Highlights
-
•
We examined the specific amino acids contributing to S2 epitopes in IBVs.
-
•
16R in S2 protein was a key amino acid mediating the antigenicity of S2 protein.
-
•
S2-derived peptides with 16R, but not those with 16 K, reacted with IBV-infected serum.
-
•
Commercial ELISAs did not react with sera harboring all types of IBVs.
-
•
S2-derived peptides with 16R could be novel antigens for anti-IBV vaccines.
Keywords: Infectious bronchitis virus, S2 protein, 16R, Peptide, Serology
Abstract
Vaccination plays a vital role in controlling diseases caused by chicken infectious bronchitis virus (IBV). The continuously variant antigenicity of IBV limits the application of current vaccine strategies and serological diagnostic systems. S2 protein is an invariant that harbors broad neutralizing epitopes. However, little is known about the key amino acids that contribute to the broad-spectrum S2 epitopes. In this study, we aimed to elucidate the specific amino acids contributing to S2 epitopes. Site mutagenesis and peptide-based enzyme-linked immunosorbent assays (ELISAs) showed that 16R in S2 protein was a key amino acid mediating the antigenicity of S2 protein. S2-derived peptides with 16R, but not those with 16 K, could react with sera against different types of IBVs. Notably, a commercial ELISA kit for detection of antibodies against IBV did not react with sera against all types of IBVs. Taken together, these data demonstrated that S2-derived peptides with 16R could be used as novel marker-based antigens for developing both broad-spectrum vaccines and serological diagnostic kits to control IBV.
1. Introduction
Infectious bronchitis (IB) is a highly contagious disease caused by IB virus (IBV). IBV infection generally causes serious respiratory and renal diseases in meat chickens and decreased egg laying in laying hens (Cavanagh, 2007), resulting in huge economic losses in the poultry industry (Bande et al., 2017). Although IBV vaccines are widely used, wild-type IBV or IBV escape mutants are frequently isolated from vaccinated chicken flocks because of the circulation of multiple serotypes in flocks and continuous mutation of the protective epitopes of IBVs. Thus, current vaccine strategies need to be re-evaluated, and more efficient, broadly protective IBV vaccines or diagnostic approaches are urgently needed.
The IBV genome encodes four major structural proteins, i.e., spike (S), small envelope (E), membrane (M), and nucleocapsid (N), as well as 15 nonstructural proteins and some accessory proteins (Lin and Chen, 2017). Among these proteins, S glycoprotein is the major protective antigen carrying neutralizing epitopes and can induce efficient immune responses against IBV (Ignjatovic and Galli, 1993, 1995; Kant et al., 1992). Notably, S protein can be cleaved into the amino-terminal S1 and carboxy-terminal S2 domains by a furine-like protease from the host (Cavanagh, 2007; Eldemery et al., 2017; Kusters et al., 1989). S1 is responsible for IBV attachment to host cells, whereas S2 contributes to viral fusion activity of IBV (Wickramasinghe et al., 2014). Both S1 and S2 play vital roles in mediating IBV infection in host cells. S1 is a major protective antigen for IBV, but is highly variable among different IBV stains (Ignjatovic and Galli, 1994). In contrast to S1, S2 is highly conserved and carries broad antigenic epitopes (Toro et al., 2014). Therefore, S2 could be a marker for developing more efficient vaccines and broad diagnostic approaches.
Previously, S2 glycoprotein from IBV was not thought to induce virus-neutralizing antibodies or chicken tracheal protection (Cavanagh et al., 1986). (Ignjatovic and Galli, 1995) found that S2 glycoprotein was the most immunogenic structural protein following vaccination with inactivated virus. S2 protein expressed from recombinant virus confers broad protection against challenge (Toro et al., 2014). Therefore, S2 protein has important roles in inducing protective immunity (Eldemery et al., 2017). Moreover, (Kusters et al., 1989) first identified a conservative immunodominant region near the N-terminus of S2 protein. Recently, several regions (e.g., 8NCPYVSYGKFCIKPDGSIST27, designated SE8-27) in S2 have been identified as broad or neutralizing epitopes for IBV (Andoh et al., 2018; Ignjatovic and Sapats, 2005). However, little is known regarding the key amino acids that contribute to broad-spectrum S2 epitopes.
