Highlights
-
•
PEDV fails to replicate in porcine Mo-DC.
-
•
PEDV does not compromise the viability of porcine Mo-DC.
-
•
PEDV activates the transcription of type I interferon.
-
•
PEDV activates the transcription of IP-10.
Keywords: PEDV, Mo-DC, IP-10, Type I interferon
Abstract
Porcine epidemic diarrhea virus (PEDV) belongs to the alphacoronavirus of the Coronaviridae. It is the major etiological agent of the recent outbreaks of piglet diarrhea and death in the US. Limited knowledge is currently available regarding the role of dendritic cells in PEDV infection. Here, we observed that PEDV did not replicate in monocyte-derived dendritic cells as evidenced by the decrease of viral gene transcript copies in infected cells by qRT-PCR and the absence of viral proteins by immunofluorescence staining as well as the absence of virus particles in infected cells by transmission electron microscopy. In addition, PEDV did not compromise cell viability at 48, 72, and 96 h after infection at either a MOI of 2.5 or 5. Interestingly, an increased transcription of type I interferon including interferon-α and β was observed in infected cells compared to mock infected cells. Surprisingly, we did not detect any interferon-β in the supernatants of infected cells. A slight increase in interferon-α protein production in the supernatants of PEDV-infected cells was observed compared to mock infected cells. We also observed a markedly increased transcription of interferon inducible protein −10 (IP-10). Overall, PEDV does not replicate in porcine Mo-DC, but activates the transcription of type I interferon and chemokine IP-10.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) is a member of the Coronaviridae and has a positive sense, single-stranded RNA genome of approximately 27 kb (Brian and Baric, 2005, Kocherhans et al., 2001). PEDV is genetically related to porcine transmissible gastroenteritis virus (TGEV), also a member of the Coronavirdae. Both PEDV and TGEV cause diarrhea and death in young piglets, which results in significant economic losses worldwide. The recent emergence of PEDV in the US swine industry poses a great concern on the control and prevention of the devastating disease in young piglets. The intentional exposure of pregnant sows to PEDV has been shown to induce durable lactogenic immunity, which can provide some protection of neonatal piglets against PEDV (Goede et al., 2015). Although two vaccines have been conditionally approved for use in diseased farms in the US, the duration of immunity induced by these vaccines in pregnant sows has not been studied in detail. Furthermore, the innate and adaptive immunity against PEDV infections are largely unknown and remain to be explored. Dendritic cells (DC) are professional antigen-presenting cells that bridge innate and adaptive immunity. Given the essential role of dendritic cells (DC) in antigen presentation and immune cell activation, a better understanding on the interaction between PEDV and DC will facilitate the design of vaccine candidates that are capable of inducing potent and long-lasting immunity.
PEDV is mainly detected in the intestinal epithelial cells in infected animals. One recent study shows that PEDV undergoes an early replication in porcine monocyte-derived dendritic cells (Mo-DC) and activates the antigen-presenting ability of Mo-DC (Gao et al., 2015). Although viral antigen was detected in the dendritic-like cells in the laminar propria of small intestine of TGEV-infected animals, it remains controversial whether TGEV actually replicate in dendritic cells in vivo and in vitro (Riffault et al., 2001). PEDV infection of neonatal piglets triggered a strong and rapid induction of type I interferon (Annamalai et al., 2015). However, the source and function of type I interferon in PEDV pathogenesis and immunity remains to be determined. DC are known to secret cytokines such as type I interferon, IL-12, IL-10, and chemokine to regulate the subsequent development of antibody based humoral response and cell-mediated immune responses (Cervantes-Barragan et al., 2007, Fitzgerald-Bocarsly and Feng, 2007). Human SARS coronavirus failed to replicate in Mo-DC, but induced type I interferon and chemokine such as monocyte chemoattractant protein 1 (MCP-1) and interferon inducible protein −10 (IP-10) (Law et al., 2005). Here, we further studied the interaction between PEDV and Mo-DC. We observed that PEDV failed to undergo a productive replication in Mo-DC. PEDV did not compromise the viability of DC and activated the transcription of type I interferon and chemokine IP-10, which may suggest the possible role of PEDV-infected DC in promoting the development of immune responses against PEDV invasion.
