Abstract
Hepatitis E virus (HEV) is a zoonotic pathogen to which several species, including human beings, pigs and rodents, are reported to be susceptible. To date, vaccines developed against HEV still need to be improved and a structural gene (ORF2), which encodes a capsid protein with high sequence conservation found across HEV genotypes, is a potential candidate. To exploit the possibility of using RNA interference (RNAi) as a strategy against HEV infection, four small interference RNA (siRNA) duplex targeting ORF2 gene were constructed. A challenge against HEV infection by RNAi was performed in A549 cells. Real-Time quantitative polymerase chain reaction (Real-Time qPCR) and Western blot assay demonstrated that four HEV specific siRNAs (si-ORF2-1, si-ORF2-2, si-ORF2-3 and si-ORF2-4) were capable of protecting cells against HEV infection with very high specificity and efficiency. The results suggest that RNAi is a potent anti-HEV infection prophylaxis strategy.
Keywords: Hepatitis E virus, RNAi, ORF2
1. Introduction
Hepatitis E virus, a member of the genus Hepevirus under the family Hepeviridae, has caused several outbreaks in developing countries and has prevalence in developed countries. HEV is transmitted predominantly by a fecal-oral route (Aggarwal and Krawczynshi, 2000, Hsieh et al., 1999); its prevalence is mainly waterborne. HEV is transmitted across species in humans, pigs, deer and rodents, and can cause experimental infection in chickens, cats and nonhuman primates (Arankalle et al., 2001, Feagins et al., 2008, Gardner and Luciw, 2008, Okamoto et al., 2004, Tei et al., 2003, Vitral et al., 2005). Evidence has proven that HEV is a zoonotic virus, with pigs as the main reservoirs of HEV in nature. Meng et al. (2002) showed that veterinarians working with swine were at increased risk of HEV infection and HEV has been found in commercially available pig livers (Feagins et al., 2007, Bouwknegt et al., 2007). Travelers in endemic countries, including India, Nepal, China, Africa and Mexico are at high risk of infection with hepatitis E virus (Abreu, 2007). Therefore, it is urgent to develop an effective strategy for prevention of HEV infection.
Given that all four HEV genotypes (1–4) correspond to one serotype; vaccines may be helpful to control for HEV infections (Tsarev et al., 1993, He et al., 1993, Gardner and Luciw, 2008). Purified polypeptide vaccine has completed a phase 3 trials (Abreu, 2007, Mushahwar, 2008). The safety and efficacy of this recombinant vaccine were tested after three vaccine doses (at months 0, 1, and 6) in subjects in the Nepalese Army (Shrestha et al., 2007). However, questions were subsequently raised, the exclusively male subjects and inconsistent data in humans and in rhesus monkeys (Krawczynski, 2007, Tacke and Trautwein, 2007). Even if all of these questions are resolved, vaccinations (three times) will take 6 months, but susceptive animals (pigs or rodents) can transmit the disease as early as the first week post-infection and can continue to do so for at least several weeks (Halbur et al., 2001, Shrestha et al., 2007). Moreover, immune escapes can also occur as new virulent HEV variants evolve, as the case with severe acute respiratory syndrome coronavirus (SARS-CoV) evolved from human coronavirus (Lin et al., 2007, Saif, 2004). In this respect, the mutational events in cell cultures that have been reported during the primary propagation and consecutive passages (p5, p10, and p13) reported in 2008 (Lorenzo et al., 2008, Tanaka et al., 2008) are particularly worrisome.
In this paper, RNA interference (RNAi) was used as a way of circumventing some of these issues regarding vaccines, such as high specificity and efficiency. RNAi is a phenomenon in which small double-stranded RNA molecules induce sequence-specific degradation of homologous single-stranded RNA (Hannon, 2002). RNAi is a powerful tool to investigate gene function through specific suppression of a particular mRNA, and has been employed in therapeutic studies of human diseases, including cancer, neurogenerative diseases and viral infectious diseases. Similar strategies were capable to reduce significantly SARS-CoV replication (Akerstrom et al., 2007) and hepatitis C virus mRNA accumulation (Liu et al., 2006), which suggests that RNAi may be one of the most useful antiviral therapy strategies currently being investigated. As the field of RNAi has expanded, much research has been reported for many genes of interest, including oncogenes and viral genes, indicating their successful silencing in both cells and animal models (Akerstrom et al., 2007, Kirchhoff, 2008, Kleinman et al., 2008, Liu et al., 2006). There have also been a number of clinical trials using RNAi strategies, including one on age-related macular degeneration (AMD) (Kleinman et al., 2008), and more applications are expected.