In this study, we analyzed 330 IBV isolates from GenBank database and evaluated effects of mutations in the SE8-27 epitope in S2 on antigenicity.
2. Materials and methods
2.1. Virus and serum samples
M41 strain (GenBank accession number: DQ834384; mass type IBV) and 4/91 strain (GenBank accession number: KF377577; 4/91 type IBV) of IBV was obtained from Sinopharm Yangzhou Vac Biological Engineering Co., Ltd. (Yangzhou, China). CK/CH/2010/JT1 (GenBank accession number: KU361187; new cluster type IBV), CK/CH/2014/QL1403 (GenBank accession number: KU361198; TC07-2 type IBV), and CK/CH/2014/FJ14 strains of IBV (GenBank accession number: MN262521; QX-like type IBV) were isolated and identified by our laboratory. SPF white leghorn chicken eggs were purchased from Beijing Merial Vital Laboratory Animal Technology Co., Ltd. (Beijing, China) and hatched out in our laboratory. All birds were kept in isolators throughout the experiments. To prepare serum against M41, 4/91, CK/CH/2010/JT1, CK/CH/2014/QL1403, and CK/CH/2014/FJ14 strains, 14-day-old SPF chickens were inoculated intraocular-nasally with 103 EID50 of different strains. Thirty days after immunization, serum from immunized chickens was collected and stored at −20℃. All experiments complied with institutional animal care guidelines and were approved by the University of Yangzhou Animal Care Committee.
2.2. Preparation of the peptide library
Amino acid sequences of the S2 protein of IBV strains, including H120, D207, and CK/CH/2010/JT1 strains, were obtained from GenBank (accession numbers: MK937831, X58003, and KU361187, respectively). Six peptides (Pep1–Pep6) with good conservation and antigenicity were selected by MegAlign and Protean software, version 7.1.0 (DNAstar, Madison, WI, USA). Pep1 is a conservative epitope for all IBV strains (Ignjatovic et al., 2005). Pep2–Pep5 were designed according to conserved peptides in the S2 gene of IBV. Peptides containing between 16 and 20 amino acid residues were synthesized by Synpeptide Co., Ltd. (Shanghai, China). The peptide sequences are listed in Table 1 .
Table 1.
Description of the synthetic peptides used in this study.
| ID | Sequences of S2 | Amino acid position |
|---|---|---|
| Pep1 | NCPYVSYGKFCIKPDGSIST | 8–27 |
| Pep2 | GKKSSYYTTFDNDVVTEQYRPKKSV | 601–625 |
| Pep3 | SQQRELATQKINECVKSQSIR | 355–375 |
| Pep4 | NLTVTDEYIQTRMDKVQINCLQYICGNSLE | 52–81 |
| Pep5 | QQYGPVCDNILSVVNSVGQKEDMELLNFYSSTKPAG | 87–122 |
| Pep6 | SCPYVSYGRFCIQPDGSIKQ | 8–27 |
| Pep1-1 | NCPYVSYGKFCIKPDG | 8–23 |
| Pep6-1 | SCPYVSYGRFCIQPDG | 8–23 |
| Pep1-1-1 | SCPYVSYGKFCIKPDG | 8–23 |
| Pep1-1-2 | NCPYVSYGRFCIKPDG | 8–23 |
| Pep1-1-3 | NCPYVSYGKFCIQPDG | 8–23 |
| Pep1-1-4 | SCPYVSYGKFCIQPDG | 8–23 |
Notes: Amino acid sequences were deduced from S2 protein of CK/CH/2010/JT1 (accession number: KU361187). The numbers represent the amino acid position of the peptide in the S2 subunit of IBV.
