Highlights
-
•
A novel genotype, designated as GVII-1, has been identified in south China over a 5-year period.
-
•
GVII-1 was derived from two recombination events that replaced the spike gene in a GI-18-like virus with an as-yet-unidentified sequence.
-
•
GVII-1 represented a novel serotype.
-
•
GVII-1 showed a low affinity to the respiratory tract in chickens.
Keywords: Infectious bronchitis virus, Genotype, Serotype, Recombination event, Affinity
Abstract
Recombination events are known to contribute to the emergence of novel infectious bronchitis virus (IBV) genotypes. In this study, we carried out detailed phylogenetic analysis and sequence comparisons based on 74 complete nucleotide sequences of the IBV S1 gene, including strain I0636/16 and 73 representative sequences from each genotype and lineage. The results showed that strain I0636/16 represented a novel genotype, designated as lineage 1 within genotype VII (GVII-1). Further comparative genomic analysis revealed at least two recombination sites that replaced the spike gene in a lineage 18 within genotype I (GI-18)-like virus with an as-yet-unidentified sequence, likely derived from another IBV strain, resulting a novel serotype with a lower affinity to the respiratory tract in chickens. To the best of our knowledge, this provides the first evidence for recombination leading to replacement of the complete spike gene and the emergence of a novel genotype/serotype with a lower affinity to the respiratory tract in chickens comparing to one of its parental virus ck/CH/LGX/111119. These results emphasize the importance of limiting exposure to novel IBVs that may serve as a source of genetic material for emerging viruses, as well as the importance of IBV surveillance in chicken flocks.
1. Introduction
Avian infectious bronchitis, caused by infectious bronchitis virus (IBV), was first described in North Dakota, USA in the 1930s (Schalk and Hawn, 1931). It is a highly contagious viral respiratory disease that is considered as one of the most important causes of heavy economic losses to the poultry industry worldwide (Cavanagh and Gelb, 2008). All IBV strains replicate primarily in the respiratory tract and cause respiratory diseases in birds. However, some virus strains are also able to replicate in many other epithelial surfaces, including kidneys and oviducts, and infection with these strains may thus result in nephritis, decreased egg production and quality, and significant mortality. Numerous different IBV strains have been reported to date, with pathologies ranging from mild respiratory symptoms to severe kidney and oviduct diseases (Cavanagh, 2007).
IBV is an avian coronavirus with a single-stranded, positive-sense, RNA genome of approximately 27 kb. The 3′ end of the genome encodes four structural proteins, including spike glycoprotein (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, and four non-structural accessory proteins, 3a, 3b, 5a, and 5b. The 5′ end of the genome encodes two polyproteins (1a and 1ab) that contain proteins necessary for RNA replication (Boursnell et al., 1987). Similar to other coronaviruses, genetic diversity in IBV is created by both recombination events (Kottier et al., 1995) and by mutations, including substitutions, deletions, and insertions (Shi et al., 2000), that occur during replication of the viral genome. The high mutation rates are attributed to the limit of replication fidelity although it has been shown that coronaviruses possess proofreading enzyme (3′-to-5′ exoribonuclease) which is essential for replication fidelity (Denison et al., 2011), while recombination events are thought to result from a unique template-switching copy-choice mechanism during RNA replication (Simon-Loriere and Holmes, 2011).
The spike protein comprises about 1145 amino acids and undergoes post-translational cleavage to form S1 and S2 subunits (Cavanagh, 1983). The S1 subunit contains virus-neutralizing epitopes and carries serotype-specific determinants. Furthermore, the S1 gene has demonstrated the highest variability in the whole viral genome (Niesters et al., 1986), with mutations and recombination events in the S1 gene considered as critically important for the emergence of new virus genotypes, serotypes, and variants. Many different IBV types have been found worldwide, and new variants continue to emerge in different parts of the world. A new classification based on analysis of the whole S1 gene has recently been proposed (Valastro et al., 2016), including grouping and naming 32 lineages, comprising six genotypes (GI-1-GI-27, GII-GVI), and a number of inter-lineage recombinants. Some of these lineages are distributed in several continents, countries, or regions, such as GI-1 (formerly Massachusetts; Mass), GI-13 (793/B, 4/91, or CR88), GI-19 (LX4 or QX), GI-16 (ck/CH/LDL/97I (LDL/97I) or Q1), GI-21 (Italy 02), and GI-23 (Var2) (de Wit et al., 2011; Valastro et al., 2016), while others are geographically confined to specific regions.
Among the widely distributed IBV lineages, GI-1, GI-13, GI-16, and GI-19 are currently circulating in China (de Wit et al., 2011; Valastro et al., 2016), while strains assigned to some of these lineages, such as GI-16 and GI-19, were also found to originate in China. In addition, IBV variants of indigenous lineages were also found to be circulating in chicken flocks in China. New lineages, GI-7 (Xu et al., 2016), GI-28, and GI-29 (Chen et al., 2017; Jiang et al., 2017), were recently found in chickens suffering from respiratory and urinary health problems. These results indicated that novel IBV lineages have been emerging continuously in China in recent years. An epidemiological study conducted in China from January 2016 to December 2017 based on the S1 gene sequence isolated and characterized a new variant, designated I0636/16, in Guangxi province in 2016 (Xu et al., 2018). Despite the fact that most heterogeneity of IBVs occurs in the S1 gene, sequence analysis of this gene alone was not sufficient to characterize this novel strain. We therefore carried out a detailed investigation of the S1 gene and the complete genomic sequence of the I0636/16 strain to clarify its genetic characteristics and gain further insights into the origin of the strain. We also investigated the antigenicity and pathogenicity of the IBV strain I0636/16 in the present study.