2. Materials and methods
2.1. Construction of plasmids
The HEV used in this study was characterized as genotype 4 HEV (GenBank accession no. EF570133) and was isolated from swine feces from the Shanghai, China. This was used as template for a reverse transcription nested polymerase chain reaction PCR (RT-nPCR) to amplify the HEV ORF2 gene. The ORF2 gene encodes the major structural or capsid protein of HEV (Riddell et al., 2000). The external forward primer and reverse primer were 5′-CGATTTTGCGCTTGAGCTTGA-3′ (P1) and 5′-TGGAGACCGAGCGCACGGCAC-3′ (P2), respectively. The internal forward primer and reverse primer were 5′-CTCGGCGGGCTCCCGACAG-3′ (P3) and 5′-AGGTGCGAGGACACCAACGGCAG-3′ (P4), respectively. RT-nPCR analysis was conducted using an AMV Reverse Transcriptase XL kit for RT-PCR (Takara, Tokyo, Japan) according to the manufacturer's directions. The RT-PCR protocol performed included a reverse transcription phase at 42 °C for 30 min and 85 °C for 15 s. Two microliters of the cDNA synthesized were then amplified by nested PCR at 94 °C for 2 min, followed by 94 °C for 30 s, 42 °C for 30 s and 72 °C for 1 min, and repeated for 29 cycles. The PCR products were detected by electrophoresis on agarose gel containing 0.5 μg/mL ethidium bromide. The amplified DNA fragment was inserted into the multicloning site of a eukaryotic expression vector pEGFP-N1 (Clontech, a gift from Dr. Wei Liu) which contains the reporter gene of enhanced green fluorescence protein (eGFP), using standard cloning procedures with the restriction sites of EcoRI and BamHI. The EGFP gene was located downstream of the target genes. The recombinant plasmid was named pEGFP-ORF2.
2.2. siRNA construction
Four 19 nt siRNAs with DNA ends corresponding to the target genes (Fig. 1 ) were designed according to Qiagen's guideline (http://www.qiagen.com). The siRNA duplexes have been designed using the Hiperformance Design Algorithm licensed from Novartis AG, integrated with a stringent in-house homology analysis tool. The highest-ranking siRNA duplexes generated by the algorithm were chosen as representing the best combination of activity and specificity. Scrambled siRNA, constructed from a random sequence heterology with the HEV sequence, served as a negative control for identifying the specificity of HEV siRNA (Table 1 ). The synthesized non-modified siRNAs were diluted with RNase-free buffer to obtain a 20 μM solution. The solution was denatured by heating at 90 °C for 1 min, incubated at 37 °C for 60 min, then either used immediately or stored at −20 °C in an RNase-free environment.
Fig. 1.
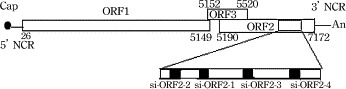
Genomic structure of HEV and position of target siRNAs.
Table 1.
Nucleotide sequences of siRNAs targeting ORF2 of HEV.
| siRNA | Nucleotide sequence |
|---|---|
| si-ORF2-1 | (6775–6795 nt) |
| Sense | 5′-r(UCC AGC AGU AUU CUA AGAC) dTdT-3′ |
| Antisense | 5′-r(AGG UCU UAG AAU ACU GCU G)dTdT-3′ |
| si-ORF2-2 | (6607–6627 nt) |
| Sense | 5′-r(UUU CCC UGA CGG CUG CCG A) dTdG-3′ |
| Antisense | 5′-r(AUU CGG CAG CCG UCA GGG A) dTdT-3′ |
| si-ORF2-3 | (6866–6886 nt) |
| Sense | 5′-r(UAA UUA UAA UAC UAC CGC C)dAdG-3′ |
| Antisense | 5′-r(CTG GCG GUA GUA UUA UAA)-dTdT-3′ |
| si-ORF2-4 | (7030–7050 nt) |
| Sense | 5′-r(UUG AGU AUC CUG CUC GAGC) dTdC-3′ |
| Antisense | 5′-r(GAG CUC GAG CAG AGU CA A) dTdT-3′ |
| Scramble siRNA | |
| Sense | 5′-r(UUC UCC GAA CGU GUC ACG U)dTdT-3′ |
| Antisense | 5′-r(ACGUGACACGUUCGGAGAA)dTdT-3′ |
2.3. siRNA and plasmid transfection
