids. The sequences of primers are listed Supplementary Table 1. To produce pseudotyped lentivirus particles, HEK293 T cells in 6-well plates were co-transfected with pTRIP-CMV-NbsEGFP-Puro, psPAX2 and pMD2.0 G plasmids using X-tremeGENE HP DNA Transfection Reagent (Roche Life Science, USA) according to the manufacturer’s instructions. Subsequently, 48 h post transfection, when the green fluorescence reached approximately 80% observed under a fluorescence microscope, cell culture supernatants were collected and cell debris was removed by brief centrifugation at 10,000 rpm for 5 min, then cleared supernatants were 0.22 μM-filtered and stored at −80 °C until use.
MDBK cells were transduced with the pseudotyped lentiviruses expressing the nanobodies and supplemented with 1 μg/ml of polybrene (Sigma, St. Louis, MO, USA); 24 h later, transduced cells were screened by adding 2 μg/ml of puromycin (Thermo Fisher Scientific, Shanghai, China). Surviving cells were monitored using a fluorescence microscopy and the expression of nanobody-EGFP fusion protein was analyzed by Western blotting. MDBK cell lines stably expressing nanobody-EGFP fusion proteins were designated MDBK-NbsEGFP.
2.7. Cell viability assay
The viability of MDBK-NbsEGFP cell lines was evaluated using a Cell Counting Kit-8 (CCK-8) assay (Beyotime, Nanjing, China) as previously described (Xiao et al., 2014). Briefly, MDBK and MDBK-NbsEGFP cell lines were added to each well of 96-well plates (1 × 104/well) and cultured at 37 °C with 5% CO2. Cells were collected and counted after digestion with trypsin, assessing three wells per cell per day. Cell proliferation curves were plotted through statistics for one week of data.
2.8. Western blotting
Western blotting was performed as previously described (Zhang et al., 2017) with minor modifications. Cells were harvested and lysed using NP-40 lysis buffer (Beyotime, Shanghai, China) supplemented with protease inhibitor phenylmethanesulfonyl fluoride (PMSF), and the protein concentration was determined using a Bicinchoninic acid Protein Assay Kit (Thermo Fisher Scientific, Shanghai, China). Then, 50 μg of total protein was subjected to 12% SDS-PAGE and then transferred to PVDF membranes (Millipore, Billerica, MA, USA). The cellular proteins were probed with one of the following primary antibodies: mouse anti-BVDV NS5B polyclonal antibodies at a 1:1000 dilution produced in our laboratory (see Supplementary), mouse anti-GFP monoclonal antibody (Tiangen, Beijing, China) at a 1:1000 dilution and anti-α-tubulin antibody (Abcam, Cambridge, UK) at a 1:5000 dilution. This was followed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG at a 1:5000 dilution (Jackson,West Grove, PA, USA) as the secondary antibody. Immunolabeled proteins were visualized using an ECL chemiluminescent detection system according to the manufacturer’s instructions (Pierce, Rockford, IL, USA).
2.9. Quantitative reverse transcriptase PCR (RT-qPCR)
Total RNA was extracted from cells or supernatants using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). After measuring the concentration, 500 ng of total RNA was reverse transcribed into cDNA using a Primescript RT Reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. RT-qPCR was conducted using the StepOne Plus real-time PCR system (Applied Biosystems, Foster City, CA, USA) and FastStart Universal SYBR green master (Roche, Basle, Switzerland). Reactions were performed in a 10 μl volume, and the reaction conditions were as follows: 95 °C 10 min, 40 cycles of 95 °C for 20 s, 55 °C for 30 s and 72 °C for 20 s. Beta-2-microglobulin (B2M) mRNA was determined simultaneously as the internal control, and the 2−ΔΔCt method was used to calculate the relative gene expression levels. Primers used for RT-qPCR amplification are listed in Supplemental Table 1.
To evaluate BVDV RNA copies in culture supernatants, a plasmid containing a 357 bp fragment of the BVDV 5′UTR was utilized to generate a standard curve, which was plotted from the results of parallel PCRs performed with serial dilutions of the standard plasmid. Absolute quantities of supernatant RNA copies were calculated according to the standard curve.
2.10. Virus titration assay
Virus titration detection was performed as described previously (Xiao et al., 2014). Briefly, MDBK cells were trypsinized and plated in 96-well cell culture plate at a density of 1.0 × 104 cells/well 24 h before infection with BVDV. Sample supernatants were 10-fold serially diluted and 100 μl was added to each well with eight replicates. Six days after infection, the 50% tissue culture infective dose (TCID50) was calculated based on the Reed-Muench method.
2.11. Co-immunoprecipitation
BVDV-infected cells were homogenized in NP40 lysis buffer (Beyotime, Nanjing, China). After centrifugation at 12,000 rpm for 10 min at 4 °C, the supernatant was incubated with GFP-Trap®_A beads (Chromotek, Germany) at 4 °C for 2 h. Then the beads were washed with PBS three times, which was followed by resuspension in SDS sample buffer and boiling for 10 min at 100 °C. Immunoprecipitated proteins were analyzed by Western blotting using a mouse anti-GFP monoclonal antibody (Tiangen, Beijing, China) and mouse anti-NS5B polyclonal antibody.
