Abstract
Orthoreoviruses infect virtually all mammalian species, causing systemic infections including mild gastrointestinal and respiratory illnesses. However, little is known about the prevalence or genetic diversity of porcine orthoreoviruses in South Korea. We examined 237 diarrheic fecal samples collected from 78 pig farms around the country. RT-PCR utilizing primers specific for the L1 gene of mammalian orthoreoviruses showed that 45 (19.0%) samples were positive. The 10 strains isolated from orthoreovirus-positive samples formed typical perinuclear cytoplasmic inclusion bodies and had an atypical hemagglutination pattern; these are characteristics of type 3 orthoreovirus. Phylogenetic analysis of the S1 gene in these 10 Korean and other strains showed that type 3 orthoreoviruses could be divided into four lineages; the 10 Korean strains were included in porcine lineage IV, along with T3/porcine/Sichuan/2006. Sequence analysis showed that strains in lineage IV had nucleotide identities of 97.0–98.1% and deduced amino acid identities of 96.4–98.2%. Sequence analysis of the σ1 protein, a viral attachment protein, revealed that the amino acid sequences associated with neurotropism (amino acids 198–204, 249I, 350D, and 419E) were highly conserved among the Korean strains, confirming that neural tropism was present. In conclusion, our findings suggest that porcine orthoreovirus infections are endemic in pig farms in South Korea and that the 10 novel Korean porcine orthoreoviruses belong to porcine lineage IV of type 3 orthoreovirus. In addition, sequence analysis of S1 genes encoding the σ1 protein showed that the 9 of 10 Korean porcine orthoreoviruses exhibited neural tropism.
Keywords: Orthoreovirus, Porcine, Prevalence, Genetic diversity
1. Introduction
Mammalian orthoreoviruses, members of the Reoviridae family, are widely distributed in humans and a number of animal species (Schiff et al., 2007). Originally, orthoreoviruses were termed respiratory enteric orphans because of repeated isolation from respiratory and enteric tracts of children with asymptomatic illnesses (Sabin, 1959). However, the viruses have now been shown to cause upper and lower respiratory tract illnesses, gastrointestinal problems, hepatitis, myocarditis, meningitis, and encephalitis in humans (Johansson et al., 1996, Schiff et al., 2007, Tyler, 2001, Tyler et al., 2004).
Orthoreoviruses are non-enveloped double-stranded RNA viruses containing 10 RNA segments; of large, medium, and small size on SDS-polyacrylamide gels (Schiff et al., 2007). Differences in the migration patterns of RNA segments among strains have allowed viral recombinants derived from different serotypes to be identified (Ramig et al., 1977). Mammalian orthoreoviruses have been grouped into three serotypes, which can be differentiated by the capacity of type-specific antisera to neutralize virus infectivity and inhibit hemagglutination. The serotypes are represented by three commonly studied prototype isolates, type 1 Lang (T1L), type 2 Jones (T2J), and type 3 Dearing (T3D) (Schiff et al., 2007). The genomic sequences of these three prototype strains confirm the serological classification (Wiener and Joklik, 1989, Breun et al., 2001, Yin et al., 2004). Specifically, study of orthoreovirus S1 gene encoding the σ1 protein, which is involved in viral attachment to cells, has been found to be useful to understand functional organization within an attachment protein (Chappell et al., 1997). The σ1 protein is a target of serotype-specific neutralizing antibodies, and the protein is responsible for cell and tissue tropism (Lee et al., 1981, Weiner et al., 1980).
Mammalian orthoreoviruses have a wide geographic distribution and can infect virtually all mammals, including humans and cattle (Tyler, 2001). However, the prevalence and genetic diversity of porcine orthoreoviruses remain largely unclear; only a single report has described a type 3 porcine orthoreovirus isolated in China (Zeng et al., 2008). Therefore, we assessed the prevalence and genetic diversity of porcine orthoreoviruses circulating in South Korea.
2. Materials and methods
2.1. Specimens
From January 2004 to December 2005, 237 fecal specimens were collected from 2- to 70-day-old diarrheic pigs housed on 78 farms throughout South Korea. The fecal samples were examined for common bacterial enteric pathogens, including Salmonella spp., Clostridium spp., and Campylobacter spp., using specific agar-based media, and suspect colonies were identified employing biochemical tests. Shiga toxin-producing Escherichia coli, Brachyspira hyodysenteriae, and Lawsonia intracellularis were detected by PCR with the specific primers (Asakura et al., 1998, Suh and Song, 2005).