A549 (human lung carcinoma, ATCC, VA, USA) cell line was used in this study and maintained as described previously (Huang et al., 1999). A549 cells were trypsinized and transferred to 500 μL antibiotic-free growth medium in 24-well plates at 0.5–2.0 × 105/well. Cells were cultured 24 h before transfection. One microgram of pEGFP-ORF2 recombinant plasmid was diluted in 50 μL of serum-free DMEM medium, and mixed gently with 0.5 μL each of the five 20 nM siRNAs and 1.5 μL Lipofectamine 2000 (Invitrogen, CA, USA) according to the manufacturer's instructions in 50 μL of serum-free DMEM medium for 20 min, and then co-transfected to the cells. Meanwhile, each siRNAs were either co-transfected with pEGFP vector or transfected alone into A549 cells to test the interferon response. The optimal combination of four parameters for each cell line, including the highest transfection efficiency, the lowest non-specific effects, the conditions for the most efficient delivery of siRNA, and the concentration of Lipofectamine 2000, was first determined in preliminary experimentation. Four to six hours post-transfection, the culture medium was replaced with fresh complete medium and the cells were incubated at 37 °C for 48 h for mRNA detection, and 72 h for protein detection. To determine transfection efficiency, the EGFP fluorescence intensity of transfected cells was monitored with an inverted fluorescence microscope (Nikon TE2000, Tokyo, Japan) before Real-Time qPCR analysis. The efficiency of suppression of the HEV ORF2 gene by various HEV specific siRNAs was evaluated by fluorescence microscopy, Real-Time qPCR and Western blot. Specificity of the inhibition was confirmed by co-transfection of the pEGFP-ORF2 recombined plasmid with scrambled siRNA.
2.4. Live virus siRNA antiviral assays
HEV has been successfully cultured in A549 cells as previously described (Huang et al., 1999). For HEV infection challenge, A549 cells were transfected with each of siRNAs (si-ORF2-1, si-ORF2-2, si-ORF2-3, si-ORF2-4, and scrambled siRNA) as described previously 24 h before the virus challenge. The viral challenge was performed as the previously described (Huang et al., 1999, Tanaka et al., 2007). The virus at a viral count of 1–2 × 105/mL as calculated by viral genomic titer determined by Real-Time quantitative PCR (Li et al., 2006, Kasorndorkbua et al., 2004) was inoculated into A549 cells, and CPEs were observed daily. The cells were harvested 48 h post-inoculation, when the CPE was observed, for Real-Time qPCR analysis (downstream of the target in ORF2), and 72 h for Western blot assay.
2.5. MTS assay
A549 cells (0.5–1 × 105) per well seeded in 96-well plates were incubated as described above except that the amounts of siRNAs and virus/pEGFP in each well were one-fourth of those in 24-well plates. At 36 h post-infection/post-transfection, cell viability was assessed by adding 20 μg/well of CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS, Promega, WI, USA) to cell cultures according to the manufacturer's instructions. After 4 h incubation along with MTS, cells were determined in OD492 nm and each performed in triplicate.
2.6. Real-Time qPCR analysis
Forty-eight hours post-inoculation with viruses, the culture medium was removed, and the cells were lysed by freeze–thaw three times. The total RNA was extracted by Trizol (Invitrogen, CA, USA) according to the manufacturer's directions and reverse transcribed into cDNA using an AMV Reverse Transcriptase XL kit. The synthesized first strand cDNA (2 μL) was added as a template for Real-Time qPCR using SYBR Premix Ex Taq™ (perfect Real-Time, Takara, Tokyo, Japan) according to the manufacturer's directions. The forward primer for ORF2 was 5′-CGCACCTCACTCTGCGCTG-3′ (6996–7008 nt), and reverse primer was 5′-ATTGGAAAGCGCAGCCCTG-3′ (7097–7079 nt). The mixtures were reacted at 95 °C for 30 s, followed by 95 °C for 5 s and 60 °C for 31 s repeated for 39 cycles. The product was expected to be 79 bp. The housekeeping gene GAPDH served as a loading control. The forward primer of GAPDH was 5′-TGGGCTACACTGAGCACCAG-3′, and reverse primer was 5′-AAGTGGTCGTTGAGGGCAAT-3′. The PCR protocol was same as the cells except repeated for 29 cycles. The Real-Time qPCR analysis was performed in the ABI PRISM 7000 Real-Time PCR System (ABI, CT, USA). All procedures were performed in triplicate and data are expressed as means ± S.D.