2. Materials and methods
2.1. Eggs and chickens
Fertile White Leghorn specific pathogen-free (SPF) eggs and chickens were purchased from Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences. Ethical approval to carry out this study was obtained from the Ethical and Animal Welfare Committee of Heilongjiang province, China (License no. HSY-IACUC-2018-186).
2.2. Virus strains
We used the virus γCoV/ck/China/I0636/16, referred to as I0636/16, in this study. This virus was previously isolated in SPF chicken eggs from a disease outbreak associated with respiratory disease in a layer flock. The virus underwent six passages in SPF eggs and was tentatively classified as a variant (Xu et al., 2018). A further eight IBV strains (H120 (GI-1) (Chen et al., 2015), 4/91 (GI-13) (Han et al., 2017), LDL/092022 (LDL/092022) (GI-19) (Liu et al., 2013), LDL/97I (GI-16) (Liu et al., 2007), ck/CH/LSC/99I (LSC/99I) (GI-22) (Liu et al., 2006), ck/CH/LGX/111119 (LGX/111119) (GI-28) (Chen et al., 2017), I0111/14 (GI-29) (Jiang et al., 2017), and I0725/17 (GVI-1) (Xu et al., 2018)), representative of different IBV lineages circulating in China in recent years, were used in cross virus-neutralization (VN) tests. Viral stock was prepared by inoculating each of the viruses into 9-day-old SPF embryonated chicken eggs by the allantoic cavity route, followed by incubation for 48–72 h and chilling at 4 °C overnight. Allantoic fluid was harvested from the inoculated eggs and clarified by low-speed centrifugation at 1500 × g for 10 min. Cleared supernatant was stored in aliquots of 0.3 ml at −70 °C until further processing. Virus titration was performed using 9-day-old SPF chicken eggs, as described previously (Liu et al., 2009). The eggs were examined for IBV lesions including curling and dwarfing up to 7 days post-inoculation. Viral titers were calculated according to Reed and Muench (1938) and expressed as the 50% egg infective dose per milliliter (EID50/ml).
2.3. RNA extraction, reverse transcription-polymerase chain reaction, sequencing, and sequence determination
Viral RNA was extracted from the allantoic fluid using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The genomic sequence was amplified by reverse transcription-polymerase chain reaction (RT-PCR) using a PrimeScript™ One-Step RT-PCR kit ver. 2 (Takara Bio Inc., Shiga, Japan) according to the manufacturer’s instructions. The strategy and primers used for amplifying, cloning, and sequencing the complete genome of strain I0636/16 have been described previously (Liu et al., 2013). The primers were previously designed for IBV complete genome sequencing (Liu et al., 2013), and if the primers failed to work due to sequence differences, new primers were designed based on the newly determined sequences flanking those genome regions. The 3′/5′ ends of the genome were determined using a 3′/5′ RACE kit (Takara Bio Inc.) according to the manufacturer’s protocol. The PCR products were cloned into the pMD18-T vector (Takara Bio Inc.) according to the manufacturer’s instructions. Each genome fragment was sequenced three times.
Chromatograms were analyzed using the program Chromas (http://technelysium.com.au/wp/chromas/) and sequences were aligned using BioEdit (http://www.mbio.ncsu.edu/bioedit.htm). Open reading frame (ORF) predictions were carried out using the ORF-finder program (https://www.ncbi.nim.nih.gov/orffinder/), and ORFs were compared with the Beaudette strain (GenBank accession number: NC_001451).
The complete genomic sequence of I0636/16 has been deposited in the GenBank database, with the accession number MH924835.
2.4. Genotyping and S1 gene sequence comparison
The full S1 gene sequence generated in this study was subjected to BLAST searches using the National Center for Biotechnology Information database, and then analyzed phylogenetically using a dataset consisting of 73 sequences (Supplemental Table 1), including representative sequences for each genotype and lineage, as recently recommended (Valastro et al., 2016; Chen et al., 2017; Jiang et al., 2017), and the GX-NN130021 strain, which was closely related to I0636/16 by BLAST analysis. The S1 gene sequences of the IBV strain I0636/16 and the selected viruses were aligned using ClustalW, and phylogenetic analysis was carried out using Mega version 6.0 software (http://www.megasoftware.net/) using the maximum likelihood method with the Tamura–Nei substitution model and 1000 bootstrap replicates to assess the robustness of the branches. Only one IBV representative was selected from each lineage based on the phylogenetic trees to calculate the percentage identities between strain I0636/16 and the representative strains at both the nucleotide and amino acid levels.