2.12. Statistical analysis
All results are based on at least three independent experiments. Statistical significance was determined using a Student’s t-test if two groups were compared and one-way analysis of variance (ANOVA) when more than two groups were compared to the control group. A p value <0.05 was considered statistically significant.
3. Results
3.1. Preparation of antigen and construction of a VHH library from an NS5B immunized camel
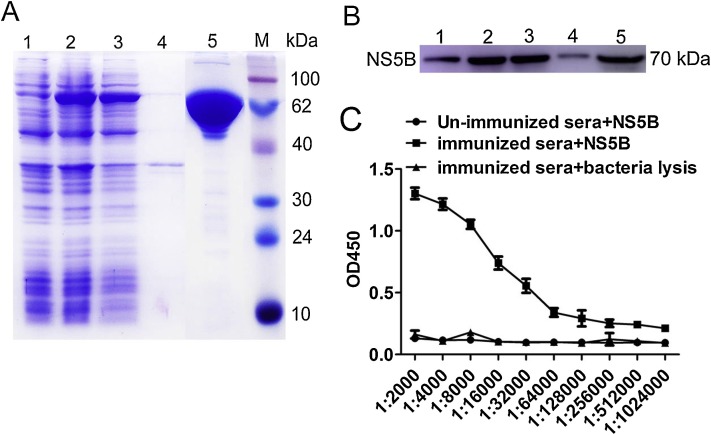
Soluble NS5B recombinant protein was expressed in E. coli Transetta (DE3) cells transformed with pET28a-NS5B vector after induction with 1 mM IPTG at 25 °C for 18 h (data not shown). Then, the NS5B-His recombinant protein was purified using a Ni-NTA chromatography column and subsequently a S200 gel filtration column. SDS-PAGE identification results showed that the molecular weight of NS5B-His recombinant protein was approximately 70 kDa (Fig. 1 A). The expressed and purified N-His recombinant protein was confirmed by Western blotting using an anti-His monoclonal antibody, which resulted in the observation of target protein bands of 70 kDa (Fig. 1B). The purified NS5B-N-His recombinant protein was then used for immunization and to screen specific-nanobodies as coating antigens. Before and after immunization, NS5B-specific antibodies in serum samples of the Alashan Bactrian camel were determined by iELISA. These results indicated that the anti-serum titer against NS5B increased to 1:128,000 after the last immunization, compared to that before immunization (Fig. 1C). A phage display VHH library, consisting of approximately 4.2 × 108 individual colonies, was built from PBMCs of the immunized camel. Positive rate analysis by colony PCR revealed that 98% of these colonies contained the correct insert corresponding to the sizes of VHH genes. Next, 50 randomly selected clones were sequenced and analyzed, and each clone was determined to contain a distinct VHH sequence, suggesting that the diversity of the antibody library was good and it had high quality.
Fig. 1.
Expression, purification, and identification of BVDV NS5B recombinant protein and detection of serum anti-NS5B antibody titer. (A) The expression and purification of NS5B recombinant protein were analyzed by SDS-PAGE. Lane 1: pET28a vector control; lane 2: induction with 1 mM IPTG; lane 3: soluble protein in supernatant after sonication; lane 4: inclusion body in precipitate after sonication; lane 5: purified NS5B protein; M: molecular weight markers, size indicated in kDa. (B) Antigenic analysis of Western blotting, lanes 1–5: same as (A); samples were reacted with anti-His antibodies. (C) Determination of serum antibody titers after immunization by iELISA.
3.2. Isolation and identification of NS5B-specific nanobodies
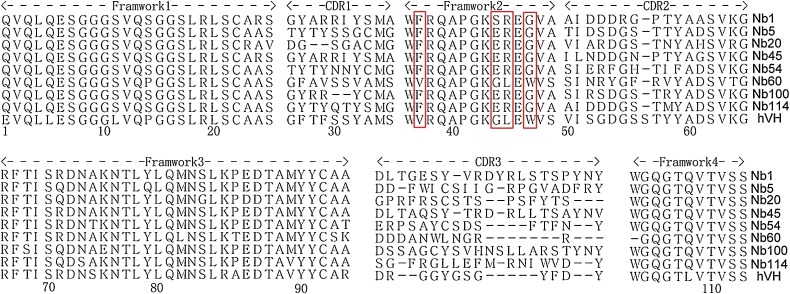
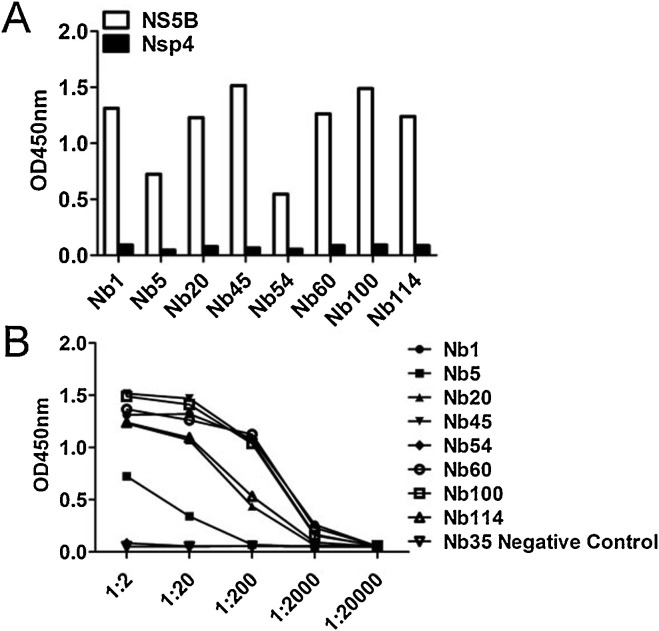
To screen NS5B-specific nanobodies, biopanning was conducted with immobilized recombinant NS5B-His protein. The NS5B-specific VHH recombinant phage was well enriched after three rounds of panning (Table 1 ). Then, 121 clones were randomly selected from the third round of screening for further iELISA detection. iELISA results revealed that 116 of the 121 clones were positive (data not shown). Next, these 116 colonies were sequenced and eight different NS5B-specific nanobodies were eventually screened based on the amino acid sequence classification of the antibody CDR3 hypervariable region; these were named Nb1, Nb5, Nb20, Nb45, Nb54, Nb60, Nb100, and Nb114. The deduced amino acid sequences were aligned against the human VH sequence, numbering and CDRs followed the methods described by Kabat et al (Kabat and Wu, 1991). The data in Fig. 2 show that except for Nb60, the other seven nanobodies had typical hydrophilic amino acid substitutions in the FR2 region. Interestingly, Nb60 also displayed notable reactivity against NS5B. In addition, iELISA results indicated that all eight nanobodies could specifically bind the NS5B recombinant protein, but did not cross-react with PRRSV-NSP4 recombinant protein (Fig. 3 A). PRRSV NSP4 recombinant protein was expressed and purified using the same vector pET-28a and method used for BVDV NS5B. This protein also contained a 6×His-Tag, excluding the possibility that these nanobodies might recognize the 6×His region. Of all nanobodies, Nb1, Nb45 and Nb100 displayed the highest affinity (Fig. 3B). Based on these results, these three nanobodies were selected for subsequent experimental studies and the negative nanobody Nb35 was selected simultaneously as a negative control.
Table 1.
Enrichment of NS5B-specific phages during three rounds of panning.
| Round of panning | Phage input (PFU/Well) | Phage output (PFU/Well) | Recovery rate | Enrichment |
|---|---|---|---|---|
| 1st round | 5 × 1010 | 2.3 × 105 | 4.6 × 10−6 | 5.8 |
| 2nd round | 5 × 1010 | 9.5 × 106 | 1.9 × 10−4 | 2.2 × 102 |
| 3rd round | 5 × 1010 | 6.1 × 107 | 1.22 × 10−3 | 1.6 × 103 |
Fig. 2.
Amino acid sequence alignment of NS5B-specific nanobodies with human VH. Numbering and CDRs are in accordance with the previously described methods of Kabat et al (9). The hallmark residues at positions 37, 44, 45, and 47 are highlighted by a red box (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Fig. 3.
Specificity and affinity of screened nanobodies against NS5B. (A) Detection of the binding of the eight unique nanobodies against NS5B using iELISA; PRRSV-Nsp4-His protein was used as a control. (B) Titration of binding the eight screened nanobodies with the NS5B protein by iELISA; Nb35 was used as a negative antibody control, 4 µg/ml NS5B protein was the coating antigen; different dilutions of nanobodies were added; E-tag as a primary antibody.
3.3. Generation of MDBK cell lines stably expressing nanobodies
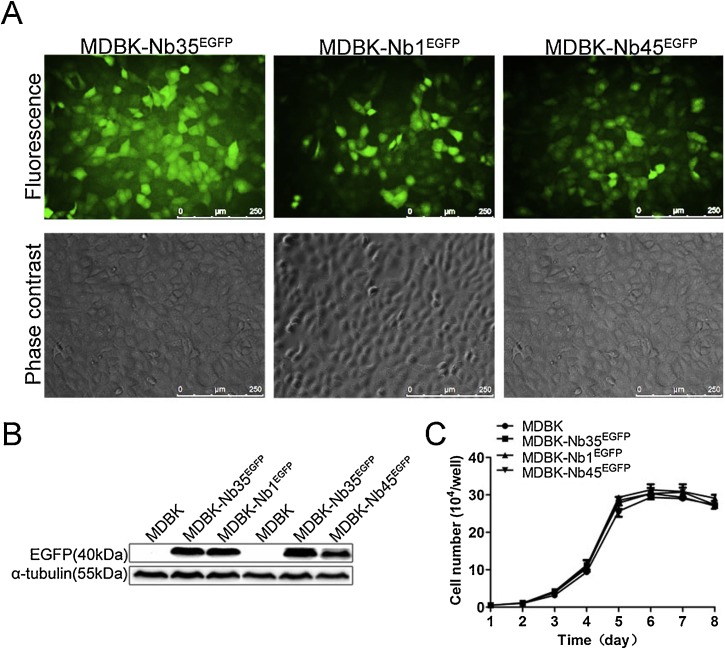
To generate cell lines stably expressing Nbs, supernatants containing the pseudotyped lentiviruses with 1 μg/ml of polybrene were incubated with MDBK cells and screened using 2 μg/ml puromycin after 24 h of transduction. Surviving cells were imaged using a fluorescence microscope and named MDBK-Nb1EGFP, MDBK-Nb45EGFP and MDBK-Nb35EGFP, respectively (Fig. 4 A). Then the cell lines were validated by Western blotting with anti-GFP antibody. Western blotting results indicated that bands of interest were detected in MDBK-Nb1EGFP, MDBK-Nb45EGFP and MDBK-Nb35EGFP cell lines (40 kDa), but not in wild-type MDBK cells (Fig. 4B), confirming that MDBK cell lines stably expressing Nbs were successfully established. Next, to determine whether the intracellular expression of nanobodies is cytotoxic, cell proliferation assays, using recombinant MDBK-NbsEGFP cell lines and wild-type MDBK cells were performed and growth curves were plotted over time. The results showed that the stable expression of intracellular nanobodies in MDBK-NbsEGFP cell lines were not cytotoxic, and no remarkable differences were observed between recombinant and the wild-type cells (Fig. 4C). Since pre-experimental results implied that the MDBK-Nb100EGFP cell line had no antiviral activity and the MDBK-Nb45EGFP cell line showed weaker antiviral activity, but the MDBK-Nb1EGFP cell line has strong antiviral effects (Fig. S1), the MDBK-Nb1EGFP cell line was utilized eventually to further explore the antiviral effect of nanobodies.