2.7. Western blot
The infected A549 cells were harvested 72 h post-inoculation and lysed with buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.02% sodium azide, 1% Triton X-100, 1 μg/mL aprotinin, and 100 μg/mL PMSF). Equivalent amounts of total protein were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a nitrocellulose membrane. After blocking non-specific-binding sites with 10% skim milk, the membrane was incubated with primary antibodies at 4 °C overnight. The primary antibodies used in this experimental were rabbit polyclonal anti-HEV (ABR, CA, USA, 1:500 dilution), and a rabbit polyclonal anti-GAPDH for loading control (Proteintech Group, CO, USA, 1:500 dilution). After washing with TBS buffer, the blots were incubated with HRP conjugated goat anti-rabbit IgG (Promega, WI, USA, 1:500 dilution) at room temperature for 2 h. The bands were then exposed to X-ray films with SuperSignal West Pico Trial Kit (Pierce, Rockford, USA).
3. Results
3.1. Construction of HEV ORF2 plasmid
The 670 bp ORF2 fragment of HEV (6327–6997 nt) was amplified by RT-nPCR and introduced into the pEGFP-N1 vector with the restriction sites of EcoRI and BamHI to yield pEGFP-ORF2. The recombinant plasmid was identified by digestion with restriction enzymes (EcoRI and BamHI) and sequence.
3.2. Expression of HEV ORF2 protein and inhibition by siRNA
A549 cells transfected with pEGFP-ORF2 recombinant plasmid with each siRNAs were observed by fluorescence microscope at 36–48 h post-transfection (Fig. 2 pEGFP-ORF2). The efficiency of transfection was confirmed by the highly expressed reporter gene EGFP protein. Fluorescence assays showed an obvious reduction effect induced by all HEV specific siRNAs except the scrambled one (Fig. 2), which suggested that a high efficiency and specificity of inhibition effects by siRNA had been achieved.
Fig. 2.
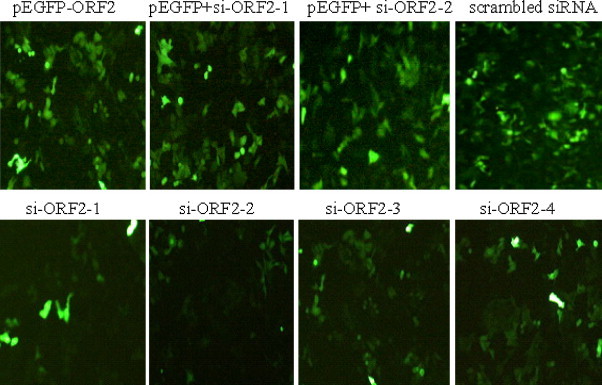
Each of siRNAs co-transfected with pEGFP-ORF2/pEGFP in A549 cells. The expression of HEV ORF2 protein was observed 48 h post-transfection. HEV specific siRNAs induced an obvious reduction, whereas the expression level of EGFP protein co-transfected with scrambled siRNA showed no significant change. Each of siRNAs (si-ORF2-1 or si-ORF2-2) was co-transfected with pEGFP vector to test the interferon response. pEGFP-ORF2: pEGFP-ORF2 recombinant plasmid transfection alone; pEGFP+ si-ORF2-1: pEGFP vector co-transfected with si-ORF2-1; pEGFP+ si-ORF2-2: pEGFP vector co-transfected with si-ORF2-2; scrambled siRNA: pEGFP-ORF2 recombinant plasmid co-transfection with scrambled siRNA; si-ORF2-1: pEGFP-ORF2 recombinant plasmid and si-ORF2-1 co-transfection; si-ORF2-2: pEGFP-ORF2 recombinant plasmid and si-ORF2-2 co-transfection; si-ORF2-3: pEGFP-ORF2 recombinant plasmid and si-ORF2-3 co-transfection; si-ORF2-4: pEGFP-ORF2 recombinant plasmid and si-ORF2-4 co-transfection. Pictures were taken at 48 h post-transfection with a Nikon TE2000 fluorescence microscope.
3.3. MTS assay
To further study the effect of siRNAs on protecting A549 cells against HEV destruction, a MTS assay was performed. The OD values (mean ± S.D.) of solutions in wells treated with each siRNAs (si-ORF2-1, si-ORF2-2, si-ORF2-3, and si-ORF2-4) were shown in Fig. 3 . For the absorbance at 492 nm is directly proportional to the number of living cells in culture, the number of living cells in a well treated with each of HEV specific siRNAs was more than that treated with scrambled siRNA or untransfected controls cells. Interferon response was not observed in A549 cells transfected with each siRNAs alone or co-transfected with pEGFP vector.
Fig. 3.