2.5. Recombination event analysis
Recombination events and the probable parental genotypes of strain I0636/16 were analyzed by selecting and downloading the complete genomes of 121 IBV strains (Supplemental Table 2) from GenBank (www.ncbi.nlm.nih.gov/genbank/), representing sequences of IBV strains from all continents and each lineage if possible, three turkey coronavirus (TCoV) strains, including North American and European strains, and the only available strain of GfCoV. The complete genomic sequences of the IBV strain I0636/16 and the other selected viruses were aligned using ClustalW, and phylogenetic trees were constructed using the above-mentioned method to reduce the database size, by collapsing the sequences into clusters with different sequence identities. Sequence representative(s), including the complete genome sequences of IBV strains CK/CH/GD/GZ14 and GX-YL9, which showed close sequence identity to the I0636/16 strain, were examined using SimPlot version 3.5.1 (http://sray.med.som.jhmi.edu/SCRopftware/simplot/) to identify likely recombination breakpoints (Lole et al., 1999).The IBV strain GX-YL9 was used as a query virus. The window width and step size were set to 500 bp and 30 bp, respectively.
To confirm these recombination breakpoints, three phylogenetic trees were constructed on the basis of the results of similarity plot (SimPlot) analysis for the nucleotide fragments 1–20,265 (5′ untranslated region (UTR) to 3′ end of Gene 1), 20,266–24,151 (3′ end of Gene 1 to 5′ end of Gene 3), and 24,152–27,629 (5′ end of Gene 3 to 3′ UTR), from nine IBV strains, including I0636/16, five strains (LGX/111119, CK/CH/GD/GZ14, γCoV/ck/China/I0114/14 (I0114/14), GX-YL9, and GX-YL5) showing close relationships with I0636/16 based on the complete genomic sequence analysis, and three Mass strains (Beaudette, H120, and M41) as an outgroup. In addition, the nucleotide identities between I0636/16 and the eight IBV strains were calculated.
Finally, BLASTn (http://blast.ncbi.nlm.nih.gove/Blast.cgi) analysis using the fragment (20,266–24,151) of the recombined region was conducted using the GenBank database.
2.6. Virus cross-neutralization test
Antisera against the viruses H120, 4/91, LDL/091022, LDL/97I, LSC/99I, LGX/111119, and I0111/14 (Gao et al., 2016; Chen et al., 2017; Jiang et al., 2017) were used in this study. Antisera against strains I0725/17 and I0636/16 were produced in this study following a standard protocol (Gao et al., 2016). Briefly, 1-month-old SPF chickens were inoculated intratracheally with approximately 105 EID50 per bird, followed by an intravenous injection of the same dose 3 weeks later. Blood samples were collected after a further 3 weeks, and serum was harvested, pooled, and inactivated for 30 min at 56 °C before being used in VN tests.
We investigated the antigenic relationships between I0636/16 and the other reference strains by reciprocal β VN tests, using a fixed concentration of virus and serial dilutions of serum (Ducatez et al., 2009). Two-fold serial dilutions of each antiserum were mixed with an equal volume of virus dilution containing 100 EID50 in 0.1 ml and incubated for 1 h at 37 °C. Each serum–virus mixture was then inoculated into embryonated SPF chicken eggs via the allantoic sac route. The eggs were evaluated for non-specific mortality 24 h after inoculation and for the presence of specific lesions at 1 week after inoculation. End points corresponded to the serum dilutions that neutralized 50% of the virus. End-point titers were calculated by the method of Reed and Muench (1938).
VN end-point titers were used to calculate the percentage of antigenic relatedness (r) according to the method of Archetti and Horsfall (1950). Isolates with r values between 50% and 100% were considered to be antigenically related (Gelb et al., 1997).
2.7. Pathogenicity studies
Forty-five 1-day-old SPF chickens were separated in three groups of 15 birds each and housed in negative pressure isolators with ad libitum food and water. Birds in groups 1 and 2 were inoculated via the ocular and nasal routes with 105 EID50/per bird of strain I0636/16 and LGX/111119 in 100 μl volume, respectively, while birds in group 3 served as a negative control. Oropharyngeal and cloacal swabs and blood were collected from all birds in all groups at 4, 8, 12, 16, 20, 24, 28 and 32 days post-inoculation. The oropharyngeal and cloacal swabs were used to recover virus using 9-day-old SPF chicken eggs via the allantoic cavity route (Liu et al., 2013) and the allantoic fluids were used for RT-PCR to detect the presence of challenge viruses (Gao et al., 2016), to measure the duration of excretion of the strains I0636/16 and LGX/111119 during the course of the experiment. The sera were used to detect specific antibodies against IBV using a commercial enzyme-linked immunosorbent assay (IDEXX Corporation, Westbrook, ME, USA) according to the manufacturer’s instructions.
Morbidity and mortality were recorded daily. Five birds from each group were selected randomly and killed humanely at 5 days post-inoculation. Trachea, lung, proventriculus, cecal tonsil, and kidney tissues were collected from these birds and used for virus titration in 9-day-old SPF chicken eggs, as described previously (Han et al., 2018). Trachea and kidney were considered as the two targeted organs for IBV strains. Lung is the representative of respiratory tract of chickens. Proventriculus and cecal tonsil were representatives of upper and lower digestive tracts which were often used for IBV isolation (Alvarado et al., 2006; Yu et al., 2001).
2.8. Statistical analysis
Data are expressed as mean ± standard deviation. Virus titers were analyzed by a Student’s t-test using GraphPad Prism for Windows version 5 (GraphPad Software, La Jolla, CA, USA). Differences were considered significant if the p value was <0.05 (*p < 0.05, **p < 0.01, ***p < 0.001).