2.3. Enzyme-linked immunosorbent assay (ELISA)
For ELISA, 96-well plates were coated with 0.1 μg/well synthetic peptide in 0.1 M carbonate buffer (pH 9.6) at 4℃ overnight (Khairy et al., 2017). After washing with phosphate-buffered saline (PBS) with Tween 20 (PBST), the plates were blocked with 300 μL of 8% rabbit serum (Lanzhou Minhai Biological Engineering Co., Ltd., China) for 3 h at 37℃. Following three washes with PBST, 100 μL serum (1:200 in PBST) was added to the wells, and the plates were incubated at 37℃ for 1 h. The plates were then washed five times and further incubated with 100 μL horseradish peroxidase (HRP)-conjugated rabbit anti-chicken IgG (Jackson ImmunoResearch Laboratories, Inc., USA) at 1:30000 dilution in blocking buffer for 1 h at 37℃. After washing five times, and the colorimetric reaction was developed by incubating the plates with 100 μL TMB substrates for 1 5 min at 37℃. Color development was stopped with 100 μL of 1% sodium dodecyl sulfate. The absorbance at OD650 was obtained using an ELISA reader (BioTek, VT, USA). There were five serum samples for each genotype IBV strain. Each experiment was repeated twice.
A commercial IBV antibody detection kit was purchased from IDEXX (USA). Briefly, all reagents were allowed to warm to 18–26℃ before use. Negative and positive controls were dispensed into duplicate wells. Samples diluted 500-fold with sample diluent were added to appropriate wells and incubated for 30 min. The plates were washed with approximately 350 μL distilled water five times. Next, 100 μL of HRP-conjugated secondary antibodies was added to each well, and plates were incubated for 30 min. After washing five times, 100 μL TMB substrate was dispensed into each well, and plates were incubated for 15 min. Color development was stopped with 100 μL stop solution. The absorbance at OD650 was then determined. Positive results were defined as S/P values over 0.2.
2.4. Sequence analysis of IBV S2 protein
All available 330 complete genome IBV strains were downloaded from the NCBI database. The sequences were analyzed using MegAlign and Protean software, version 7.1.0 (DNAstar).
2.5. Statistical analysis
All the data were graphed, and statistical analyses were performed using Prism 5 software(GrapghPad, La Jolla, CA). One-way ANOVA with repeated measures was used to evaluate the reactivity of peptide antigens with sera against IBV. The differences were considered statistically significant at p values of <0.01.
3. Results
3.1. Variations in the broad-spectrum region (amino acids 8–27) in the S2 protein of IBV
The region containing 8NCPYVSYGKFCIKPDGSIST27 in S2 was downloaded from GenBank. After analysis of complete genome sequences of 330 IBV isolates, we found that almost all IBV strains contained a K at amino acid position 16 (16 K) in S2 before the 1990s. Subsequently, strains with an R at amino acid position 16 (16R) emerged and became the dominant pandemic IBV strains. Interestingly, 16 K IBV strains were mainly associated with respiratory tract diseases, and the 16R IBV strains were mainly associated with renal pathogenicity. Based the region with 8NCPYVSYGKFCIKPDGSIST27 in S2, IBV strains could be divided into two groups: Group Ⅰ containing 16 K, and Group Ⅱ containing 16R (Table 2 ). Two strains, i.e., B1648 (accession number: KR231009) and gamaCoV/ck/China/I0636/16 (accession number: MH924835), contained T and Q as the 16th amino acids (group I-like), respectively.
Table 2.