2. Materials and methods
2.1. Cells and virus
Porcine Mo-DC were prepared as described previously (Wang et al., 2007). PEDV-CO was grown in Vero-76 cells and titered before use in the experiments as described previously (Wang and Nelson, 2016).
2.2. Immunofluorescence staining
Mo-DC and Vero-76 cells were either mock infected or infected with PEDV-CO at an MOI of 0.1. At 18 and 72 h post infection, Mo-DC were first stained with mouse anti-porcine MHC II monoclonal antibody (VMRD), followed by Alex546 labeled goat-anti-mouse monoclonal antibody (Invitrogen) to show the expression of MHC II on Mo-DC. FITC labeled monoclonal antibody specific to the nucleocapsid protein of PEDV was used to detect virus in infected cells. DAPI was used to stain the nuclei of cells. Vero-76 cells were stained with FITC labeled monoclonal antibody specific for the nucleocapsid protein of PEDV at 72 h post infection and served as a positive control. Images were taken using Fluoview FV1200 IX81 confocal microscope (Olympus, Center Valley, PA).
2.3. Cytotoxicity assay
Cell Counting Kit − 8 was used per manufacturer instructions (Sigma-Aldrich, St Louis, MO). Mo-DC were plated in a 96-well plate at a concentration of 5 × 104 cells/well and incubated for 24 h at 37 °C in a 5% CO2 atmosphere. Cells were then infected with either an MOI of 2.5 or 5 of PEDV-CO or mock infected in triplicate. At 24, 48, and 72 h post infection, we added 10 μL of CCK-8 solution to each well and incubated at 37 °C in a 5% CO2 atmosphere for 3 h. Absorbance was then measured at 450 nm with the Synergy 2 plate reader (Biotek, Winooski, VT).
2.4. QRT-PCR
Mo-DC were seeded in 6-well plates at a concentration of 1 × 106 per well. Cells were then infected with 0.1 MOI of PEDV-CO, Poly I:C at 50 μg/mL, or mock-infected, and incubated at 37 °C in a 5% CO2 atmosphere. Supernatant and cells were then collected at 24, 48, 72, 96 h post-infection and stored at −80 °C until use in ELISA and qRT-PCR.
Total RNAs were extracted from the PEDV-infected DCs by using the RNeasy mini kit (Qiagen, Valencia, CA) per manufacturer’s instructions. RNA concentrations for all samples were determined, and equal amounts of RNA for each sample went through reverse transcription using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Carlsbad, CA) according to the manufacturer’s instructions. The following primers were synthesized by IDT Inc. PEDV N gene, (FP: 5′ −GAA TTC CCA AGG GCG AAA AT- 3′; RP: 5′ −TTT TCG ACA AAT TCC GCA TCT- 3′), IFN-α (FP: 5′ −ACT CCA TCC TGG CTG TGA GGA AAT- 3′; RP: 5′ −ATC TCA TGA CTT CTG CCC TGA CGA- 3′), IFN-β (FP: 5′ −TGC AAC CAC CAC AAT TCC AGA AGG- 3′; RP: 5′ −TCT GCC CAT CAA GTT CCA CAA GGA- 3′), MCP-1 (FP: 5′ —CCA GCA GCA AGT GTC CTA AA- 3′; RP: 5′ −TTC TTG AGG CTT ATG- 3′), IP-10 (FP: 5′ −TCT CCT CGA ACA CAG AGA GAA- 3′; RP: 5′ −AGG GCT TGA TGT ATG GTG TAT G- 3′) and GADPH, the reference gene, (FP: 5′-AGG TCA TCC ATG ACA ACT TCG GCA- 3′; RP: 5′ −AGC ACC AGT AGA AGC AGG GAT GAT- 3′) (Integrated DNA Technologies, Coralville, IA). Real-time PCR was performed using the Brilliant II SYBR Green QRT-PCR Master Mix (Agilent Technologies, Santa Clara, CA) and the ABI 7500HT Real-Time Thermocycler (Applied Biosystems, Foster City, CA). The fold changes of gene transcripts relative to mock-infected cells for cytokines and chemokine was determined by the ΔΔCt method after normalization using the reference gene GAPDH as described previously. Fold of change in viral RNA copies relative to the 24 h post infection was determined by the ΔΔCt method after normalization using the reference gene GAPDH.