2.2. RNA extraction and RT-PCR
RNA was extracted from 200 μl aliquots of fecal supernatants and from lysates of orthoreovirus-infected fetal rhesus monkey TF-104 kidney cells (a cloned derivative of MA-104 monkey kidney cells) using an SV Total RNA Isolation System (Promega Corporation, Madison, WI) according to the manufacturer's instructions.
Mammalian orthoreoviruses were detected by RT-PCR using a primer pair specific to the mammalian orthoreovirus L1 gene (Nicola et al., 2005) (Table S1). RT-PCR and/or nested PCR assays employing different primer sets were also performed to determine whether samples were also infected with porcine group A–C rotaviruses (GARVs-GCRVs), porcine sapovirus (PSaV), porcine norovirus (PNoV), transmissible gastroenteritis coronavirus (TGEV), and/or porcine epidemic diarrhea coronavirus (PEDV) (Table S1), using standard one-step RT-PCR as described (Jeong et al., 2007). All amplification products were analyzed by electrophoresis on 1.5% or 2% (w/v) agarose gels, which were next stained with ethidium bromide and visualized under UV light.
2.3. Virus isolation
Porcine orthoreoviruses were isolated on monolayers of TF-104 cells grown for 3-4 days in six-well plates, as described (Virgin et al., 1988). The isolated porcine orthoreoviruses were adapted and passaged eight times in TF-104 cells and the identity thereof was confirmed using a direct immunofluorescence assay (IFA), employing a mouse monoclonal antibody directed against type 3 orthoreovirus σ1 protein (Abcam, Cambridge, MA) and by RT-PCR using a primer pair specific for the L1 gene.
2.4. Detection of intracytoplasmic viral inclusion body
The morphology and staining features of intracytoplasmic viral inclusion bodies were assessed using a Diff-Quick commercial kit (Dade Behring Inc., Newark, NJ). Briefly, confluent TF-104 cells grown on eight-well chamber slides were infected with orthoreovirus at a multiplicity of infection (MOI) of 0.1 for 24 h. After removal of medium, slides were fixed with 100% methanol and allowed to dry in air. The slides were immersed for 10–20 s in dye solution and rapidly dipped in water to remove excess dye. After drying in air, slides were examined under a bright field microscope.
2.5. Transmission electron microscopy (TEM)
Virus-infected cells were frozen and thawed three times, and the supernatants were clarified by centrifugation at 6000 × g for 20 min in a refrigerated centrifuge, with the clear supernatants next being centrifuged at 10,000 × g for 40 min. The resultant pellets were resuspended in a few drops of distilled water, placed on formvar/carbon-coated electron microscope grids, and stained with 2% (w/v) sodium phosphotungstate at pH 6.8. A minimum of five grid squares were examined using a Philips 201 electron microscope (Whorwell et al., 1976).
2.6. Hemagglutination (HA) assay
Purified orthoreovirus virions, consisting of the cryolysate obtained from the eighth passage of TF-104 cells, were serially diluted in 50 μl volumes of PBS (pH 7.4) in 96-well round-bottomed microtiter plates (Corning-Costar, USA). Fifty microliter amounts of standardized suspensions of porcine RBC, bovine RBC, or type O human RBC, prepared as described (WHO, 2002), were added to each well. The plates were incubated for 2 h at 4 °C and the HA patterns read.
2.7. DNA sequencing and molecular analysis
To obtain genomic data on the porcine orthoreoviruses isolated from the diarrheic fecal samples of pigs, extracted RNA was served to RT-PCR with primer pair specific to S1 gene (Table S1). The RT-PCR products of the full-length S1 gene (1416 bp) were purified using a QIAEX II gel extraction kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. DNA sequencing was carried out using an ABI system 3700 automated DNA sequencer (Applied Biosystems Inc., Foster City, CA).
The nucleotide and deduced amino acid sequences of the S1 gene (1380 bp, not including primer sequences) were compared with those of other mammalian orthoreoviruses using the DNA Basic module (DNAsis MAX, Alameda, USA) (Table S2). Phylogenetic analyses based on nucleotide and amino acid alignments were performed using the neighbor-joining method and UPGMA Molecular Evolutionary Genetics analysis (MEGA version 4.0) employing pair-wise distance comparisons (Tamura et al., 2007). A sequence similarity search of the mammalian orthoreovirus S1 gene was performed using the LALIGN Query program of the GENESTREAM network server at the Institut de Génétique Humaine, Montpelier, France (http://www.eng.uiowa.edu/∼tscheetz/sequence-analysis/examples/LALIGN/lalign-guess.html).