Sequence analysis of amino acids 8–27 in S2 protein from 314 IBV strains.
| Genotypes | Sequences of amino acids 8–27 in S2 protein | |
|---|---|---|
| Group Ⅰ (136/328) | GI-1, GI-2, GI-3, GI-4, GI-5, GI-6, GI-8, GI-9, GI-11, GI-12, GI-13, GI-14, GI-16, GI-21, GI-22, GI-26, GII-1, GIV-1, GVI-1 | NCPYVSYGKFCIKPDGSIST |
| NCPYVSYGKFCIKPDGSIAT | ||
| NCPYVSYGKFCIKPDGSLFI | ||
| NCPYVSYGKFCIKPDGSVSP | ||
| NCPYVSYGKFCIKPDGSISV | ||
| NCPYVSYGKFCIKPDGSVSE | ||
| NCPYVSYGKFCIKPDGLIAT | ||
| HCPYVSYGKFCIKPDGDLSV | ||
| SCPYVSYGKFCIKPDGSIST | ||
| QCPYVSYGKFCIKPDGDLSV | ||
| RCPYVSYGKFCIKPDGSLKT | ||
| Group Ⅱ (192/328) | GI-7, GI-19, GI-22 | SCSYVSYGRFCIEPDGSLKM |
| SCPYVSYGRYCIEPDGSLKQ | ||
| SCSYVSYGRFCIQPDGSIKQ | ||
| SCPYVSYGRFCIEPDGSLKM | ||
| SCPYVSYGRFCIQPDGSIKQ | ||
| SCSYVSYGRFCIEPDGSLKL | ||
| SCSHVSYGRFCIEPDGSLKM | ||
| SCHYVSYGRFCIQPDGSIKQ | ||
| SCPYVTYGRFCIQPDGSIKQ | ||
| SCNYVSYGRFCIQPDGSIKQ | ||
| SCPFVSYGRFCIEPDGSLKT | ||
| TCPFVSYGRFCIKPDGLVSE | ||
| SCPYVSYGRYCIEPDGSLKK | ||
| SCSYVSYGRFCIEPDGSLKP |
Notes: Red highlighting indicates differences in amino acid 16, which were used for grouping.
3.2. Pep6, not Pep1, showed broad reaction with sera from different types of IBVs
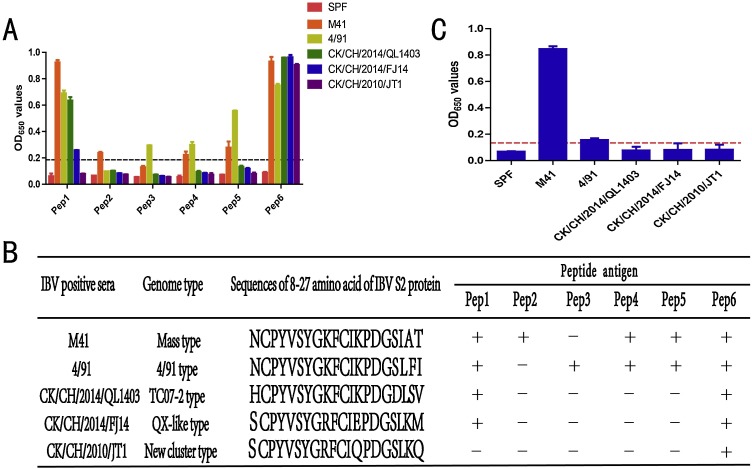
To further evaluate and identify the broad antigenic epitopes in S2, Pep1(epitope SE8-27) and Pep6 (amino acids 8–27 from S2 of Group Ⅰ and Group Ⅱ, respectively) and Pep2–5 (derived from conserved peptides of IBV S2 protein; Table 1) were synthesized and used as peptide antigens for testing crossreaction with sera from different genotypes of IBVs, including mass, QX-like, TC07-2, 4/91, and new cluster types. We analyzed the reactivity between the peptides and different genotypes of IBVs. Only Pep1 showed good reactions with most sera. However, Pep1 did not react with the sera from CK/CH/2010/JT1 strains. After comparing and analyzing amino acid sequences from different genotype IBV strains in this region, we found that there were some differences in amino acids among different genotype strains.