2.5. Transmission electron microscopy (TEM)
Vero-76 cells were grown in DMEM media supplemented with 10% fetal bovine serum and 1% PSG. Mo-DC were prepared and grown as described previously (Wang et al., 2007). Prior to virus infection, Vero-76 cells were resuspended in freshly prepared DMEM media containing 1% PSG and 1.3 μg/mL TPCK-treated trypsin (Sigma). Both vero-76 cells and Mo-DC were infected with PEDV-CO at a MOI of 1. At 24 and 48 h after virus infection, cells were washed with PBS and fixed in phosphate-buffered 2.5% glutaraldehyde. Cells were then processed as described previously for thin sectioning (Wang et al., 2003). The thin sections were placed onto copper grids, stained with 1.0% uranyl acetate and lead citrate, and examined and photographed by using a Hitachi 7100 electron microscope.
2.6. ELISA for IFN-α and IFN-β
Supernatants collected from experiments described in Section 2.4 were used to quantify the amount of type I interferon. For the measurement of IFN-α, a protocol developed in the lab was used and has been described previously (Zhang et al., 2012). A quantitative swine IFN-β ELISA kit was purchased from MyBioSource and used by following the manufacturer’s instructions. The absorbance at 450 nm was measured in Synergy 2 Multi-mode microplate reader (BioTek) and the concentration of IFN-α and IFN-β was calculated based on the standard curve generated from each experiment.
2.7. Statistical analysis
Statistical significance was determined with the student’s t-test. P values of equal or less than 0.05 were considered as significantly different.
3. Results
3.1. PEDV fails to replication in porcine Mo-DC
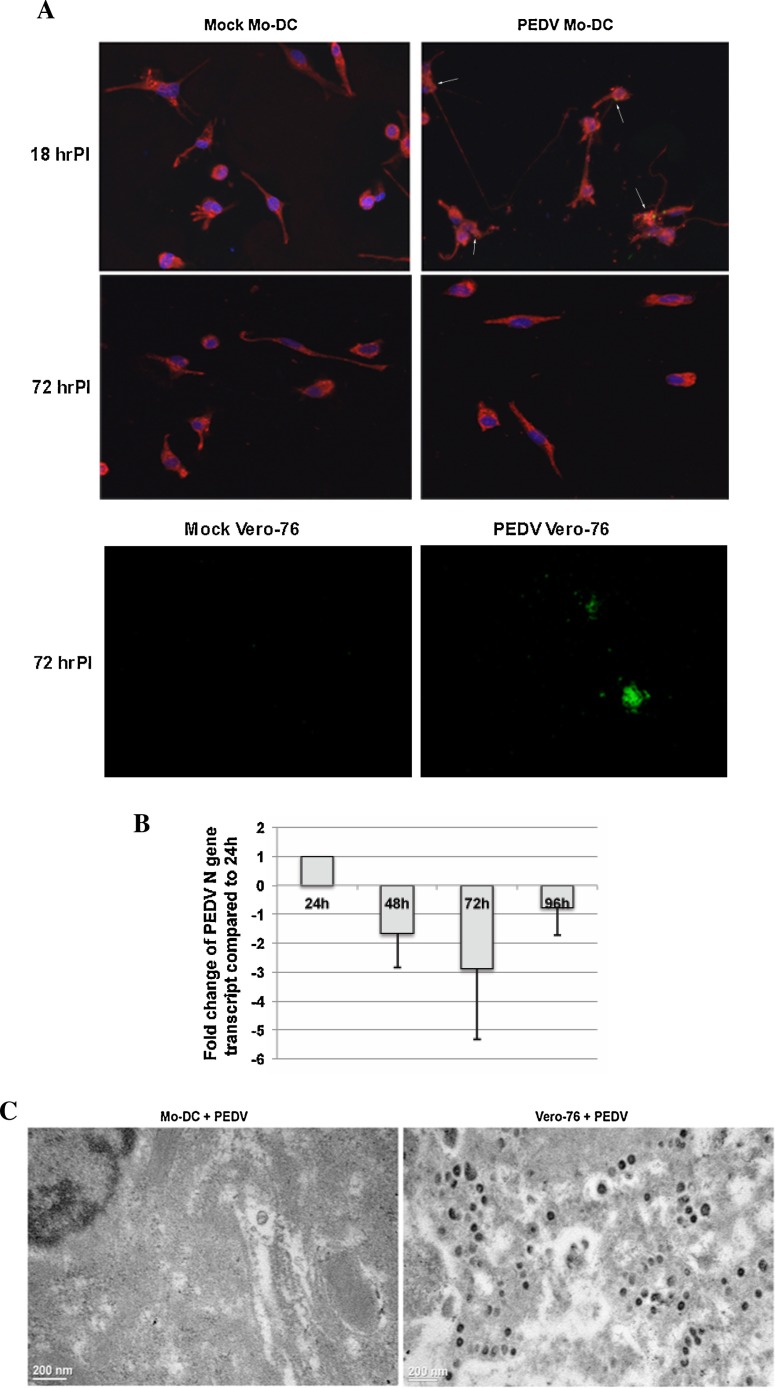
To determine whether PEDV is capable of replicating in porcine Mo-DC, we first used immunofluorescence staining to examine the presence of virus proteins in infected cells. As shown in Fig. 1 A, PEDV was observed in Mo-DC at 18 h post-infection. However, no virus positive staining was observed in Mo-DC at 72 h post infection. Cells appear to be healthy with a typical Mo-DC morphology. In contrast, syncytia and virus were detected in Vero-76 cells at 72 h post infection. The results suggest that virus may enter the Mo-DC, but fails to undergo a productive replication.