3. Results
3.1. IBV strain I0636/16 represented a novel genotype GVII-1
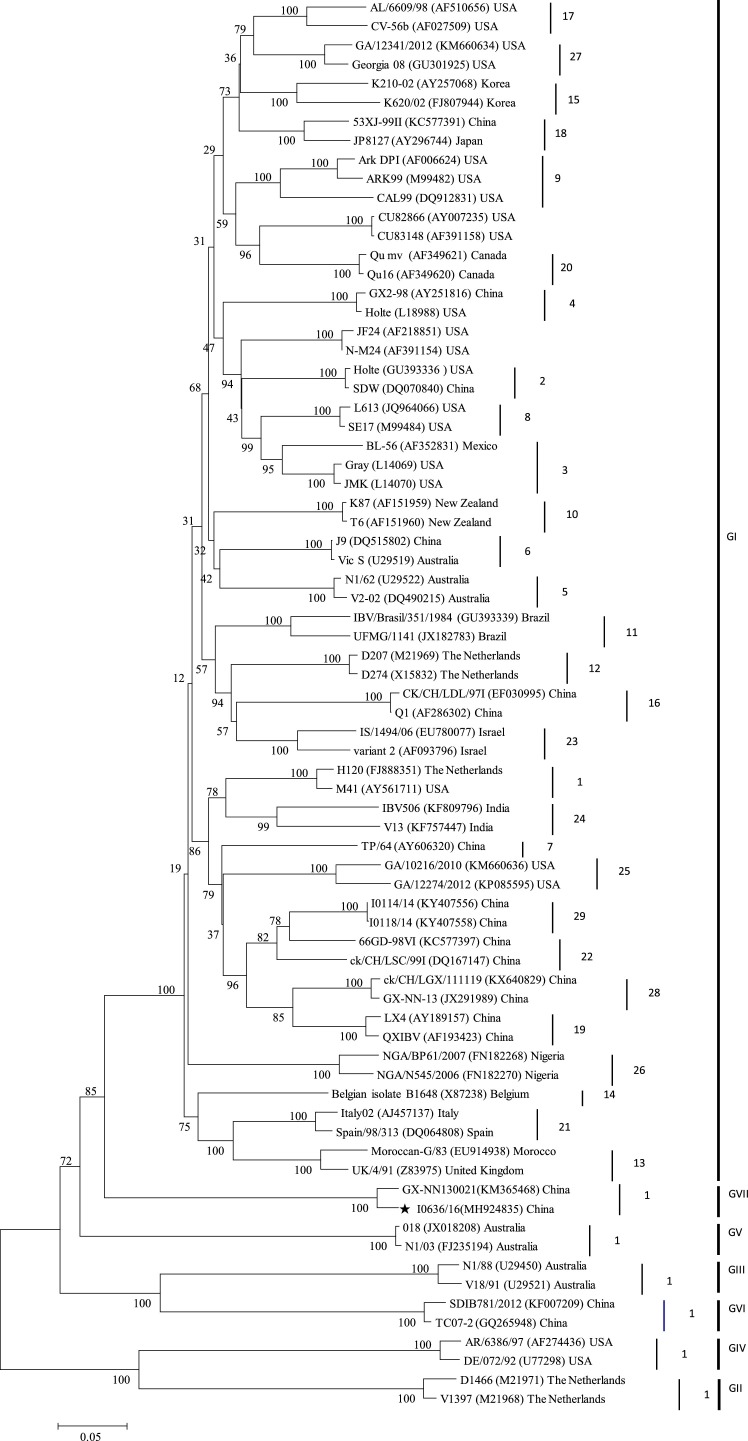
Based on BLAST analysis using the complete nucleotide sequence of the S1 gene, the I0636/16 strain was found to be closely genetically related to GX-NN130021, which was isolated in Guangxi Province in 2013 (Zhang et al., 2016). A phylogenetic tree comparing 74 reference strains representing the well-established genotypes and lineages, strain GX-NN130021 and the current strain I0636/16 are illustrated in Fig. 1 . The IBV strain I0636/1, which was isolated in Guangxi Province in 2016, was closely related to the reference strain GX-NN130021. Strains I0636/16 and GX-NN130021 were clearly separated from other IBV reference strains, including the six well-established genotypes and 29 lineages.
Fig. 1.
Phylogenetic relationships between strain I0636/16 and reference strains. Phylogenetic relationships between strain I0636/16 and 73 representatives of well-established lineages based on the complete S1 gene sequence. The unrooted tree was created by the maximum likelihood method with the Tamura–Nei substitution model and 1000 bootstrap replicates. Strain I0636/16 is highlighted by a star.
Homology analysis of the IBV strains from the different lineages revealed similarities between isolate I0636/16 and the established lineages of 59.0%–71.7% and 51.3%–69.6% at the nucleotide and deduced amino acid levels, respectively. The deduced amino acid substitutions were scattered across the S1 subunit of the spike proteins in I0636/16, compared with the other lineages (data not shown). However, strains I0636/16 and GX-NN130021 shared 97% and 96.9% nucleotide and amino acid identities, respectively, with each other. Alignment of the S1 deduced amino acid sequences identified 18 substitutions between I0636/16 and GX-NN130021, of which only two were located in the hypervariable region (HVR) (Supplemental Fig. 1A).