Fig. 4.
MDBK cell lines stably expressing nanobody-EGFP fusion proteins. (A) Characterization of MDBK cell lines were performed by fluorescent microscopy. The scale bar indicates 250 μm. (B) Expression of nanobody-EGFP proteins was detected by western blotting using an anti-GFP monoclonal antibody. (C) Growth curves of MDBK-NbsEGFP cell lines. Data are shown as the mean ± SD representative of three independent experiments performed in triplicate.
3.4. Intracellularly-expressed Nb1 suppresses BVDV infection and replication
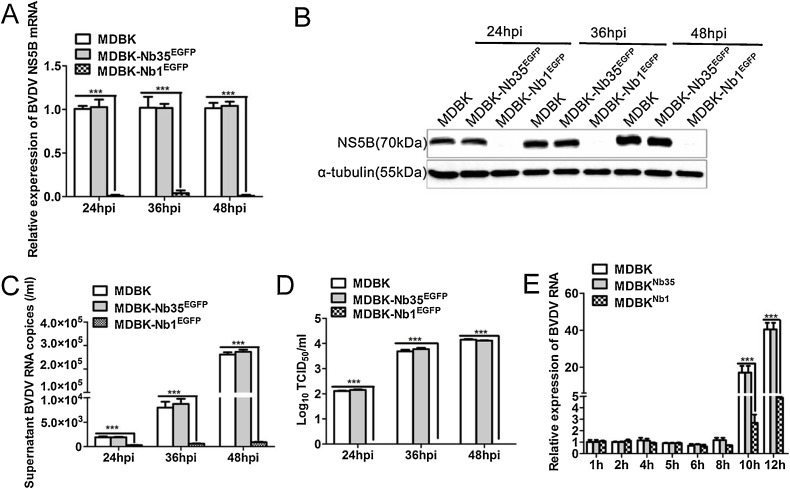
To evaluate the effect of intracellular Nb1 expression on BVDV infection, MDBK, MDBK-Nb35EGFP and MDBK-Nb1EGFP cells were infected with 0.05 MOI of BVDV, and cells and supernatants were harvested at 24, 36 and 48 h post-infection (hpi). RT-qPCR and Western blotting were performed to detect NS5B mRNA and protein expression, whereas TCID50 assays were applied to detect supernatants virus titers.
RT-qPCR results showed that BVDV NS5B mRNA and protein expression was almost undetectable at 24, 36 and 48 h after BVDV infection in MDBK-Nb1EGFP cells (Fig. 5 A and B), and that the number of BVDV RNA copies in the supernatant from these cells was also decreased significantly (Fig. 5C). However, NS5B mRNA, protein levels and the number of virus copies in MDBK-Nb35EGFP cells as a control group was not significantly different from those in MDBK cells (Fig. 5A–C). Furthermore, an evaluation of progeny virus titers in the supernatant revealed similar results; that is, the virus titer was 0 at different time points after BVDV infection in MDBK-Nb1EGFP cells (Fig. 5D). These results imply that the intracellular expression of Nb1 in MDBK cells can almost completely interfere with the replication of 0.05 MOI BVDV.
Fig. 5.
Intracellularly-expressed Nb1 inhibits BVDV replication. MDBK, MDBK-Nb35EGFP and MDBK-Nb1EGFP cells were infected with 0.05 MOI BVDV for 24, 36, and 48 h, and culture supernatants and cells were collected. Expression in cell lysates was detected by RT-qPCR (A), and western blotting (B) using an anti-NS5B polyclonal antibody or anti-α-tubulin antibody. Extracellular BVDV RNA was analyzed (C) and the virus titers in the culture supernatants were measured based on the TCID50 value (D). MDBK, MDBK-Nb35EGFP, and MDBK-Nb1EGFP cells were infected with 1 MOI BVDV for 1, 2, 4, 5, 6, 8, 10, and 12 h, and cells were collected for RT-qPCR (E). Data are shown as the mean ± SD representative of three independent experiments. p values were calculated using ANOVA. Significant differences, as compared to control values are denoted as ***(p < 0.001).