Fig. 1.
PEDV fails to replicate in Mo-DC. A: Immunofluorescence staining of PEDV-infected cells with PEDV N protein specific monoclonal antibody. Red: MHC II; Green: PEDV N; Blue: DAPI. Arrows indicate PEDV positive staining in Mo-DC. B: Kinetics of viral N gene transcription of PEDV-infected Mo-DC by qRT-PCR. Fold of change relative to the 24 h post infection is shown. One representative result of three independent experiments is shown. C: TEM of PEDV-infected Mo-DC and Vero-76 cells.
To confirm the above observation, we performed qRT-PCR to quantify the viral nucleocapsid (N) gene transcription at defined time points after PEDV infection of Mo-DC. As expected, we detected viral gene transcript at all time points after virus infection, indicating that PEDV may enter or adhere to the Mo-DC. However, the viral gene transcript showed steady decrease when compared to 24 h post infection (Fig. 1B). This supports our earlier observation that PEDV fails to replicate in Mo-DC. To further validate the observation, we examine the presence of virus particles in Mo-DC by TEM. We did not observe any virus particles in Mo-DC, but abundant virus particles were detected in Vero-76 cells (Fig. 1C). Taken together, this evidence clearly suggests that PEDV does not undergo a productive replication in Mo-DC.
3.2. PEDV does not compromise the viability of Mo-DC
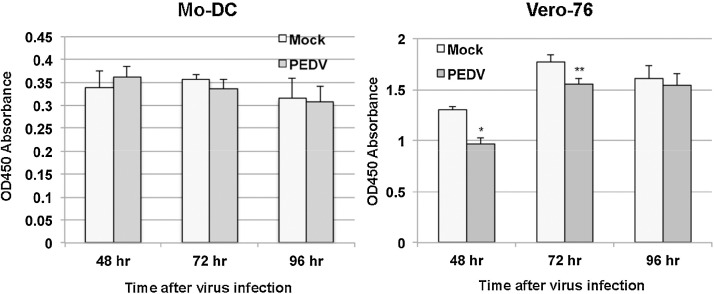
As described earlier, PEDV infection of Mo-DC does not appear to affect cell morphology and appearance under the microscope. To determine whether PEDV causes cell death of Mo-DC, we seeded the cells in a 96-well plate at 5 × 104 per well and infected the cells with PEDV-CO at MOIs of 2.5 or 5 or mock-infected in triplicate. After incubation at 37 °C in a 5% CO2 atmosphere for 48, 72 and 96 h, 10 μL of CCK-8 solution (Sigma 96992 Cell Counting Kit −8) was added to each well and incubated at 37 °C in a 5% CO2 atmosphere for 3 h. Absorbance was then measured at 450 nm with the Synergy 2 plate reader (Biotek, Winooski, VT). As shown in Fig. 2 , there is no significant difference in cell death between PEDV-infected cells and mock-infected cells at all time points examined. This indicates that PEDV does not cause either apoptotic or necrotic cell death of Mo-DC.
Fig. 2.