3. Results
A one-step RT-PCR assay, amplifying a 416 bp fragment of the L1 gene of mammalian orthoreoviruses, showed that porcine orthoreovirus was present in 45 of 237 (19.0%) diarrheic fecal samples obtained from 40 of 78 (51.2%) pig farms throughout South Korea. Of the 45 porcine orthoreovirus-positive fecal specimens, 42 (93.3%) also tested positive for other enteric pathogens, including porcine GARV, GBRV, GCRV, PSaV, E. coli, Salmonella spp., and swine dysentery; whereas only 3 (6.7%) tested positive for the porcine orthoreovirus alone (Table 1 ). Of the other enteric pathogens, GARVs were the most common, being present in 24 fecal samples. In addition, of the 192 specimens that tested negative for porcine orthoreovirus, 149 (63.0%) specimens were positive for other enteric pathogens (Table 1). In contrast, no enteric pathogen was detected in the remaining 43 (18.2%) fecal samples.
Table 1.
Summary of the enteric pathogens present in the fecal samples obtained from pigs with diarrhea 2004–2005.
| Enteric pathogens presenta | No. of farms (%)b | No. of samples (%)c |
|---|---|---|
| MRV alone | 3 (3.8) | 3 (1.3) |
| MRV plus GARV | 19 (24.3) | 24 (10.1) |
| MRV plus GCRV | 1 (1.3) | 1 (0.4) |
| MRV plus PSaV | 2 (2.6) | 2 (0.8) |
| MRV, GARV plus GBRV | 1 (1.3) | 1 (0.4) |
| MRV, GARV plus GCRV | 1 (1.3) | 1 (0.4) |
| MRV, GARV plus PSaV | 4 (5.1) | 4 (1.7) |
| MRV, GARV, GBRV plus PSaV | 1 (1.3) | 1 (0.4) |
| MRV, GARV, GCRV plus PSaV | 2 (2.6) | 2 (0.8) |
| MRV plus salmonellosis | 3 (3.8) | 3 (1.3) |
| MRV, GARV plus salmonellosis | 1 (1.3) | 1 (0.4) |
| MRV, salmonellosis plus swine dysentery | 1 (1.3) | 1 (0.4) |
| Other enteric pathogens detected | 1 (1.3) | 1 (0.4) |
| No enteric pathogens detected | 21 (26.9) | 149 (63.0) |
| No enteric pathogens detected | 17 (21.8) | 43 (18.2) |
| Total | 78 (100) | 237 (100) |
MRV: Mammalian orthoreovirus; GARV, GBRV, GCRV: Groups A, B, C rotaviruses; PSaV: Porcine sapovirus.
Number of positive farms.
Number of positive fecal samples.
We isolated 10 porcine orthoreovirus strains from the 45 porcine orthoreovirus-positive fecal samples. After the second or third passage of cells inoculated with viruses from diarrheic piglets, a cytopathic effect (CPE) was observed, characterized by rounded and detached cells that formed clusters. Virus-infected cells showed typical perinuclear cytoplasmic inclusion bodies, porcine orthoreovirus-specific cytoplasmic fluorescence by the direct IF test, and icosahedral, non-enveloped viral particles by TEM (Fig. 1A–C). The 10 strains hemagglutinated porcine (1%, v/v) and bovine (1%, v/v) erythrocytes, but not type O human erythrocytes (1%, v/v), a finding characteristic of type 3 mammalian orthoreoviruses (Tyler, 2001).
Fig. 1.