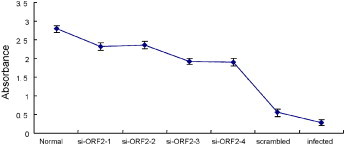
Protection of A549 cells from HEV infection by siRNAs. RNAi inhibited A549 cells growth as determined by MTS assay. Cells transfected with HEV specific siRNAs, scrambled siRNA, and untransfected (infected) cells were served as controls. Absorbance was read at 492 nm and the results were obtained in triplicate wells. OD value shown is the mean ± S.D.
3.4. siRNA decreased the accumulation of HEV mRNA was dose-dependent
The reduction of mRNA level of HEV was evaluated by Real-Time qPCR. The percentage of HEV mRNA in cells previously transfected with siRNA over that in cells inoculated with HEV alone was calculated according to Pfaffl method (Pfaffl, 2001). The suppression of HEV mRNA 48 h post-inoculation was decreased about 17.4-fold, 19.7-fold, 13.7-fold and 11.5-fold in cells transfected with 10 nM/well siRNAs (si-ORF2-1, si-ORF2-2, si-ORF2-3 and si-ORF2-4, respectively), while no significant changes were noted in cells transfected with scrambled siRNA (Fig. 4A). The GAPDH transcription level was rarely changed in either untransfected or transfected cells (data not shown).
Fig. 4.
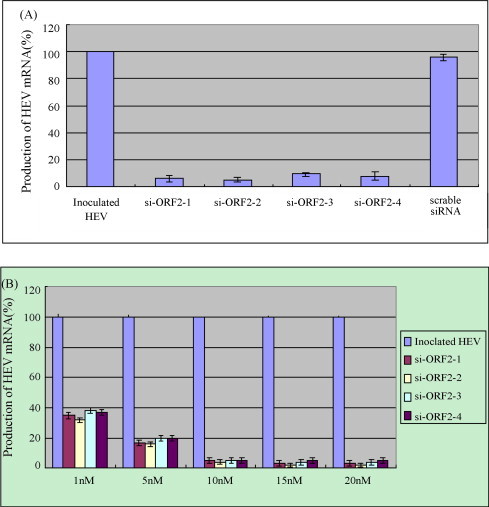
The suppression of HEV mRNA expression was dose-dependent. (A) The percentage of HEV mRNAs in A549 cells transfected with each siRNAs was normalized against the values obtained with GAPDH mRNA. The mRNA in cells inoculated with HEV alone served as control, and the mRNA in inoculated cells transfected with scrambled siRNA was used to identify the specificity. All procedures were performed in triplicate and data are expressed as means (±S.D.). (B) The inhibition of HEV mRNA expression by HEV specific siRNAs was dose-dependant.
To optimize the inhibitory effect of siRNA on HEV transcription, a viral mRNA yield reduction assay was conducted by transfection A549 cells with each siRNAs at indicated doses. As shown in Fig. 4B, transfection of 1 nM/well of each siRNAs just induced an approximately 60% reduction in viral mRNA. However, with the dosage of siRNAs increasing, the HEV mRNA decreased. When the amount of siRNA reached 10 nM/well, the highest inhibition of HEV transcription was obtained. The data indicated that the suppression of HEV transcription by siRNA was dose-dependent.
3.5. Western blot
To analyze further the efficiency of suppression of the HEV protein synthesis by siRNA, A549 cells were collected for Western blot analysis. Only the cells infected with HEV expressed the expected ∼71 kDa protein band (Fig. 5-HEV), while uninfected A549 cells showed no protein band reacting with anti-HEV antibody. Western blot showed that HEV putative structural protein expression was reduced significantly in cells transfected with HEV specific siRNAs (Fig. 5-si-ORF2-1 to si-ORF2-4) compared with cells infected with HEV alone or transfected with scrambled siRNA (Fig. 5-HEV and -scrambled). GAPDH was used as a loading control in this experiment.
Fig. 5.
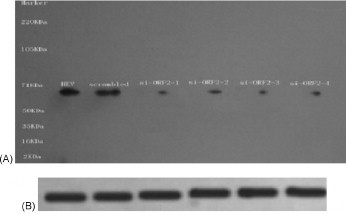
Inhibitory effect of HEV protein expression by siRNAs. Western blot was performed on equal amounts of protein harvested from transfected HEV specific siRNAs (10 nM) A549 cells at 72 h post-inoculation. GAPDH was used as a loading control. M: Marker; HEV: HEV inoculated alone; scrambled: scrambled siRNA transfection; si-ORF2-1: si-ORF2-1 transfection; si-ORF2-2: si-ORF2-2 transfection; si-ORF2-3: si-ORF2-3 transfection; si-ORF2-4: si-ORF2-4 transfection.