To elucidate which single amino acid was essential for the antigen reaction of CK/CH/2010/JT1 strain in this region, Pep6 was synthesized according to the same region in S2 in the CK/CH/2010/JT1 strain. Pep6, which contained the epitope of Group Ⅱ, reacted with all sera against the five different types of IBVs, whereas Pep1, which contained the epitope of Group Ⅰ, only reacted with four of five different types of IBVs (Fig. 1 A, 1B). Pep1 could not efficiently react with sera against IBV CK/CH/2010/JT1 (new cluster type IBV). Pep4 and Pep5 could react with sera against M41 type and 4/91 type, whereas Pep2 and Pep3 only reacted with sera against M41 type and 4/91 type, respectively. Notably, a commercial ELISA kit for detection of antibodies against IBV was also used to detect sera against different types of IBVs. The commercial kit could only react with sera against M41 and 4/91 strains, but not with sera against CK/CH/2014/QL1403, CK/CH/2014/FJ14, and CK/CH/2010/JT1 strains (Fig. 1C).
Fig. 1.
Reactivity of Pep1–Pep6 with sera against different IBV strains. The black/red dotted lines represent critical values. There were five serum samples for each genotype. Each experiment was repeated twice. The results show the mean values. (A) Reactivity of Pep1–Pep6 with sera against different IBV strains. (B) Sequences of amino acids 8–27 in S2 protein from different IBV strains and reactivities of Pep1–Pep6 in sera against different IBV strains. “+” represents peptides that reacted with sera harboring IBV; “-” represents peptides that did not react with sera harboring IBV. (C) Reactivity of a commercial ELISA kit with sera against different IBV strains. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. 16R was identified as a key amino acid for S2 broad antigenicity
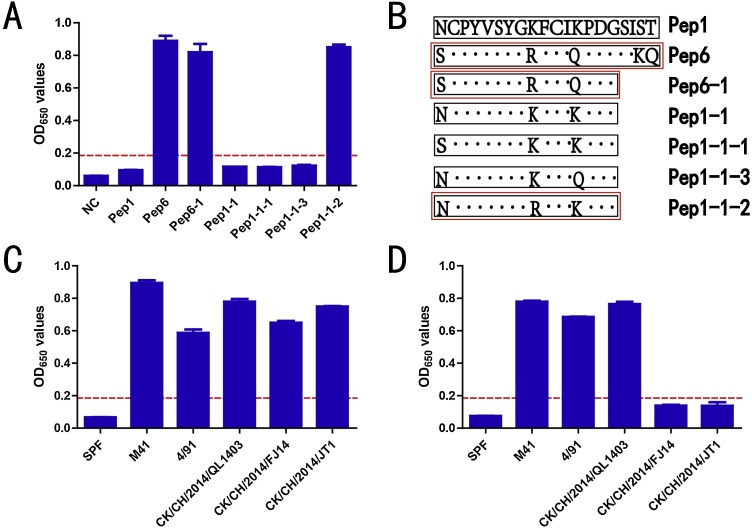
To identify the critical amino acids contributing to the broad-spectrum antigenicity of Pep6, the last four amino acids were deleted in other peptides because they showed high variation in different genotype or the same genotype of IBV. We used Pep1-1 as a prototype and mutated N8S, K16R, and K20Q, yielding Pep1-1-1, Pep1-1-2, and Pep1-1-3, respectively. The results demonstrated that 16R in Pep6-1, not 8S or 20Q, contributed to the broad-spectrum antigenicity of this epitope (Fig. 2 A). Pep1-1-2 with 16R (similar to Pep6) could react with all sera against different types of IBVs (Fig. 2C). Pep1-1-4, in which amino acid 16 (R) was mutated to K, similar to Pep6-1, did not react with positive sera against CK/CH/2014/FJ14 and XK/XH/2010/JT1. Taken together, these results showed that 16R was the only determinant of the broad-spectrum antigenicity of Pep6.
Fig. 2.