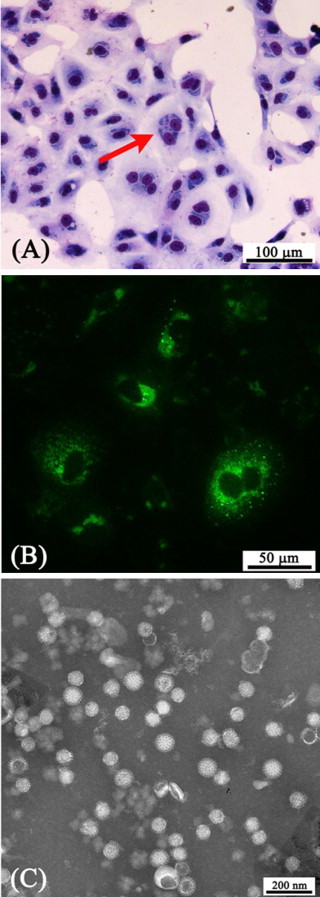
Characterization of the 10 isolated Korean porcine orthoreovirus strains. (A) Histopathologic examination, showing intracytoplasmic inclusion bodies in the perinuclear region (arrow) of TF-104 cells. Diff-Quick stain. Bar = 100 μm. (B) Immunofluorescence, showing that a positive reaction (green) was confined to the cytoplasm. Bar = 50 μm. (C) Transmission electron micrograph, showing icosahedral, non-enveloped virus particles. Negative staining with 2% (w/v) sodium phosphotungstate at pH 6.8. Bar = 200 nm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Utilizing the 1380 bp sequence of the full-length S1 gene, we phylogenetically compared the 10 newly isolated Korean porcine orthoreovirus strains with other mammalian orthoreoviruses. Alignments showed that the new strains were type 3 mammalian orthoreoviruses. Type 3 strains can be further divided into four lineages, and our 10 Korean porcine strains formed a novel lineage IV containing one other porcine strain, T3/porcine/Sichuan/2006 (Fig. 2 ). In comparison, lineage I contains the human strain T3/Human/Colorado/96, isolated in 1996; whereas lineages II and III consist of bovine and human strains isolated in 1950 and 1960, respectively. No newly isolated Korean porcine orthoreovirus was closely related to any previously described strain (Fig. 2).
Fig. 2.
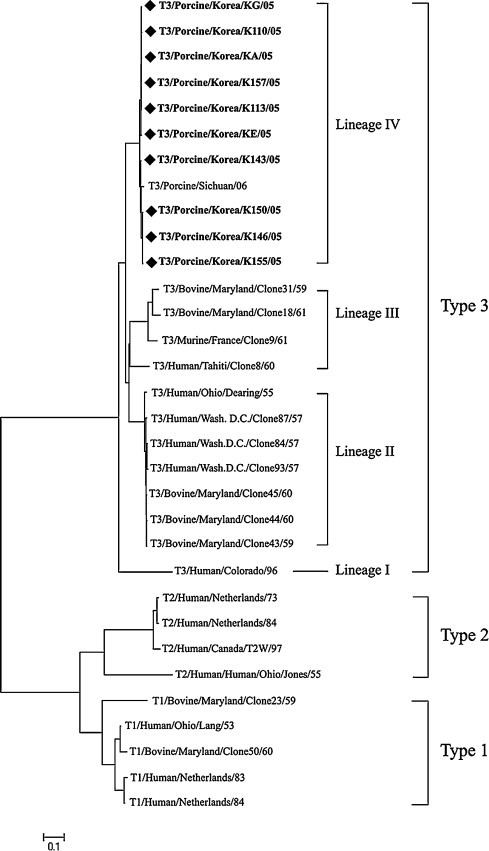
Phylogenetic tree of the σ1 protein of mammalian orthoreovirus strains.
The nucleotide and deduced amino acid sequences of the S1 genes of all 10 Korean porcine orthoreoviruses and other type 1–3 strains are shown in Table 2 . All 10 Korean porcine orthoreoviruses showed the highest nucleotide (97.0–98.1%) and deduced amino acid (96.4–98.2%) identities with T3/porcine/Sichuan/2006, but lower nucleotide (70.7–84.2%) and deduced amino acid (74.8–90.5%) identities with other type 3 strains originating from humans, cattle, and mice. In contrast, the 10 Korean strains showed very low nucleotide and deduced amino acid identities with types 1 (39.7–41.5% and 21.3–26.5%, respectively) and 2 (39.8–42.0% and 24.2–27.2%, respectively) strains (Table 2 and Fig. 3 ).
Table 2.
Nucleotide and deduced amino acid sequence comparison of the S1 gene of the Korean porcine orthoreoviruses with that of the other strains.