In conclusion, the results demonstrated that highly efficient inhibition of HEV transcription by HEV specific siRNAs in A549 cells was obtained.
4. Discussion
HEV is a major cause of enterically transmitted acute hepatitis of adults and is highly lethal in pregnant women. The HEV ORF2 gene is highly conserved in four genotypes and is the most important structural region of the virus. Evidence from epidemiological, animal transmission and vaccine studies had indicated that the ORF2 gene is a suitable region for HEV treatment studies. Therefore, the HEV ORF2 gene was chosen as the target for siRNAs gene silencing in the current study.
RNA interference has showed great promise as an antiviral therapy since its discovery in mammalian cells (Elbashir et al., 2001). Its application in vitro and in vivo has shown it to be a new and efficient approach for inhibition of viral infection. However, as also shown in this study, different siRNA sequences have different interference efficacies, which depending on the characteristics of the target RNA, including local RNA folding and the accessibility of the siRNA-binding site on the target RNA (Kurreck, 2006, Shao et al., 2007). The highest potential efficiency of interference is considered a high knockdown that avoids the degradation of untargeted genes (off-target effects) with the fewer siRNAs (Ladunga, 2006).
RNA interference in viral infection has been the focus of numerous studies (Liu et al., 2006, Mungall et al., 2008), and an effective and specific interference in HEV transcription was obtained in A549 cells in the current study. RNAi, its exquisite specificity in seeking targets, which determines the exclusive protection against hepatitis E virus. HEV specific siRNAs were capable of preventing HEV infection in A549 cells. Antiviral activity was demonstrated both in mRNA transcription level by Real-Time qPCR and in protein expression level by Western blot. These findings indicate that this protocol may be an effective antiviral strategy for protecting host cells against viral invasion.
The siRNA is very effective in vitro; however, there are many obstacles to application of RNAi in vivo as a therapy. The main obstacles are prevention of degradation of siRNA and avoidance of side effects to the host induced by siRNA itself. Couzin (2006) and Grimm and Kay (2007) mentioned the toxicity of both siRNA or shRNA expression vectors in vitro and in vivo, and cell apoptosis was observed when vectors were introduced at a high concentration. Although the RNAi pathway is a promising treatment for cancer, virus and hepatitis therapy, there are serious safety issues that first need to be addressed (McBride et al., 2008). There needs to be a focus on decreasing the toxicity that accompanies siRNA treatment.
In conclusion, the present study demonstrated that HEV specific siRNAs could specifically and effectively suppress HEV transcription and translation in A549 cells. These results indicate that RNAi can be a potential antiviral therapy for suppression HEV infection. Further study is required to determine the effectiveness and the safety of RNA interference as a means of protection against HEV infection in vivo.
Acknowledgments
This work was supported by Key Project of Shanghai Science and Technology committee of China under Grant No. 063919121. We would like to thank Dr. Wei Liu and Dr. Yibo Shi for their generous help.
References
- Abreu C. Viral hepatitis in travelers. Acta Med. Port. 2007;20:557–566. [PubMed] [Google Scholar]
- Aggarwal R., Krawczynshi K. Hepatitis E: an overview and recent advances in clinical and laboratory research. J. Gastroenterol. Hepatol. 2000;15:9–20. doi: 10.1046/j.1440-1746.2000.02006.x. [DOI] [PubMed] [Google Scholar]
- Akerstrom S., Mirazimi A., Tan Y.J. Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and S. Antivir. Res. 2007;73:219–227. doi: 10.1016/j.antiviral.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arankalle V.A., Joshi M.V., Kulkarni A.M., Gandhe S.S., Chobe L.P., Rautmare S.S., Mishra A.C., Padbidri V.S. Prevalence of anti-hepatitis E virus antibodies in different Indian animal species. J. Viral Hepat. 2001;8:223–227. doi: 10.1046/j.1365-2893.2001.00290.x. [DOI] [PubMed] [Google Scholar]