It is known that as an RdRp, NS5B plays a critical role in BVDV RNA synthesis. To determine the effect of Nb1 on BVDV replication in the early stages of infection, RT-qPCR was performed within 12 h of inoculation. Results showed that Nb1 had no significant effect on BVDV replication in the early stages of infection except for at 10 and 12 hpi, implying that it mainly exerts its antiviral effect during the late stage of BVDV infection (10, 12, 24, 36, and 48 hpi, etc), and not at early stage of infection (0–10 hpi) (Fig. 5E).
3.5. Nb1 suppresses BVDV replication of different MOIs
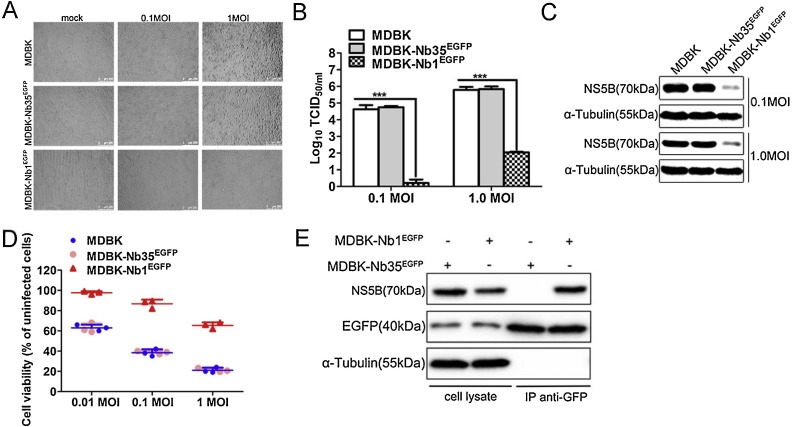
To further verify the effect of Nb1 intrabodies on viral replication, MDBK-NbsEGFP cells were infected with BVDV at MOIs of 0.1 and 1. Then, BVDV-induced CPEs were monitored at 48 hpi. Results indicated that BVDV-infected MDBK and MDBK-Nb35EGFP cells showed significant CPEs, whereas those in MDBK-Nb1EGFP cells were not as obvious (Fig. 6 A). In addition, cells and supernatants were harvested to assess NS5B protein expression and viral titers, respectively. Western blotting results showed that Nb1 could markedly abrogated NS5B protein expression at different MOIs (Fig. 6C). Similarly, titration assay results suggested that Nb1 could significantly prohibit BVDV replication at 48 hpi, resulting in approximately 10000-fold and 1000-fold decreases in viral titers at MOIs of 0.1 and 1, respectively, compared to those in MDBK and MDBK-Nb35EGFP cells (Fig. 6B). These results suggest that Nb1 intrabodies can obviously inhibit BVDV replication of different MOIs.
Fig. 6.
Nb1 suppresses BVDV replication of different MOIs and interacts with BVDV-encoded NS5B protein. MDBK, MDBK-Nb35EGFP and MDBK-Nb1EGFP cell lines were infected with BVDV at MOIs of 0.1 and 1 for 48 hpi. BVDV-induced CPEs were monitored under an inverted microscope (A). The scale bar indicates 250 μm. BVDV production was analyzed based on TCID50 values (B). BVDV-NS5B protein levels were measured by western blotting with α-tubulin as the control (C). Cell viability was assessed using the CCK-8 method at 48 hpi (D). Interaction between intracellularly expressed Nb1 and BVDV-encoded NS5B in MDBK cells infected with BVDV. MDBK-Nb1EGFP and MDBK-Nb35EGFP cell lines were infected with 1 MOI of BVDV, and cells were collected at 72 hpi. Immunoprecipitated proteins were analyzed by western blotting using a mouse anti-GFP monoclonal antibody and a mouse anti-NS5B polyclonal antibody (E). Data are shown as the mean ± SD representative of three independent experiments. p values were calculated using ANOVA. Significant differences, as compared to control values are denoted as ***(p < 0.001).
To further evaluate the antiviral capacity of Nb1 intrabodies, the aforementioned three cell lines were infected with BVDV at MOIs of 0.01, 0.1 and 1, and cell viability was evaluated at 48 hpi using CCK-8 assays. Results indicated that Nb1 intrabodies promoted cell survival at all three MOIs compared to that in MDBK and MDBK-Nb35EGFP cells (Fig. 6D), further confirming the anti-BVDV effects of Nb1 intrabodies.
3.6. Intracellularly expressed Nb1 interacts with BVDV-encoded NS5B protein
To confirm the interaction between intracellularly-expressed Nb1 and BVDV-encoded NS5B in BVDV-infected cells, co-immunoprecipitation (Co-IP) assays were performed on lysates of BVDV-infected MDBK cell lines stably expressing nanobodies. MDBK-Nb1EGFP and MDBK-Nb35EGFP cell lines were infected with 1 MOI of BVDV, and cells were collected at 72 hpi for Co-IP assays. Results revealed that Nb1, but not Nb35, could interact with BVDV-encoded NS5B in BVDV-infected cells (Fig. 6E). These results suggest that intracellularly-expressed Nb1 might recognize the functional epitope of NS5B and inhibit the replication of viral genes, thereby inhibiting the replication of BVDV in MDBK cells.