| Strain | Origin | % Identity with strainsa | |
|---|---|---|---|
| 10 Korean strains |
|||
| nt | aa | ||
| T1L/53 | Human | 41.0–41.4 | 26.0–26.5 |
| T1N83 | Human | 40.8–41.2 | 25.2–26.2 |
| T1N84 | Human | 41.2–41.5 | 25.4–26.5 |
| T1C23 | Bovine | 39.7–40.1 | 21.3–21.9 |
| T1C50 | Bovine | 41.0–41.3 | 25.4–26.5 |
| T2 J/55 | Human | 41.7–42.0 | 26.5–27.2 |
| T2N73 | Human | 39.8–40.5 | 24.2–24.7 |
| T2N84 | Human | 39.8–40.5 | 24.9–25.7 |
| T2W97 | Human | 39.8–40.4 | 24.9–25.7 |
| T3D/55 | Human | 83.4–84.2 | 88.2–90.2 |
| T3C84 | Human | 83.0–83.9 | 88.4–90.0 |
| T3C87 | Human | 83.2–84.0 | 88.2–90.2 |
| T3C93 | Human | 82.7–83.6 | 87.4–89.5 |
| T3C8 | Human | 82.5–83.1 | 86.9–88.7 |
| T3C96 | Human | 70.7–71.0 | 74.8–75.1 |
| T3C43 | Bovine | 83.1–84.1 | 88.4–90.5 |
| T3C31 | Bovine | 78.7–79.7 | 85.1–86.9 |
| T3C45 | Bovine | 83.1–84.0 | 88.2–90.2 |
| T3C44 | Bovine | 83.1–84.1 | 88.2–90.2 |
| T3C18 | Bovine | 78.5–79.7 | 85.1–86.9 |
| T3C9 | Murine | 80.1–81.2 | 84.8–86.6 |
| T3S06 | Porcine | 97.0–98.1 | 96.4–98.2 |
The classification of Korean MRV strains into 10 is based on the phylogenetic data in which they clustered on the separate branches (Fig. 2).
Fig. 3.
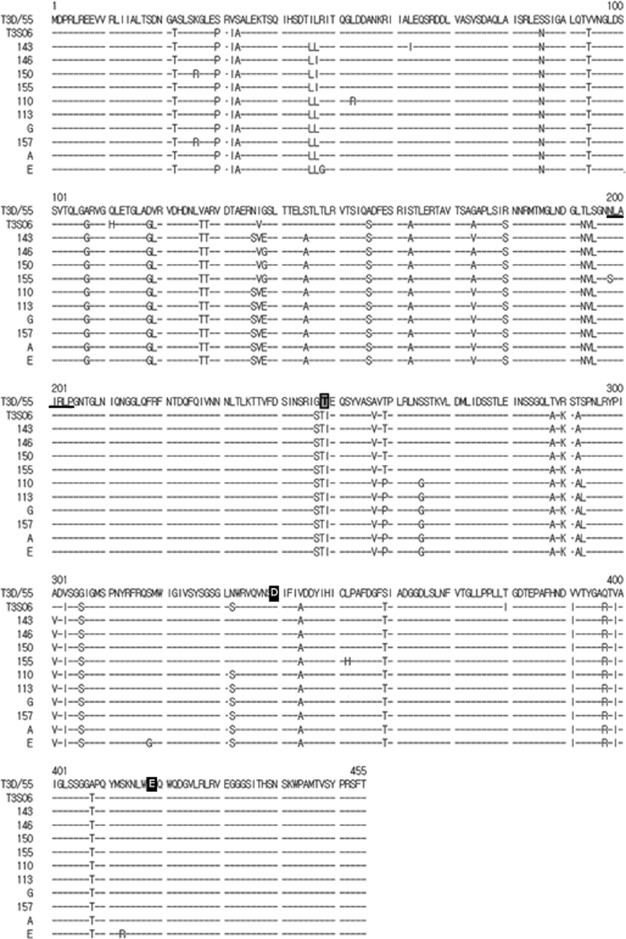
Deduced amino acid sequences of the σ1 protein of the 10 Korean porcine orthoreovirus strains, and the T3D/55 and T3S06 strains. The predicted sialic acid-binding domain is underlined; the sequence associated with sensitivity to cleavage by intestinal proteases is shown in a light-grey box; the sequences associated with neuronal tropism are in black boxes (Tyler et al., 2004).
When we compared the deduced amino acid sequences of the σ1 proteins of our 10 Korean porcine orthoreoviruses with those of the T3D55 and T3/porcine/Sichuan/2006 strains, we found that the sequence NLATRLP, representing amino acids 198–204 and constituting a binding site for sialic acid, was conserved in 9 of the Korean strains, with strain KPR-155 showing a change in amino acid 198 (198N → 198S). Polymorphisms at amino acid 249 have been found to affect the susceptibility of type 3 σ1 protein to cleavage by intestinal proteases (Chappell et al., 1998). All 10 Korean porcine orthoreoviruses encoded an isoleucine residue at amino acid 249, which is characteristic of all type 3 orthoreovirus strains with protease-resistant σ1 proteins (Chappell et al., 1998). Two amino acid residues (350D and 419E) in the σ1 head domain (amino acids 340-419) have been implicated in orthoreovirus neurotropism (Kaye et al., 1986, Bassel-Duby et al., 1986); these residues were conserved in all 10 Korean porcine orthoreoviruses.