3.2. Strain I0636/16 emerged as a result of recombination events
A consensus sequence of strain I0636/16 was obtained and the full-length genome consisted of 27,629 nucleotides excluding the 3′ poly A tails. ORF analysis predicted ten ORFs, resulting in a typical IBV genome organization of 5′UTR-1a-1b-S-3a-3b-3c(E)-M-5a-5b-N-3′UTR. In addition, there was also an ORF with the potential to code for a protein of 94 amino acids between gene M and 5a of strain I0636/16 which was identified previously in some IBV strains (Bentley et al., 2013).
Phylogenetic analysis of 121 complete genomes was conducted to investigate the relationships between the obtained strain I0636/16 and different avian coronaviruses downloaded from GenBank. Strain I0636/16 was grouped together with all the IBV strains used in this study, which had been isolated from different regions at various times but was grouped apart from coronaviruses isolated from other avian species (Fig. 1B). Within IBV strains, I0636/16 showed close relationships with IBV strains LGX/111119 (93.5%), CK/CH/GD/GZ14 (93.1%), I0114/14 (92.8%), CK/CH/2010/JT-1 (92.8%), GX-YL9 (92.4%), and GX-YL5 (92.1%), most of which were isolated from South China, particularly from Guangxi province.
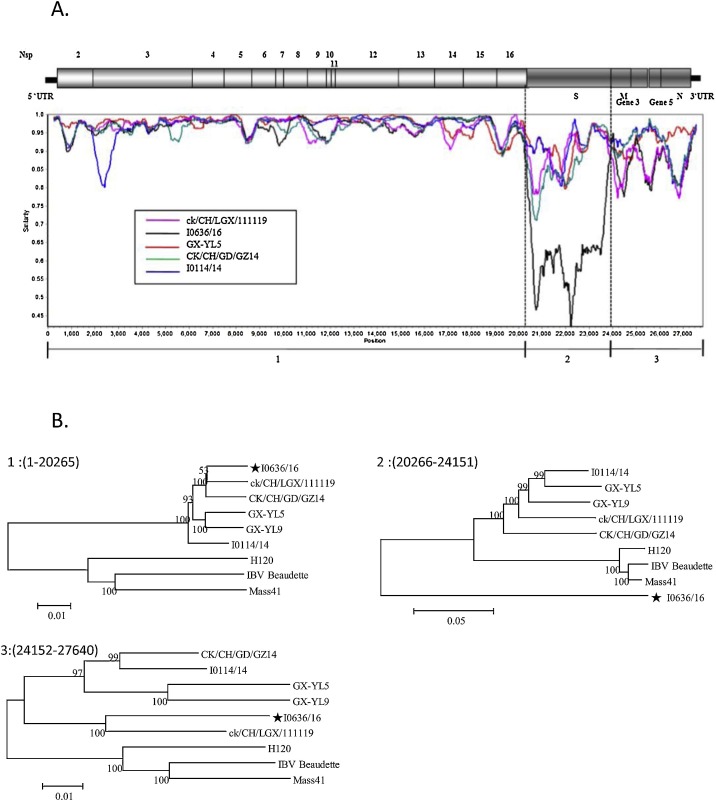
We investigated the impact of probable recombination events on the origin of strain I0636/16 by conducting an analysis using SimPlot software (Fig. 2 A). Two major recombination events were observed, one in the 3′ end of Gene 1 (at approximately nucleotide 20,265) and another in the 5′ end of Gene 3 (at approximately nucleotide 24,152). The phylogenetic trees (Fig. 2B) clearly supported the SimPlot results, with strain I0636/16 showing identities of >96.6% with LGX/111119, CK/CH/GD/GZ14, I0114/14, CK/CH/2010/JT-1, GX-YL9, and GX-YL5 for the genomic region 1–20,265, compared with an identity of <86.9% with the Mass-type strains. Similar to this genomic region, I0636/16 showed an identity of 92.5% with LGX/111119 for the genomic region 24,152–27,640, compared with <85.2% with the Mass-type strains. These results suggested that the LGX/111119 (GI-28 lineage) virus, circulating in chicken flocks in China (Chen et al., 2017), was a potential parent of recombinant I0636/16. In contrast, in the second phylogenetic tree, the branch (genomic region 20,266–24,151) that included the spike gene separated the strain I0636/16 from all reference strain sequences, and shared no more than 75% nucleotide identity with any of the reference strains, making difficult to assess the origin of the fragment 24,152–27,629. BLASTn analysis using this fragment of I0623/16 showed that this strain was closely related to strain GX-NN130021 (97% nucleotide identity), but shared no more than 75% nucleotide identity with any other IBV strain in the GenBank database.
Fig. 2.
Recombination events in the genome of strain I0636/16. Simplot analysis to detect recombination and estimate recombination breakpoints within the I0636/16 genome (A). Complete genomic sequence of the strain GX-YL9 was used as the query sequence in SimPlot analysis. Vertical dotted lines show the deduced recombination breakpoint. Phylogenetic trees constructed using different nucleotide fragments based on the results of SimPlot analysis (B).
Overall, these data suggest that GVII-1 viruses emerged as a result of replacement of the spike gene in strain LGX/111119-like viruses (GI-28) through recombination.