4. Discussion
Since the advent of hybridoma technology in 1975, monoclonal antibodies have been widely used in scientific research and medical diagnosis and treatment (Liu, 2014). However, despite their huge potential for development, monoclonal antibodies still have apparent limitations, such as complex structure, large molecular weights, high development and production costs, which limit their applications in related fields. With the development of genetic engineering technology, the emerging nanobody has greatly improved antibody performance, with its unique advantages such as small size (15 kDa), high specificity and affinity, and recognition of specific antigenic epitopes (Dmitriev et al., 2016; Hassanzadeh-Ghassabeh et al., 2013). In addition, according to a previous report, nanobodies are more soluble due to typical hydrophilic amino acid substitutions (Vu et al., 1997); thus, we speculate that the substitution of hydrophilic amino acid to hydrophobic amino acid in this study might further increase the hydrophilicity of NS5B-specific nanobodies (Fig. 2).
Intrabodies represent an important class of therapeutic drugs with potential uses for microbial diseases therapy, and they can interrupt early and late events of the viral life cycle (Marasco, 2001; Rondon and Marasco, 1997). Recent studies have shown that several camel single domain intrabodies targeting the HIV-1 Rev protein, the IFV nucleoprotein, and the HCV envelope protein could exert antiviral activity (Ashour et al., 2015; Boons et al., 2014; Serruys et al., 2009), indicating that intracellularly-expressed Nbs are very effective in protecting the host against viral infection. BVD is of great concern worldwide to the bovine industry, due to its complex genetic variability and lack of effective therapeutic drugs that can be used to date (Curti and Jaeger, 2013). Whether intracellularly-expressed nanobodies exert an antiviral effect during BVDV infection and whether they can be used for BVD treatment were not clear. Thus, it is necessary to determine the effect of intracellularly-expressed nanobodies on BVDV infection.
To investigate such effects, MDBK cell lines were constructed to stably express intracellular Nbs against NS5B. NS5B has been shown to possess RdRp activity and plays a pivotal role in the transcription and replication of the BVDV genome (Choi et al., 2004). Therefore, NS5B-specific nanobodies were screened using phage display in this study to explore their anti-BVDV effect. Here, to facilitate this study, stable cell lines were generated by first packaging the lentivirus and then transducing it into MDBK cells, which not only avoided problems associated with individual transfections but also resulted in stable and efficient expression of Nbs fusion proteins. Further, to clarify that nanobodies were successfully delivered to the cells, a C-terminal EGFP was fused to the Nbs via an IgA-hinge linker to act as a marker and facilitate Co-IP detection (Fig. 4, Fig. 6). It has been reported that fluorescent protein-nanobody fusion proteins of the HIV-1 capsid protein can recognize and dynamically trace HIV-1 morphogenesis in living cells, and this novel fluorescent biosensor provides a robust tool for cell-based HIV research (Helma et al., 2012). This suggests that the NS5B-specific nanobodies identified here might also have the potential to act as a tool for the direct and dynamic visualization of NS5B in BVDV-infected living cells. Results of subsequent virus infection experiments showed that a novel nanobody, named Nb1, which specifically targets BVDV NS5B, significantly abolished viral replication (Fig. 5, Fig. 6), demonstrating that intracellularly-expressed nanobodies can suppress BVDV infection in vitro.
In this study, three NS5B-specific Nbs with high affinity were selected to explore their anti-BVDV activity. Unexpectedly, even though all three Nbs showed similar in vitro affinities for NS5B, the degree of inhibition of viral replication was completely different. Nb100 had no antiviral activity and Nb45 showed weaker antiviral activity, whereas Nb1 exhibited a strong antiviral effect (Fig. S1). We speculate that this is because different Nbs might bind different NS5B epitopes, and that Nb1, but not Nb100, could bind the key epitope of NS5B, inhibiting its RdRp activity and thus exerting strong antiviral effects. The NS5B protein is mainly distribute in the cytoplasm of BVDV-infected cells, and the replication and transcription of viral genes also primarily occurs in the cytoplasm, implying that Nbs might exert antiviral activity mainly by targeting NS5B protein in the cytoplasm. Surprisingly, our IFA and Western blotting results both clearly indicated that the Nbs were mainly distributed in the nucleus, and only a small portion were identified in the cytoplasm (Fig. S2). In view of these inconsistent results inconsistency in the subcellular localization of NS5B protein and Nbs, the anti-viral molecular mechanism of Nbs needs to be further researched, and we are currently working on this.
For RNA viruses, the genome has high variability, and the neutralizing epitope regions of viral structural proteins tend to have high variability due to immune pressure, whereas the functional regions of non-structural proteins located in the cytoplasm are relatively conserved (Chand et al., 2012). The BVDV-NS5B protein, which is highly conserved in the genus Pestivirus and has its own unique function as a non-structural proteins, plays an important role in viral replication, as it has RdRp activity (Choi et al., 2004). This provides an ideal target for the development of anti-BVDV drugs. Due to advantages that are unmatched by traditional antibodies, Nbs have broad value for clinical application (Pardon et al., 2014; Siontorou, 2013). However, The biggest problem associated with intracellular antibodies is the delivery of related expression genes into cells, while several possible strategies have been developed to overcome this problem, such as nanobody gene delivery into cells via an intrabody gene therapy delivery strategy (Southwell et al., 2009) and a trans-activating transduction (TAT) peptide Nb6 fusion to promote protein entry into cells (Wang et al., 2019). These reports provide a useful reference for research on and the application of intracellular nanobodies against BVDV, and this represents the ongoing work in our laboratory.