4. Discussion
To the best of our knowledge, the prevalence of porcine orthoreovirus has not previously been examined, and only one type 3 porcine orthoreovirus strain, isolated in China in 2008, has been characterized in molecular detail (Zeng et al., 2008). Thus, the present work is the first molecular epidemiological study of porcine orthoreoviruses in diarrheic pigs. We found that 19.0% of diarrheic fecal samples from pigs tested positive for porcine orthoreovirus, indicating that infection with such viruses is endemic in piglets with diarrhea in South Korea. Among orthoreovirus-positive fecal samples, only 6.7% tested positive for porcine orthoreoviruses alone, whereas the remainder were also positive for other enteric pathogens including the GARVs, GBRVs, GCRVs, PSaVs, E. coli, Salmonella spp., and swine dysentery, suggesting that concurrent infections with other enteric pathogens may increase the clinical signs and intestinal pathology caused by porcine orthoreovirus infection (Chang et al., 1999).
The 10 strains of porcine orthoreovirus isolated from fecal samples were identified as type 3 orthoreoviruses by RT-PCR, HA capacity, and molecular and phylogenetic analyses of the S1 gene. Phylogenetically, type 3 orthoreoviruses can be classified into four lineages, with lineage I consisting of a human strain isolated in 1996; lineages II and III of bovine, murine, and human strains isolated in the 1950s and 1960s; and lineage IV of porcine strains isolated in the 2000s, including the 10 Korean porcine orthoreovirus strains of the present work and a Chinese porcine strain isolated in 2008. The classification of type 3 lineages is assisted by comparison of deduced amino acid sequences, in that strains of lineage IV have the highest nucleotide (97.0–98.1%) and deduced amino acid (96.4–98.2%) identities, but much lower nucleotide (70.7–84.2%) and deduced amino acid (74.8–90.2%) identities when compared to strains of other lineages. This indicates that the porcine orthoreovirus strains belong to a different genetic lineage, and may have evolved on a pathway distinct from that of the human, bovine, and murine strains. Because the sequences of recently isolated human, bovine, and murine strains, and those of other porcine strains, are not yet available, the detailed classification remains unclear. In-depth molecular analysis using strains isolated from other species and from various geographic areas is needed to understand viral diversity and evolution and to accurately classify the strains.
The type 3 orthoreovirus S1 gene, which encodes the viral cell attachment protein, is strongly associated with tissue tropism (Schiff et al., 2007). For example, the key amino acid residues of the σ1 protein implicated in type 3 neural tropism and neurovirulence have been shown to be conserved (Kaye et al., 1986, Bassel-Duby et al., 1986). In all known mammalian orthoreoviruses, the σ1 protein contains a binding site for sialic acid at amino acids 198–204 (NLATRLP) (Chappell et al., 1997, Rubin et al., 1992). The conservation of this sequence suggests that binding to sialic acid may be important for neurovirulence (Chappell et al., 1997, Chappell et al., 2000, Dermody et al., 1990), enhancing the ability of an orthoreovirus to migrate from the murine intestine to the CNS (Barton et al., 2003). Interestingly, the NLATRLP sequence was conserved in 9 of our 10 Korean porcine orthoreoviruses except KPR-155 strain (198N → 198S). In addition, the isoleucine at position 249 of the σ1 protein has been linked to the capacity of type 3 orthoreovirus strains to infect the murine intestine and to spread therefrom to the CNS (Chappell et al., 1998). All 10 Korean porcine orthoreovirus strains contained an isoleucine residue at position 249 of the σ1 protein. A protease-resistant σ1 protein may be required both for efficient viral growth in the intestine and migration to secondary sites of replication, including the CNS (Barton et al., 2003). Moreover, two amino acids (350D and 419E) in the σ1 head domain, associated with neurotropism, were conserved in all 10 Korean porcine orthoreovirus strains, suggesting that these 9 of 10 strains may be neurovirulent in animals. Thus, we are currently studying both enterotropism and CNS tropism of the novel viruses, as well as replication in other organs and tissues of pigs and mice.