- Bouwknegt M., Lodder-Verschoor F., van der Poel W.H., Ruties S.A., De Roda Husan A.M. Hepatitis E virus RNA in commercial porcine livers in The Netherlands. J. Food Prot. 2007;70:2889–2895. doi: 10.4315/0362-028x-70.12.2889. [DOI] [PubMed] [Google Scholar]
- Couzin J. Molecular biology. RNAi safety comes under security. Science. 2006;312:1121. doi: 10.1126/science.312.5777.1121a. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Feagins A.R., Opriessnig T., Guenette D.K., Halbur P.G., Meng X.J. Detection and characterization of infectious Hepatitis E virus from commercial pig liver sold in local grocery stores in the USA. J. Gen. Virol. 2007;88:912–917. doi: 10.1099/vir.0.82613-0. [DOI] [PubMed] [Google Scholar]
- Feagins A.R., Opriessnig T., Huang Y.W., Halbur P.G., Meng X.J. Cross-species infection of specific-pathogen-free pigs by a genotype 4 strain of human hepatitis E virus. J. Med. Virol. 2008;80:1379–1386. doi: 10.1002/jmv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.B., Luciw P.A. Macaque models of human infectious disease. ILAR J. 2008;49:220–255. doi: 10.1093/ilar.49.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D., Kay M.A. Therapeutic application of RNAi: is mRNA targeting finally ready for prime time? J. Clin. Invest. 2007;117:3633–3641. doi: 10.1172/JCI34129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbur P.G., Kasorndorkbua C., Gilbert C., Guenette D., Potters M.B., Purcell R.H., Emerson S.U., Toth T.E., Meng X.J. Comparative pathogenesis of infection of pigs with hepatitis E virus recovered from a pig and a human. J. Clin. Microbiol. 2001;39:918–923. doi: 10.1128/JCM.39.3.918-923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- He J., Tam A.W., Yarbough P.O., Reyes G.R., Carl R. Expression and diagnostic utility of hepatitis E virus putative structural protein expressed in insect cells. J. Clin. Microbiol. 1993;31:2167–2173. doi: 10.1128/jcm.31.8.2167-2173.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh S.Y., Meng X.J., Wu Y.H., Liu S.T., Tam A.W., Lin D.Y., Liaw Y.F. Identity of a novel swine hepatitis E virus in Taiwan forming a monophyletic group with Taiwan isolates of human hepatitis E virus. J. Clin. Microbiol. 1999;37:3828–3834. doi: 10.1128/jcm.37.12.3828-3834.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Li D., Wei S., Li Q., Yuan X., Geng L., Li X., Liu M. Cell culture of sporadic hepatitis E virus in China. Clin. Diagn. Lab. Immunol. 1999;6:729–733. doi: 10.1128/cdli.6.5.729-733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasorndorkbua C., Guenette D.K., Huang F.F., Thomas P.J., Meng X.J., Halbur P.G. Routes of transmission of swine hepatitis E virus in pigs. J. Clin. Microbiol. 2004;42:5047–5052. doi: 10.1128/JCM.42.11.5047-5052.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff F. Silencing HIV in vivo. Cell. 2008;134:566–568. doi: 10.1016/j.cell.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Kleinman M.E., Yamada K., Takeda A., Chandrasekaran V., Nozaki M., Baffi J.Z., Albuquerque R.J., Yamasaki S., Itaya M., Pan Y., Appukuttan B., Gibbs D., Yang Z., Kariko K., Ambati B.K., Wilgus T.A., Dipietro L.A., Sakurai E., Zhang K., Smith J.R., Taylor E.W., Ambati J. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczynski K. Hepatitis E vaccine-ready for prime time? N. Engl. J. Med. 2007;356:949–951. doi: 10.1056/NEJMe068311. [DOI] [PubMed] [Google Scholar]
- Kurreck J. siRNA efficiency: structure or sequence—that is the question. J. Biomed. Biotechnol. 2006;2006:83757. doi: 10.1155/JBB/2006/83757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladunga I. More complete gene silencing by fewer siRNA: transparent optimized design and biophysical signature. Nucleic Acids Res. 2006;35:433–440. doi: 10.1093/nar/gkl1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.T., Zhang J.S., Su N., Xu J.G., Wang N., Chen J.T., Chen X., Liu Y.X., Gao H., Jia Y.P., Liu Y., Sun R.H., Wang X., Yu D.Z., Hai R., Gao Q., Ning Y., Wang H.X., Li M.C., Kan B., Dong G.M., An Q., Wang Y.Q., Han J., Qin C., Yin W.D., Dongs X.P. Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antivir. Ther. 2007;12:1107–1113. [PubMed] [Google Scholar]
- Li X., Kamili S., Krawczynski K. Quantitative detection of hepatitis E virus RNA and dynamics of viral replication in experimental infection. J. Viral Hepat. 2006;13:835–839. doi: 10.1111/j.1365-2893.2006.00754.x. [DOI] [PubMed] [Google Scholar]