Determination of the key amino acid sites mediating broad-spectrum antigenicity in Pep1 and Pep6. The red dotted line represents the critical value. There were five serum samples for each genotype. Each experiment was repeated twice. The results show the mean values. (A) Broad-spectrum antigenicity of the synthetic peptides. (B) Strategy for the identification of key amino acids. The black dots represent the same amino acid. The red box represent peptides that could react with positive serum against CK/CH/2010/JT1 strain. (C) Broad-spectrum antigenicity of Pep1-1-2. (D) Broad-spectrum antigenicity of Pep1-1-4. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Distribution of Group Ⅱ strains carrying 16R in the epitope
Next, we downloaded and analyzed all complete genome sequences of IBVs reported in the NCBI database (https://www.ncbi.nlm.nih.gov/). The information on the geographical origin and time of isolation of these sequences were also obtained from the NCBI database. Sequence analysis further revealed that IBV strains carrying 16R in the epitope of Group Ⅱ were first isolated in the 1990s, and the proportion then increased thereafter (Fig. 3 A). IBV strain KM91, which was isolated in 1991, was the first IBV with such an epitope. The incidence of 16R-containing IBV strains increased over time. Variations in key amino acids in the conserved S2 protein suggested that the IBVs had undergone considerable variation and evolution, necessitating new vaccine strategies and diagnostics to prevent and control IBV. Currently, the epitope of Group Ⅱ is widely present in IBV isolates from Asia, America, Europe, and Africa (Fig. 3A, 3B), consistent with the prevalence of the lineage GI-19, the so-called QX-type IBV, in China and worldwide (Zhao et al., 2017). Our results demonstrated that the epitope of the QX type IBV dominant in chicken flocks in China all belonged to this group of IBV strains containing 16R.
Fig. 3.
Epidemic distribution of strains carrying the 16R epitope. “16 K” indicates that the 16th amino acid of S2 protein is a K, whereas “16R” indicates that the 16th amino acid of S2 protein is an R. (B) “Asia” represents the number of published IBV complete genome sequences in all Asian countries excluding China.
4. Discussion
IBV is a typical coronavirus that is constantly recombining and mutating its antigenicity, leading to the presence of multiple serotypes and genotypes of IBV strains and causing major challenges in disease prevention and control (Cavanagh, 2007). As an important antigenic protein, S1 has been used in vaccine development (Hodgson et al., 2004; Wickramasinghe et al., 2014; Zou et al., 2015). Notably, S2 protein also can induce protective immune responses (Eldemery et al., 2017; Ignjatovic and Sapats, 2005). In contrast to S1, S2 protein is thought to be invariant and contain neutralizing epitopes (Wickramasinghe et al., 2014). Of these epitopes in S2, the epitope containing amino acids 8–27 has been identified as a broad antigenic epitope for IBVs. Our data demonstrated that this epitope in S2 protein from different types of IBVs was also not highly conserved in different IBV serotypes and could be divided into two groups, designated Group Ⅰ (8NCPYVSYGKFCIKPDGSIST27-like) and Group Ⅱ(8SCPYVSYGRFCIQPDGSIKQ27-like). The two strains, B1648 (accession number: KR231009) and gamaCoV/ck/China/I0636/16 (accession number: MH924835), were classified as Group I because their 16th amino acids were T and Q, respectively.
To identify the key amino acid in Pep6, the last four amino acid of Pep1 and Pep6 were removed, as previously described (Kusters et al., 1989). Our results also showed that the last amino acid had minor effects on antigenicity. Further mutational analysis confirmed that mutation of the 16th amino acid(K16R) was the key for the broad-spectrum antigenicity of this epitope.