PEDV does not compromise the viability of Mo-DC. No significant difference (P > 0.05) in cell viability between mock infected and PEDV-infected cells (MOI of 5) was observed at 48, 72, and 96 h post infection. Averages and standard deviations of three independent experiments are shown.
3.3. PEDV activates the transcription of type I interferon and IP-10 chemokine
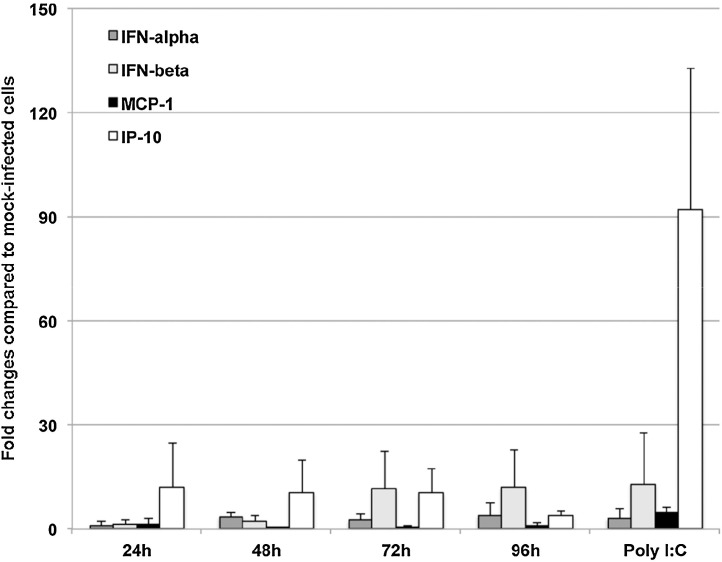
To determine whether the exposure of Mo-DC to PEDV induces the activation of type I interferon and chemokine, we first used qRT-PCR to quantify the transcript level of IFN-α, IFN-β, MCP-1, and IP-10. As shown in Fig. 3 , PEDV activated the transcription of IFN-α, IFN-β, MCP-1, and IP-10 as early as 24 h post infection. IP-10 was the most abundant chemokine induced and was maintained at a high level until 72 h post infection. The highest IFN-α induction was at 48, 72 and 96 h post infection. In contrast, IFN-β was the highest at 72 and 96 h post infection. MCP-1 showed a slight increase in transcription after PEDV infection at all time points examined. As expected, Poly I:C treatment induced the activation all four cytokines and chemokine examined.
Fig. 3.
PEDV up-regulates the transcription of type I interferon and IP-10 chemokine. IFN-β and IP-10 are the most abundantly induced cytokine and chemokine by PEDV. Averages and standard deviations of three independent experiments are shown.
Next, we used ELISA to assess the secretion of IFN-α and IFN-β into the culture supernatants of infected cells. Only a slight increase in IFN-α was detected in PEDV-infected group (data not shown) compared to mock infected cells. Surprisingly no IFN-β was detected in the PEDV infected group, despite the high gene transcript level. Overall, PEDV does activate the transcription of type I interferon and IP-10 in Mo-DC.
4. Discussion and conclusion
PEDV is known to infect the epithelial cells of the intestine to cause diarrhea and even death in young piglets (Jung et al., 2015, Madson et al., 2014). Although macrophages infiltrated in the lamina propria also contained viral antigen as revealed by immunohistochemistry staining (Lee et al., 2000), it remains to be determined whether PEDV actually productively infected the intestinal macrophages. Porcine alveolar macrophages have been shown to support PEDV replication (Park and Shin, 2014). One recent study suggests that PEDV only replicates in Mo-DC prior to 24 h after infection, but not at later time points, as determined by virus titration of culture supernatants (Gao et al., 2015). However, our data suggest that PEDV fails to replicate in Mo-DC as evidenced by the decrease of viral gene transcript over time and the absence of virus proteins and virus particles in the infected cells as examined by immunofluorescence staining and TEM respectively. Although we did not examine the gene transcript level prior to 24 h post infection, the absence of virus particles in infected cells strongly indicate that the virus did not undergo productive replication in Mo-DC. Additionally, we did not detect an increase in virus titer in the supernatants of infected cells (data not shown). Our observation is further supported by the absence of cell death after PEDV exposure. Similarly, conflicting results have been published regarding the susceptibility of Mo-DC to TGEV, a close relative of PEDV (Guzylack-Piriou et al., 2006, Zhao et al., 2014). Further research is needed to determine the underlying reasons for the discrepancy results.