- Liu M., Ding H., Zhao P., Qin Z.L., Gao J., Cao M.M., Luan J., Wu W.B., Qi Z.T. RNA interference effectively inhibits mRNA accumulation and protein expression of hepatitis C virus core and E2 genes in human cells. Biosci. Biotechnol. Biochem. 2006;70:2049–2055. doi: 10.1271/bbb.60001. [DOI] [PubMed] [Google Scholar]
- Lorenzo F.R., Tanaka T., Takahashi H., Ichiyama K., Hoshino Y., Yamada K., Inoue J., Takahashi M., Okamoto H. Mutational events during the primary propagation and consecutive passages of hepatitis E virus strain JE03-1760F in cell culture. Virus Res. 2008;137:86–96. doi: 10.1016/j.virusres.2008.06.005. [DOI] [PubMed] [Google Scholar]
- McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polosky B., Carter B.J., Davidson B.L. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.J., Wiseman B., Elvinger F., Guenette D.K., Toth T.E., Engle R.E., Emerson S.U., Purcell Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J. Clin. Microbiol. 2002;40:117–122. doi: 10.1128/JCM.40.1.117-122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall B.A., Schopman N.C., Lambeth L.S., Doran T.J. Inhibition of Henipavirus infection by RNA interference. Antivir. Res. 2008;80:324–331. doi: 10.1016/j.antiviral.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushahwar I.K. Hepatitis E virus: molecular virology, clinical features, diagnosis, transmission, epidemiology, and prevention. J. Med. Virol. 2008;80:646–658. doi: 10.1002/jmv.21116. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Takahashi M., Nishizawa T., Usui R., Kobayashi E. Presence of antibodies to hepatitis E virus in Japanese pet cats. Infection. 2004;32:57–58. doi: 10.1007/s15010-004-3078-0. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell M.A., Li F., Anderson D.A. Identification of immunodominant and conformational epitopes in the capsid protein of hepatitis E virus by using monoclonal antibodies. J. Virol. 2000;74:8011–8017. doi: 10.1128/jvi.74.17.8011-8017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif L.J. Animal coronavirus vaccines: lessons for SARS. Dev. Bio. (Base 1) 2004;119:129–140. [PubMed] [Google Scholar]
- Shao Y., Chan C.Y., Maliyekkel A., Lawrence C.E., Roninson I.B., Ding Y. Effect of target secondary structure on RNAi efficiency. RNA. 2007;13:1631–1640. doi: 10.1261/rna.546207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha M.P., Scott R.M., Joshi D.M., Mammen M.P., Thapa G.B., Thapa N., Myint K.S., Fourneau M., Kuschner R.A., Shrestha S.K., David M.P., Seriwatana J., Vaughn D.W., Safary A., Endy T.P., Innis B.L. Safety and efficacy of a recombinant hepatitis E vaccine. N. Engl. J. Med. 2007;356:895–903. doi: 10.1056/NEJMoa061847. [DOI] [PubMed] [Google Scholar]
- Tacke F., Trautwein C. Efficient recombinant hepatitis E virus vaccine: mission accomplished? Hepatology. 2007;46:941–943. doi: 10.1002/hep.21909. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Takahashi M., Kusano E., Okamoto H. Development and evaluation of an efficient cell-culture system for Hepatitis E virus. J. Gen. Virol. 2007;88:903–911. doi: 10.1099/vir.0.82535-0. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Takahashi H., Ichiyama K., Hoshino Y., Yamada K., Inoue J., Takahashi M., Okamoto H. Mutational events during the primary propagation and consecutive passages of hepatitis E virus strain JE03-1760F in cell culture. Virus Res. 2008;137:86–96. doi: 10.1016/j.virusres.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Tei S., Kitajima N., Takahashi K., Mishiro S. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet. 2003;362:371–373. doi: 10.1016/S0140-6736(03)14025-1. [DOI] [PubMed] [Google Scholar]
- Tsarev S.A., Tsareva T.S., Emerson S.U., Kapikian A.Z., Ticehurst J., London W., Purcell R.H. ELISA for antibody to hepatitis E virus (HEV) based on complete open-reading frame-2 protein expressed in insect cells: identification of HEV infection in primates. J. Infect. Dis. 1993;168:369–378. doi: 10.1093/infdis/168.2.369. [DOI] [PubMed] [Google Scholar]
- Vitral C.L., Pinto M.A., Lewis-Ximenez L.L., Khudyakov Y.E., dos Santos D.R., Gaspar A.M. Serological evidence of hepatitis E virus infection in different animal species from the Southeast of Brazil. Mem. Inst. Oswaldo Cruz. 2005;100:117–122. doi: 10.1590/s0074-02762005000200003. [DOI] [PubMed] [Google Scholar]