Serological assays further showed that the S2-derived epitope in Group Ⅱ, but not that in Group Ⅰ, could react with all sera against different types of IBVs. Moreover, 16R was identified as a key amino acid for the broad antigenicity of Group Ⅱ. Valastro et al. (2016) performed a phylogenetic analysis based on the S1 gene of 1286 strains to derive a new and coherent classification scheme for IBVs. They divided the IBV strains into six genotypes, labeled GI, GII, GIII, GIV, GV, and GVI. GI contained 27 lineages, whereas GII, GIII, GIV, GV, and GVI contained only one lineage each. Our grouping method was based on S2 protein of IBV. We further compared two different grouping methods and found that GI-7, GI-19, and GI-22 belonged to Group Ⅱ, whereas the other strains belonged to Group Ⅰ. Notably, widespread QX genotype, new cluster genotype, TW-I type, LDT3 type, and BJ type IBVs all carried 16R in the S2 protein. This suggested that peptides with 16R derived from S2 protein could be used as novel marker-based antigens for developing broad-spectrum vaccines against circulating IBVs. Our grouping method could distinguish strains causing respiratory tract disease and from those causing renal disease. We found that all QX, LDT3, and BJ genotype IBV strains prevalent in China in recent years contained the 16R mutation, which may partly explain the poor efficacy of conventional vaccines against new epidemic IBV strains. Thus, our results may provide guidance for vaccine selection.
To detect IBV antibodies as accurately and comprehensively as possible, many ELISA antibody detection methods have been established (Chen et al., 2011; Ding et al., 2015; Lei et al., 2017). All of these methods have good efficacy, but still have some limitations. In our study, we found that the IDEXX IBV antibody detection kit, as a common antibody detection tool, could not detect all serum antibodies for all IBV genotypes. Interestingly, Pep6 found in this study could cross react with sera against different genotypes of IBV. Therefore, peptides with 16R derived from S2 could be used as novel peptide-based antigens for developing diagnostic kits for IBV serological detection.
Taken together, this study showed that S2-derived peptides with 16R showed broad reactions with sera against containing different types of infectious bronchitis viruses. It could be used as novel marker-based antigens for developing both broad-spectrum vaccines and serological diagnostic kits to control IBV. Whether the 16R in the IBV S2 protein playing a role in tissue tropism and induced immune response remains to be investigated in further.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgements
This study was supported by the National Key Research and Development Program of China (grant no. 2016YFD0500800), Special Foundation for State Basic Research Program of China (grant no. 2013FY113300-4), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Jiangsu Co-innovation Centre for the Prevention and Control of Important Animal Infectious Diseases and Zoonoses.
References
- Andoh K., Ashikaga K., Suenaga K., Endo S., Yamazaki K. Identification of novel linear epitopes located in the infectious bronchitis virus spike S2 region. Avian Dis. 2018;62:210–217. doi: 10.1637/11796-011518-Reg.1. [DOI] [PubMed] [Google Scholar]
- Bande F., Arshad S.S., Omar A.R., Hair-Bejo M., Mahmuda A., Nair V. Global distributions and strain diversity of avian infectious bronchitis virus: a review. Anim. Health Res. Rev. 2017;18:70–83. doi: 10.1017/S1466252317000044. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Darbyshire J.H., Peters R.W. Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or haemagglutination-inhibiting antibody, or induce chicken tracheal protection. J. Gen. Virol. 1986;67:1435–1442. doi: 10.1099/0022-1317-67-7-1435. [DOI] [PubMed] [Google Scholar]
- Chen H.W., Wang C.H., Cheng I.C. A type-specific blocking ELISA for the detection of infectious bronchitis virus antibody. J. Virol. Methods. 2011;173:7–12. doi: 10.1016/j.jviromet.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M.D., Wang H.N., Cao H.P., Fan W.Q., Ma B.C., Xu P.W., Zhang A.Y., Yang X. Development of a multi-epitope antigen of S protein-based ELISA for antibodies detection against infectious bronchitis virus. Biosci. Biotechnol. Biochem. 2015;79:1287–1295. doi: 10.1080/09168451.2015.1025692. [DOI] [PubMed] [Google Scholar]