3.3. Strain I0636/16 is antigenically different from viruses of other lineages
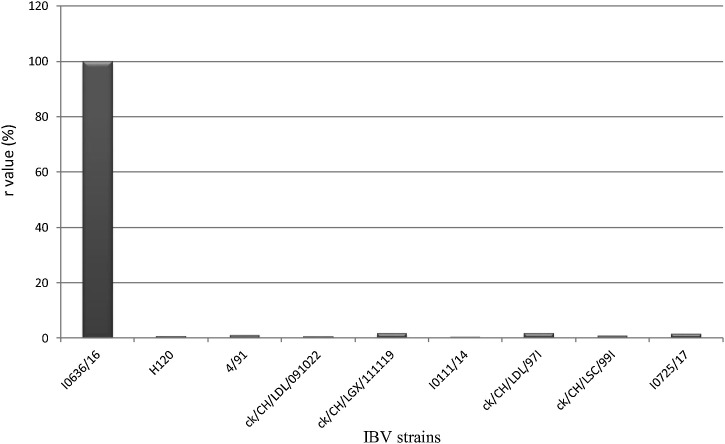
The antigenic properties of I0636/16 were compared to those of reference strains by VN tests. The calculated antigenic relatedness values, r, of strain I0636/16 against homologous and heterologous strains are listed in Fig. 3 . The r values confirmed the absence of relationships between I0636/16 and H120, 4/91, LDL/091022, LDL/97I, LSC/99I, LGX/111119, I0111/14, and I0725/17(all <5%). The value of r for LGX/111119 was slightly higher but did not reach the 50% threshold for antigenic relatedness between strains, suggesting that the I0636/16 strain represented a novel serotype.
Fig. 3.
The calculated antigenic relatedness values, r, of strain I0636/16 against homologous and heterologous strains.
3.4. Strain I0636/16 is pathogenic to SPF chickens
Respiratory signs started on day 4 in four birds post-challenge, followed by the development of symptoms in nearly all birds. The respiratory signs varied from mild respiratory distress to severe rales. Generally, the respiratory signs of chickens challenged with strain LGX/111119 were more severe than those of chickens challenged with I0636/16. One bird died on day 9 after challenge with strain I0636/16. In contrast, three birds died on days 4, 8 and 16 after challenge with strain LGX/111119. Macroscopic lesions in the dead birds were mainly confined to the kidneys, which showed pale, swollen, and mottled kidney parenchyma, and slightly distended tubules and urethras with uric acid crystals. Petechiae were also observed in the trachea, pharynx, and larynx in most challenged birds. The clinical signs continued up to day 17 in one bird that showed mild respiratory distress. No clinical signs were observed in the control birds throughout the experiment.
No birds seroconverted before day 8 post-challenge, 33.3% and 50% birds seroconverted on day 12 after challenge with I0636/16 and LGX/111119, respectively. Nearly all birds seroconverted on day 16 after challenge with the two virus strains. As expected, no birds in the negative control group showed an antibody response against IBV.
3.5. Strain I0636/16 is distributed in different tissues in SPF birds
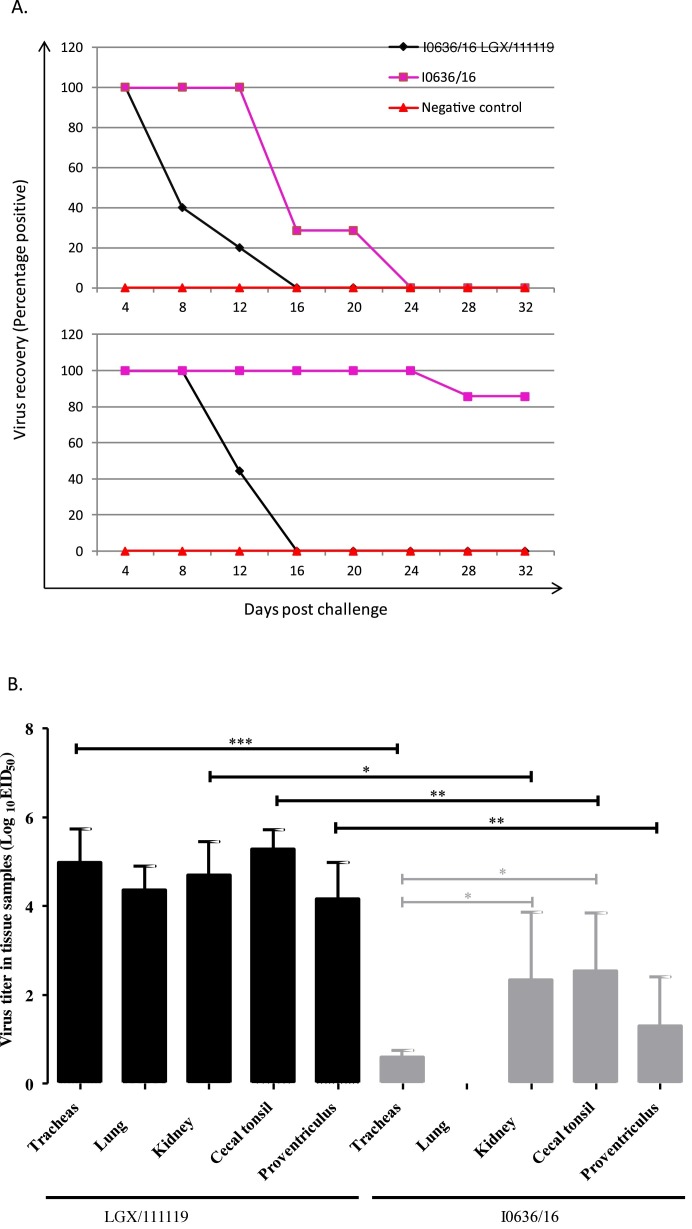
Virus recovery from oropharyngeal swabs using 9-day-old SPF chicken eggs challenged with I0636/16 revealed that 100% of birds were positive on day 4, 40% were positive on day 8, and 22.2% were positive on day 12 post-challenge. In contrast, using cloacal swabs, 100% were positive on days 4 and 8, and 44.4% were positive on day 12 post-challenge. No birds tested positive on day 16 or thereafter post-challenge. Comparing with those of chickens challenged with I0636/16, prolonged virus shedding were observed from both the respiratory tract and cloaca of chickens challenged with the LGX/111119 strain (Fig. 4 A). No birds in the negative control group tested positive for virus recovery at any timepoint.