We observed that PEDV exposure to Mo-DC greatly activated the transcription of IP-10. IP-10 plays a role in effector T cell generation and trafficking (Dufour et al., 2002). IP-10 has also been shown to exhibit antimicrobial activity in vitro and in vivo (Schutte et al., 2016). SARS coronavirus also induces the induction of IP-10 from monocyte-derived dendritic cells in the absence of a productive virus replication (Law et al., 2005). The molecular mechanisms by which SARS and PEDV trigger the activation of chemokine IP-10 in the absence of an active virus replication remain to be determined. Viral protein such as human immunodeficiency virus (HIV) tat protein has been shown to activate IP-10 induction (Kukkonen et al., 2014). It is likely that viral proteins such as the spike (S) protein of PEDV may mediate this effect. Furthermore, whether IP-10 is induced in vivo is not clear. The implication of IP-10 in PEDV pathogenesis and immunity remains to be elucidated. Future studies should focus on the implications of IP-10 in PEDV pathogenesis and immunity and uncover the mechanism by which Mo-DC is resistant to PEDV replication.
We also observed the activation of IFN-β transcription by PEDV on Mo-DC. However, PEDV did not induce the activation of IFN-β mRNA in Marc-145 cells, a known susceptible cell line for PEDV (Zhang et al., 2016). Nsp1 and other viral proteins of PEDV have been shown to antagonize the type I interferon induction and signaling in vitro (Ding et al., 2014, Zhang et al., 2016). Neonatal piglets that exhibit severe diseases after PEDV infection showed an early and strong induction of type I interferon compared to post-weaned piglets (Annamalai et al., 2015). These conflicting data suggest that PEDV behaves very differently in vivo and in vitro in terms of type I interferon induction and action. Similar phenomenon has been observed for other viruses such as porcine reproductive and respiratory syndrome virus and murine hepatitis virus (Roth-Cross et al., 2007, Wang and Christopher-Hennings, 2012). A previous study shows that the interferon producing cells are exclusively located in gut-mucosal related lymphoid tissues with dendritic cell-like properties in TGEV-infected animals (Riffault et al., 2001). More in vivo studies are needed to determine the role of type I interferon in PEDV pathogenesis and immunity.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
We thank Ms. Karen Bentley for taking the transmission electron microscopic images. The authors thank Drs. Sabrina Swenson and Melinda Jenkins-Moore of the National Animal Disease Laboratories for providing the cell culture adapted PEDV-CO isolate. This study was supported by grants from USDA NRI (2012-35204-05079), Hatch (10001514), Hatch Multi State (1010908), and by the South Dakota Agricultural Experiment Station.
References
- Annamalai T., Saif L.J., Lu Z., Jung K. Age-dependent variation in innate immune responses to porcine epidemic diarrhea virus infection in suckling versus weaned pigs. Vet. Immunol. Immunopathol. 2015;168:193–202. doi: 10.1016/j.vetimm.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Zust R., Weber F., Spiegel M., Lang K.S., Akira S., Thiel V., Ludewig B. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour J.H., Dziejman M., Liu M.T., Leung J.H., Lane T.E., Luster A.D. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J. Immunol. 2002;168:3195–3204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Bocarsly P., Feng D. The role of type I interferon production by dendritic cells in host defense. Biochimie. 2007;89:843–855. doi: 10.1016/j.biochi.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Zhao S., Qin T., Yin Y., Yang Q. Effects of porcine epidemic diarrhea virus on porcine monocyte-derived dendritic cells and intestinal dendritic cells. Vet. Microbiol. 2015;179:131–141. doi: 10.1016/j.vetmic.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Goede D., Murtaugh M.P., Nerem J., Yeske P., Rossow K., Morrison R. Previous infection of sows with a mild strain of porcine epidemic diarrhea virus confers protection against infection with a severe strain. Vet. Microbiol. 2015;176:161–164. doi: 10.1016/j.vetmic.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Guzylack-Piriou L., Piersma S., McCullough K., Summerfield A. Role of natural interferon-producing cells and T lymphocytes in porcine monocyte-derived dendritic cell maturation. Immunology. 