5. Conclusion
In conclusion, BVDV NS5B-specific nanobodies were isolated from a camel VHH library by phage display technology. Further, one of the nanobodies, Nb1, was shown to significantly suppress BVDV replication by interacting with BVDV-encoded NS5B. These results suggest that intracellularly expressed Nb1 has the potential to be developed as an anti-BVDV agent.
Declaration of Competing Interest
None.
Acknowledgements
This study was partially funded by the National Key R&D Project [grant number 2017YFD0501102]; and The Youth Innovation Team of Shaanxi Universities.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.vetmic.2019.108449.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Ashour J., Schmidt F.I., Hanke L., Cragnolini J., Cavallari M., Altenburg A., Brewer R., Ingram J., Shoemaker C., Ploegh H.L. Intracellular expression of camelid single-domain antibodies specific for influenza virus nucleoprotein uncovers distinct features of its nuclear localization. J. Virol. 2015;89:2792–2800. doi: 10.1128/JVI.02693-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boons E., Li G., Vanstreels E., Vercruysse T., Pannecouque C., Vandamme A.M., Daelemans D. A stably expressed llama single-domain intrabody targeting Rev displays broad-spectrum anti-HIV activity. Antiviral Res. 2014;112:91–102. doi: 10.1016/j.antiviral.2014.10.007. [DOI] [PubMed] [Google Scholar]
- Chand R.J., Trible B.R., Rowland R.R. Pathogenesis of porcine reproductive and respiratory syndrome virus. Curr. Opin. Virol. 2012;2:256–263. doi: 10.1016/j.coviro.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Choi K.H., Groarke J.M., Young D.C., Kuhn R.J., Smith J.L., Pevear D.C., Rossmann M.G. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4425–4430. doi: 10.1073/pnas.0400660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett M.S., Wiskerchen M., Welniak E., Belzer S.K. Bovine viral diarrhea virus genomic organization. Arch. Virol. Suppl. 1991;3:19–27. doi: 10.1007/978-3-7091-9153-8_3. [DOI] [PubMed] [Google Scholar]
- Curti E., Jaeger J. Residues Arg283, Arg285, and Ile287 in the nucleotide binding pocket of bovine viral diarrhea virus NS5B RNA polymerase affect catalysis and fidelity. J. Virol. 2013;87:199–207. doi: 10.1128/JVI.06968-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detalle L., Stohr T., Palomo C., Piedra P.A., Gilbert B.E., Mas V., Millar A., Power U.F., Stortelers C., Allosery K., Melero J.A., Depla E. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob. Agents Chemother. 2016;60:6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev O.Y., Lutsenko S., Muyldermans S. Nanobodies as probes for protein dynamics in vitro and in cells. J. Biol. Chem. 2016;291:3767–3775. doi: 10.1074/jbc.R115.679811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- Hassanzadeh-Ghassabeh G., Devoogdt N., De Pauw P., Vincke C., Muyldermans S. Nanobodies and their potential applications. Nanomedicine. 2013;8:1013–1026. doi: 10.2217/nnm.13.86. [DOI] [PubMed] [Google Scholar]
- Helma J., Schmidthals K., Lux V., Nuske S., Scholz A.M., Krausslich H.G., Rothbauer U., Leonhardt H. Direct and dynamic detection of HIV-1 in living cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat E.A., Wu T.T. Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J. Immunol. 1991;147:1709–1719. [PubMed] [Google Scholar]
- Larios Mora A., Detalle L., Gallup J.M., Van Geelen A., Stohr T., Duprez L., Ackermann M.R. Delivery of ALX-0171 by inhalation greatly reduces respiratory syncytial virus disease in newborn lambs. mAbs. 2018;10:778–795. doi: 10.1080/19420862.2018.1470727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang Y., Duan H., Zhang A., Liang C., Gao J., Zhang C., Huang B., Li Q., Li N., Xiao S., Zhou E.M. An intracellularly expressed Nsp9-specific nanobody in MARC-145 cells inhibits porcine reproductive and respiratory syndrome virus replication. Vet. Microbiol. 2015;181:252–260. doi: 10.1016/j.vetmic.2015.10.021. [DOI] [PubMed] [Google Scholar]
- Liu J.K. The history of monoclonal antibody development - Progress, remaining challenges and future innovations. Ann. Med. Surg. 2014;3:113–116. doi: 10.1016/j.amsu.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco W.A. Intrabodies as antiviral agents. Curr. Top. Microbiol. Immunol. 2001;260:247–270. doi: 10.1007/978-3-662-05783-4_13. [DOI] [PubMed] [Google Scholar]
- Muyldermans S., Baral T.N., Retamozzo V.C., De Baetselier P., De Genst E., Kinne J., Leonhardt H., Magez S., Nguyen V.K., Revets H., Rothbauer U., Stijlemans B., Tillib S., Wernery U., Wyns L., Hassanzadeh-Ghassabeh G., Saerens D. Camelid immunoglobulins and nanobody technology. Vet. Immunol. Immunopathol. 2009;128:178–183. doi: 10.1016/j.vetimm.2008.10.299. [DOI] [PubMed] [Google Scholar]