- Eldemery F., Joiner K.S., Toro H., van Santen V.L. Protection against infectious bronchitis virus by spike ectodomain subunit vaccine. Vaccine. 2017;35:5864–5871. doi: 10.1016/j.vaccine.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson T., Casais R., Dove B., Britton P., Cavanagh D. Recombinant infectious bronchitis coronavirus Beaudette with the spike protein gene of the pathogenic M41 strain remains attenuated but induces protective immunity. J. Virol. 2004;78:13804–13811. doi: 10.1128/JVI.78.24.13804-13811.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignjatovic J., Galli L. Structural proteins of avian infectious bronchitis virus: role in immunity and protection. Adv. Exp. Med. Biol. 1993;342:449–453. doi: 10.1007/978-1-4615-2996-5_71. [DOI] [PubMed] [Google Scholar]
- Ignjatovic J., Galli L. The S1 glycoprotein but not the N or M proteins of avian infectious bronchitis virus induces protection in vaccinated chickens. Arch. Virol. 1994;138:117–134. doi: 10.1007/BF01310043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignjatovic J., Galli U. Immune responses to structural proteins of avian infectious bronchitis virus. Avian Pathol. 1995;24:313–332. doi: 10.1080/03079459508419072. [DOI] [PubMed] [Google Scholar]
- Ignjatovic J., Sapats S. Identification of previously unknown antigenic epitopes on the S and N proteins of avian infectious bronchitis virus. Arch. Virol. 2005;150:1813–1831. doi: 10.1007/s00705-005-0541-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant A., Koch G., van Roozelaar D.J., Kusters J.G., Poelwijk F.A., van der Zeijst B.A. Location of antigenic sites defined by neutralizing monoclonal antibodies on the S1 avian infectious bronchitis virus glycopolypeptide. J. Gen. Virol. 1992;73:591–596. doi: 10.1099/0022-1317-73-3-591. [DOI] [PubMed] [Google Scholar]
- Khairy W.O.A., Qian K., Shao H.X., Ye J.Q., Qin A.J. Identification of two conserved B-cell epitopes in the gp90 of reticuloendothelial virus using peptide microarray. Vet. Microbiol. 2017;211:107–111. doi: 10.1016/j.vetmic.2017.10.009. [DOI] [PubMed] [Google Scholar]
- Kusters J.G., Jager E.J., Lenstra J.A., Koch G., Posthumus W.P., Meloen R.H., van der Zeijst B.A. Analysis of an immunodominant region of infectious bronchitis virus. J. Immunol. 1989;143:2692–2698. [PubMed] [Google Scholar]
- Lei J., Shi T.T., Sun D.N., Mo K.K., Yan Y., Jin Y.L., Liao M., Zhou J.Y. Development and application of nsp5-ELISA for the detection of antibody to infectious bronchitis virus. J. Virol. Methods. 2017;243:182–189. doi: 10.1016/j.jviromet.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Chen H.W. Infectious bronchitis virus variants: molecular analysis and pathogenicity investigation. Int. J. Mol. Res. Sci. 2017;18 doi: 10.3390/ijms18102030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro H., Zhao W., Breedlove C., Zhang Z., Yu Q., Van Santen V. Infectious bronchitis virus S2 expressed from recombinant virus confers broad protection against challenge. Avian Dis. 2014;58:83–89. doi: 10.1637/10641-081613-Reg.1. [DOI] [PubMed] [Google Scholar]
- Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G., Monne I. S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect. Genet. Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe I.N., van Beurden S.J., Weerts E.A., Verheije M.H. The avian coronavirus spike protein. Virus Res. 2014;194:37–48. doi: 10.1016/j.virusres.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Gao M., Xu Q., Xu Y., Zhao Y., Chen Y., Zhang T., Wang Q., Han Z., Li H., Chen L., Liang S., Shao Y., Liu S. Origin and evolution of LX4 genotype infectious bronchitis coronavirus in China. Vet. Microbiol. 2017;198:9–16. doi: 10.1016/j.vetmic.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou N., Xia J., Wang F., Duan Z., Miao D., Yan Q., Cao S., Wen X., Liu P., Huang Y. Two novel neutralizing antigenic epitopes of the s1 subunit protein of a QX-like avian infectious bronchitis virus strain Sczy3 as revealed using a phage display peptide library. Vet. Immunol. Immunopathol. 2015;168:49–55. doi: 10.1016/j.vetimm.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]