2006;118:78–87. doi: 10.1111/j.1365-2567.2006.02343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Annamalai T., Lu Z., Saif L.J. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet. Microbiol. 2015;178:31–40. doi: 10.1016/j.vetmic.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23:137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkonen S., Martinez-Viedma Mdel P., Kim N., Manrique M., Aldovini A. HIV-1 Tat second exon limits the extent of Tat-mediated modulation of interferon-stimulated genes in antigen presenting cells. Retrovirology. 2014;11:30. doi: 10.1186/1742-4690-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law H.K., Cheung C.Y., Ng H.Y., Sia S.F., Chan Y.O., Luk W., Nicholls J.M., Peiris J.S., Lau Y.L. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106:2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.M., Lee B.J., Tae J.H., Kweon C.H., Lee Y.S., Park J.H. Detection of porcine epidemic diarrhea virus by immunohistochemistry with recombinant antibody produced in phages. J. Vet. Med. Sci. 2000;62:333–337. doi: 10.1292/jvms.62.333. [DOI] [PubMed] [Google Scholar]
- Madson D.M., Magstadt D.R., Arruda P.H., Hoang H., Sun D., Bower L.P., Bhandari M., Burrough E.R., Gauger P.C., Pillatzki A.E., Stevenson G.W., Wilberts B.L., Brodie J., Harmon K.M., Wang C., Main R.G., Zhang J., Yoon K.J. Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3-week-old weaned pigs. Vet. Microbiol. 2014;174:60–68. doi: 10.1016/j.vetmic.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Park J.E., Shin H.J. Porcine epidemic diarrhea virus infects and replicates in porcine alveolar macrophages. Virus Res. 2014;191:143–152. doi: 10.1016/j.virusres.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffault S., Carrat C., van Reeth K., Pensaert M., Charley B. Interferon-alpha-producing cells are localized in gut-associated lymphoid tissues in transmissible gastroenteritis virus (TGEV) infected piglets. Vet. Res. 2001;32:71–79. doi: 10.1051/vetres:2001111. [DOI] [PubMed] [Google Scholar]
- Roth-Cross J.K., Martinez-Sobrido L., Scott E.P., Garcia-Sastre A., Weiss S.R. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J. Virol. 2007;81:7189–7199. doi: 10.1128/JVI.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte K.M., Fisher D.J., Burdick M.D., Mehrad B., Mathers A.J., Mann B.J., Nakamoto R.K., Hughes M.A. Escherichia coli pyruvate dehydrogenase complex is an important component of CXCL10-Mediated antimicrobial activity. Infect. Immun. 2016;84:320–328. doi: 10.1128/IAI.00552-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Christopher-Hennings J. Post-transcriptional control of type I interferon induction by porcine reproductive and respiratory syndrome virus in its natural host cells. Viruses. 2012;4:725–733. doi: 10.3390/v4050725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Nelson E. Ultrastructure and morphogenesis of PEDV in PEDV infected Vero-76 cells. Curr. Top. Virol. 2016;13:41–46. [Google Scholar]
- Wang X., Messerle M., Sapinoro R., Santos K., Hocknell P.K., Jin X., Dewhurst S. Murine cytomegalovirus abortively infects human dendritic cells, leading to expression and presentation of virally vectored genes. J. Virol. 2003;77:7182–7192. doi: 10.1128/JVI.77.13.7182-7192.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Eaton M., Mayer M., Li H., He D., Nelson E., Christopher-Hennings J. Porcine reproductive and respiratory syndrome virus productively infects monocyte-derived dendritic cells and compromises their antigen-presenting ability. Arch. Virol. 2007;152:289–303. doi: 10.1007/s00705-006-0857-1. [DOI] [PubMed] [Google Scholar]
- Zhang H., Guo X., Nelson E., Christopher-Hennings J., Wang X. Porcine reproductive and respiratory syndrome virus activates the transcription of interferon alpha/beta (IFN-alpha/beta) in monocyte-derived dendritic cells (Mo-DC) Vet. Microbiol. 2012;159:494–498. doi: 10.1016/j.vetmic.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Gao Q., Qin T., Yin Y., Lin J., Yu Q., Yang Q. Effects of virulent and attenuated transmissible gastroenteritis virus on the ability of porcine dendritic cells to sample and present antigen. Vet. Microbiol. 2014;171:74–86. doi: 10.1016/j.vetmic.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]