- Pardon E., Laeremans T., Triest S., Rasmussen S.G., Wohlkonig A., Ruf A., Muyldermans S., Hol W.G., Kobilka B.K., Steyaert J. A general protocol for the generation of Nanobodies for structural biology. Nat. Protoc. 2014;9:674–693. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet C., Czaplicki G., Dion E., Dal Pozzo F., Kurz A., Saegerman C. First results in the use of bovine ear notch tag for bovine viral diarrhoea virus detection and genetic analysis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0164451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon I.J., Marasco W.A. Intracellular antibodies (intrabodies) for gene therapy of infectious diseases. Annu. Rev. Microbiol. 1997;51:257–283. doi: 10.1146/annurev.micro.51.1.257. [DOI] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Tillib S., Nowak D., Schermelleh L., Gahl A., Backmann N., Conrath K., Muyldermans S., Cardoso M.C., Leonhardt H. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- Serruys B., Van Houtte F., Verbrugghe P., Leroux-Roels G., Vanlandschoot P. Llama-derived single-domain intrabodies inhibit secretion of hepatitis B virions in mice. Hepatology. 2009;49:39–49. doi: 10.1002/hep.22609. [DOI] [PubMed] [Google Scholar]
- Siontorou C.G. Nanobodies as novel agents for disease diagnosis and therapy. Int. J. Nanomed. 2013;8:4215–4227. doi: 10.2147/IJN.S39428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell A.L., Ko J., Patterson P.H. Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington’s disease. J. Neurosci. 2009;29:13589–13602. doi: 10.1523/JNEUROSCI.4286-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda Y., Murakami S., Horimoto T. Bovine viral diarrhea virus non-structural protein NS4B induces autophagosomes in bovine kidney cells. Arch. Virol. 2018 doi: 10.1007/s00705-018-4045-x. [DOI] [PubMed] [Google Scholar]
- Tautz N., Elbers K., Stoll D., Meyers G., Thiel H.J. Serine protease of pestiviruses: determination of cleavage sites. J. Virol. 1997;71:5415–5422. doi: 10.1128/jvi.71.7.5415-5422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincke C., Gutierrez C., Wernery U., Devoogdt N., Hassanzadeh-Ghassabeh G., Muyldermans S. Generation of single domain antibody fragments derived from camelids and generation of manifold constructs. Methods Mol. Biol. 2012;907:145–176. doi: 10.1007/978-1-61779-974-7_8. [DOI] [PubMed] [Google Scholar]
- Vu K.B., Ghahroudi M.A., Wyns L., Muyldermans S. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol. Immunol. 1997;34:1121–1131. doi: 10.1016/s0161-5890(97)00146-6. [DOI] [PubMed] [Google Scholar]
- Wang L., Zhang L., Huang B., Li K., Hou G., Zhao Q., Wu C., Nan Y., Du T., Mu Y., Lan J., Chen H., Zhou E.M. A nanobody targeting viral nonstructural protein 9 inhibits porcine reproductive and respiratory syndrome virus replication. J. Virol. 2019;93 doi: 10.1128/JVI.01888-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Zhang A., Zhang C., Ni H., Gao J., Wang C., Zhao Q., Wang X., Wang X., Ma C., Liu H., Li N., Mu Y., Sun Y., Zhang G., Hiscox J.A., Hsu W.H., Zhou E.M. Heme oxygenase-1 acts as an antiviral factor for porcine reproductive and respiratory syndrome virus infection and over-expression inhibits virus replication in vitro. Antiviral Res. 2014;110:60–69. doi: 10.1016/j.antiviral.2014.07.011. [DOI] [PubMed] [Google Scholar]
- Xue W., Zhao Q., Li P., Zhang R., Lan J., Wang J., Yang X., Xie Z., Jiang S. Identification and characterization of a novel nanobody against duck hepatitis A virus type 1. Virology. 2019;528:101–109. doi: 10.1016/j.virol.2018.12.013. [DOI] [PubMed] [Google Scholar]
- Yarnall M.J., Thrusfield M.V. Engaging veterinarians and farmers in eradicating bovine viral diarrhoea: a systematic review of economic impact. Vet. Rec. 2017;181:347. doi: 10.1136/vr.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Zhao L., Li N., Duan H., Liu H., Pu F., Zhang G., Zhou E.M., Xiao S. Carbon monoxide inhibits porcine reproductive and respiratory syndrome virus replication by the cyclic GMP/Protein kinase g and NF-kappaB signaling pathway. J. Virol. 2017;91 doi: 10.1128/JVI.01866-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., He L., Sun S., Qiu H., Tai W., Chen J., Li J., Chen Y., Guo Y., Wang Y., Shang J., Ji K., Fan R., Du E., Jiang S., Li F., Du L., Zhou Y. A novel nanobody targeting middle east respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain has potent cross-neutralizing activity and protective efficacy against MERS-CoV. J. Virol. 2018:92. doi: 10.1128/JVI.00837-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W., Gutshall L.L., Del Vecchio A.M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J. Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T., Dembele F., Beugnet A., Sengmanivong L., de Marco A., Li M.H. Nanobody-functionalized polymersomes. J. Control. Release. 2015;213:e79–80. doi: 10.1016/j.jconrel.2015.05.132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.