Fig. 4.
Infection of chickens with strains LGX/111119 and I0636/16. Virus recovery from oropharyngeal (upper) and cloacal (lower) swabs from chickens challenged with IBV strains LGX/111119 and I0636/16 (A). Virus recovery was performed by inoculating 9-day-old embryonated, specific pathogen-free eggs through the allantoic route with supernatant from the swabs. Replication of strains LGX/111119 and I0636/16 in trachea, lung, kidney, cecal tonsil, and proventriculus in chickens (B). One-day-old SPF layer chickens in groups 1 and 2 were inoculated via the ocular and nasal routes with 105 × EID50/per bird of strains LGX/111119 and I0636/16 in 0.1 ml, respectively, and trachea, lung, kidney, cecal tonsil, and proventriculus tissues were collected from five birds at 5 days post-challenge for virus titration in eggs. Data are expressed as mean ± standard deviation. Virus titers were analyzed by a Student’s t-test using GraphPad Prism for Windows version 5 (GraphPad Software, La Jolla, CA, USA). Differences were considered significant if the p value was <0.05 (*p < 0.05, **p < 0.01, ***p < 0.001).
The titers of the challenge viruses in trachea, lung, kidney, proventriculus, and cecal tonsil tissues were determined in five challenged chickens from each of the three groups, at 5 days post-inoculation using 9-day-old SPF chicken embryos (Han et al., 2018). Infection with strain LGX/111119 resulted in significantly higher production of infectious viruses in trachea, lung, kidney, proventriculus, and cecal tonsil tissues than those of corresponding tissues of chickens infected with strain I0636/16 (Fig. 4B). Comparatively, infection with strain I0636/16 resulted in lower production of infectious virus in the trachea than those in the kidneys and cecal tonsils. No infectious viruses were detected in the lungs in the five chickens challenged with strain I0636/16. No viruses were detected in any tissues in the five control chickens.
4. Discussion
Some studies have reported that genotyping based on sequencing the HVRs of the S1 gene or partial sequencing of the S1 gene is representative of the grouping based on the complete S1 gene (Lee et al., 2003); however, others disagree with these findings (Moreno et al., 2017; Schikora et al., 2003), and this discrepancy is considered to be due to the presence of recombination events involving the S1 HVR. In the present study, we used the complete S1 gene for IBV typing, as suggested previously (Manswr et al., 2018; Valastro et al., 2016), and found no evidence of recombination events in the I0636/16 S1 gene sequence. We did identify a novel genotype consisted by IBV isolate I0636/1 and the reference strain GX-NN130021, which clustered together in the phylogeny and did not fall within established lineages. In agreement with this result, the two strains shared high nucleotide and amino acid identities (97% and 96.9%), compared with low identities with representative IBV strains from different lineages. Based on these results, these two IBV strains should be grouped into a novel genotype, designated as GVII-1, although the designation of lineage/genotypes should be assigned to monophyletic groups of at least three viruses sampled from at least two different outbreaks (Valastro et al., 2016). Generally, it is considered that the chance of having the same serotype is higher between strains with the same genotype (Wang and Huang, 2000), though these data showed that this relationship was not very strong, given that a change in a small percentage of the amino acids in the S1 protein could result in a change of serotype (Cavanagh et al., 1992). The current virus cross-neutralization results showed that GVII-1 was not only a novel genotype, but also antigenically different from the currently tested genotype lineages. Strain GX-NN130021 was isolated in Guangxi province, China, in 2013 (Zhang et al., 2016), suggesting that the novel GVII-1 had been circulating in China over a 5-year period. However, the source of this new virus introduction into the chicken population in China remains unknown.