In summary, we found that porcine orthoreoviruses were endemic in Korean piglets with diarrhea and that the viruses isolated belonged to a newly identified lineage IV of type 3 orthoreoviruses. Sequencing of the S1 genes of the 9 Korean porcine orthoreoviruses showed that the viruses were neurotropic.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (No. 2011-0002234), KRIBB Research Initiative Program, the Korea Science and Engineering Foundation (KOSEF) Grant (2009-0081752), and Chonnam National University, Republic of Korea. H.J. Kim, J.G. Park and K.Y. Son acknowledge a graduate fellowship from the Korean Ministry of Education and Human Resources Development through the Brain Korea 21 project.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.vetmic.2011.12.032.
Appendix A. Supplementary data
References
- Asakura H., Makino S., Shirahata T., Tsukamoto T., Kurazono H., Ikeda T., Takeshi K. Detection and genetical characterization of Shiga toxin-producing Escherichia coli from wild deer. Microbiol. Immunol. 1998;42:815–822. doi: 10.1111/j.1348-0421.1998.tb02356.x. [DOI] [PubMed] [Google Scholar]
- Barton E.S., Youree B.E., Ebert D.H., Forrest J.C., Connolly J.L., Valyi-Nagy T., Washington K., Wetzel J.D., Dermody T.S. Utilization of sialic acid as a coreceptor is required for orthoreovirus -induced biliary disease. J. Clin. Invest. 2003;111:1823–1833. doi: 10.1172/JCI16303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel-Duby R., Spriggs D.R., Tyler K.L., Fields B.N. Identification of attenuating mutations on the orthoreovirus type 3 S1 double-stranded RNA segment with a rapid sequencing technique. J. Virol. 1986;60:64–67. doi: 10.1128/jvi.60.1.64-67.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breun L.A., Broering T.J., McCutcheon A.M., Harrison S.J., Luongo C.L., Nibert M.L. Mammalian orthoreovirus L2 gene and λ2 core spike protein sequences and whole-genome comparisons of orthoreoviruses Type 1 Lang, Type 2 Jones, and Type 3 Dearing. Virology. 2001;287:333–348. doi: 10.1006/viro.2001.1052. [DOI] [PubMed] [Google Scholar]
- Chang K.O., Nielsen P.R., Ward L.A., Saif L.J. Dual infection of gnotobiotic calves with bovine strains of group A and porcine-like group C rotaviruses influences pathogenesis of the group C rotavirus. J. Virol. 1999;73:9284–9293. doi: 10.1128/jvi.73.11.9284-9293.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J.D., Gunn V.L., Wetzel J.D., Baer G.S., Dermody T.S. Mutations in type 3 orthoreovirus that determine binding to sialic acid are contained in the fibrous tail domain of viral attachment protein sigma1. J. Virol. 1997;71:1834–1841. doi: 10.1128/jvi.71.3.1834-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J.D., Barton E.S., Smith T.H., Baer G.S., Duong D.T., Nibert M.L., Dermody T.S. Cleavage susceptibility of orthoreovirus attachment protein sigma1 during proteolytic disassembly of virions is determined by a sequence polymorphism in the sigma1 neck. J. Virol. 1998;72:8205–8213. doi: 10.1128/jvi.72.10.8205-8213.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J.D., Duong J.L., Wright B.W., Dermody T.S. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 orthoreoviruses. J. Virol. 2000;74:8472–8479. doi: 10.1128/jvi.74.18.8472-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody T.S., Nibert M.L., Bassel-Duby R., Fields B.N. A sigma 1 region important for hemagglutination by serotype 3 orthoreovirus strains. J. Virol. 1990;64:5173–5176. doi: 10.1128/jvi.64.10.5173-5176.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C., Park S.I., Park S.H., Kim H.H., Park S.J., Jeong J.H., Choy H.E., Saif L.J., Kim S.K., Kang M.I., Hyun B.H., Cho K.O. Genetic diversity of porcine sapoviruses. Vet. Microbiol. 2007 doi: 10.1016/j.vetmic.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P.J., Sveger T., Ahlfors K., Ekstrand J., Svensson L. Orthoreovirus type 1 associated with meningitis. Scand. J. Infect. Dis. 1996;28:117–120. doi: 10.3109/00365549609049060. [DOI] [PubMed] [Google Scholar]
- Kaye K.M., Spriggs D.R., Bassel-Duby R., Fields B.N., Tyler K.L. Genetic basis for altered pathogenesis of an immune-selected antigenic variant of orthoreovirus type 3 Dearing. J. Virol. 1986;59:90–97. doi: 10.1128/jvi.59.1.90-97.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.W.K., Hayes E.C., Joklik W.K. Protein σ1 is the orthoreovirus cell attachment protein. Virology. 1981;108:156–163. doi: 10.1016/0042-6822(81)90535-3. [DOI] [PubMed] [Google Scholar]
- Nicola D., Marco C., Costantina D., Dominga R., Michele C., Eleonora L., Gabriella E., Antonio L., Vito M., Canio B. Virological and molecular characterization of a mammalian orthoreovirus type 3 strain isolated from a dog in Italy. Vet. Microbiol. 2005;109:19–27. doi: 10.1016/j.vetmic.2005.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramig R.F., Cross R.K., Fields B.N. Genome RNAs and polypeptides of orthoreovirus serotypes 1, 2 and 3. J. Virol. 1977;22:726–733. doi: 10.1128/jvi.22.3.726-733.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin D.H., Wetzel J.D., Williams W.V., Cohen J.A., Dworkin C., Dermody T.S. Binding of type 3 orthoreovirus by a domain of the sigma 1 protein important for hemagglutination leads to infection of murine erythroleukemia cells. J. Clin. Invest. 1992;90:2536–2542. doi: 10.1172/JCI116147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin A.B. Orthoreoviruses: a new group of respiratory and enteric viruses formerly classified as ECHO type 10 is described. Science. 1959;130:1387–1389. doi: 10.1126/science.130.3386.1387. [DOI] [PubMed] [Google Scholar]
- Schiff L.A., Nibert M.L., Tyler K.L. Orthoreoviruses and their replication. In: Knipe D.M., Griffin D.E., Lamb R.A., Straus S.E., Howley P.M., Martin M.A., Roizman B., editors. Fields Virology. fifth edition. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 1853–1915. [Google Scholar]
- Suh D.K., Song J.C. Simultaneous detection of Lawsonia intracellularis, Brachyspira hyodysenteriae and Salmonella spp. In swine intestinal specimens by multiplex polymerase chain reaction. J. Vet. Sci. 2005;6:231–237. [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tyler K.L. Mammalian orthoreoviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. fourth edition. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 1729–1745. [Google Scholar]
- Tyler K.L., Barton E.S., Ibach M.L., Robinson C., Campbell J.A., O’Donnell S.M., Valyi-Nagy T., Clarke P., Wetzel J.D., Dermody T.S. Isolation and molecular characterization of a novel type 3 orthoreovirus from a child with meningitis. J. Infect. Dis. 2004;189:1664–1675. doi: 10.1086/383129. [DOI] [PubMed] [Google Scholar]
- Virgin H.W., 4th, Bassel-Duby R., Fields B.N., Tyler K.L. Antibody protects against lethal infection with the neurally spreading orthoreovirus type 3 (Dearing) J. Virol. 1988;62:4594–4604. doi: 10.1128/jvi.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorwell P.J., Baldwin R.C., Wright R. Ferritin in Crohn's disease tissue: detection by electron microscopy. Gut. 1976;17:696–699. doi: 10.1136/gut.17.9.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H.L., Adult K.A., Fields B.N. Interaction of orthoreovirus with cell surface receptors. I. Murine and human lymphocytes have a receptor for the hemagglutinin of orthoreovirus type 3. J. Immunol. 1980;124:2143–2148. [PubMed] [Google Scholar]
- Wiener J.R., Joklik W.K. The sequences of the orthoreovirus serotype 1, 2, and 3 L1 genome segments and analysis of the mode of divergence of the orthoreovirus serotypes. Virology. 1989;169:194–203. doi: 10.1016/0042-6822(89)90055-x. [DOI] [PubMed] [Google Scholar]
- Yin P., Keirstead N.D., Broering T.J., Arnold M.M., Parker J.S., Niber M.L., Coombs K.M. Comparisons of the M1 genome segments and encoded mu2 proteins of different orthoreovirus isolates. Virol. J. 2004;23:6–22. doi: 10.1186/1743-422X-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z.Y., Guo W.Z., Xu Z.W., Liang H.Y., Song Z.H., Yin H.P., Wang X., Wang X.Y. Isolation of orthoreovirus SC-A strain from fecal of diarrhea pigs and sequence analysis of σ2 gene. Chinese J. Vet. Sci. 2008;7 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