Full-length genome analysis showed that the emergence of the I0636/16 strain involved a double crossover event that replaced the spike gene by that from an as-yet-unidentified IBV strain. However, the remaining parts of the genome originated from commonly known GI-28 IBV strains, suggesting that the I0636/16 strain was a mosaic, and that one of its putative parents was descended from GI-28 lineage viruses. The antigenic differences among IBV strains have been investigated, and previous work showed that S1 sequences had a stronger correlation with protective relatedness than with antigenic relatedness between strains (Ladman et al., 2006). The as-yet-unidentified parental virus of strain I0636/16, which shared the same spike gene with strain I0636/16, might thus be antigenically different from vaccine strains used in China, such as H120 and 4/91; therefore vaccination with these live vaccines might not offer complete protection against infection with the parental virus of strain I0636/16. Similarly, considering the high prevalence of the GI-28 lineage in south China (Chen et al., 2017) where the novel GVII-1 viruses were isolated, it is reasonable to speculate that the GI-28 lineage may co-infect birds more frequently than other lineages, although the current commonly used vaccines cannot offer complete protection against GI-28 viruses (Chen et al., 2017). Vaccination with these vaccines is therefore likely to induce non-sterilizing immunity, which may allow prolonged replication, shedding, and circulation of both the deduced parental strains of GVII-1 viruses. Consequently, although vaccination with attenuated vaccines is carried out intensively in China, this procedure may produce an environment where co-infection with GI-28 and the as-yet-unidentified parental viruses can enhance the likelihood of recombination. Theoretically, the short persistence and shedding, and limited numbers of I0636/16-like viruses isolated so far in China suggest that the probability of recombination between the as-yet-unidentified parental virus and GI-28 viruses may not be high, similar to the hypothesis suggesting that the shorter persistence and shedding of Mass-based vaccines (Bijlenga et al., 2004) and the low percentage of M41-like viruses (Chen et al., 2015; Moreno et al., 2017) in birds supported the absence of recombination for this genotype. However, recombination between Mass-like viruses and other lineages, and among Mass-like viruses, have been reported (Chen et al., 2015; Liu et al., 2013).
The IBV genome undergoes mutations and recombination, and the exchange of a long region of the genome by recombination may allow viruses to rapidly explore ample areas of the sequence space, potentially leading to the emergence of novel strains with different features in terms of virulence and disease pathogenesis, antigenicity, and cell and tissue tropisms (Cavanagh et al., 1992; Kant et al., 1992; Simon-Loriere and Holmes, 2011). The genome of strain I0636/06 was closely related to GI-28 viruses, except in the spike gene. This phenomenon was similar to that of TCoV, the genome of which was also closely related to IBV, except in the spike gene. Replacement of the spike gene of TCoV was responsible for the host shift from chickens to turkeys and a pathogenicity shift from upper-respiratory disease to enteric disease (Jackwood et al., 2010). Even though the recombination events that replaced the spike gene did not result in a tissue tropism shift in the kidneys of chickens after challenge, the I0636/16 strain showed low affinity to the respiratory tract compared with the parental virus LGX/111119 (Chen et al., 2017). In contrast, the ability of I0636/16 to reach the cecal tonsils and subsequently to replicate in this tissue was slightly higher compared with that of respiratory tract. The replicase gene of avian coronavirus IBVs is known to be a determinant of pathogenicity (Armesto et al., 2009), and we were therefore unable to compare the pathogenicity of I0636/16 with one of its deduced parental virus until a source has been identified for the replicase gene because it is as-yet-unidentified, though another deduced parental virus (LGX/111119) showed more severe pathogenicity than I0636/16 in SPF chickens. However, we cannot conclude that this difference in pathogenicity was due to replacement of the spike gene until the source of the GVII-1 spike gene has been identified, and alternate explanations for the observed differences between I0636/16 and GI-28 spike should be considered.
The frequency of successful inter-typic genetic changes may depend on the viability of the recombinant progeny able to establish in the host population, and on the increase in fitness of the recombinant offspring in the host population compared with their non-recombination parents (Smits et al., 2003). The recombinant I0636/16 was only detected sporadically and persisted for a short time, and thus probably represented strains that were not sufficiently competitive with respect to one of their progenitor strains, LGX/111119-like viruses (Chen et al., 2017). However, compared with the unidentified parental virus, I0636/16-like strains may be sufficiently competitive, and the parental virus may have been circulating at below cut-off levels for detection, due to its lower replicative capacity and lower percentage.
We isolated the I0636/16 IBV strain and passaged it in SPF chicken eggs (Xu et al., 2016). The number of passages of the virus in eggs was high (six passages), and the possibility that some genetic changes were introduced as a result of its propagation in the eggs cannot be ruled out. No complete genomic sequences of GVII-1 lineage viruses are currently available in the public database. The present results thus provide valuable information on the characteristics of the I0636/16 strain and contribute to a better understanding of the origin of GVII-1 viruses. Further studies on these isolates will provide information on the emergence, evolution, and effectiveness of current vaccines against these isolates.
Competing interest
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (2017YFD0500105), the China Agriculture Research System (No. CARS-40-K18), National "Twelfth Five-Year" Plan for Science & Technology Support (2015BAD12B03) and the Provincial Supported Science Foundation of Heilongjiang Province for The National Key Technology R&D Program (GX16B003).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.vetmic.2019.01.020.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Alvarado I.R., Villegas P., El-Attrache J., Jackwood M.W. Detection of Massachusetts and Arkansas serotypes of infectious bronchitis virus in broilers. Avian Dis. 2006;50:292–297. doi: 10.1637/7458-101805R.1. [DOI] [PubMed] [Google Scholar]
- Archetti I., Horsfall F.L., Jr. Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. J. Exp. Med. 1950;92:441–462. doi: 10.1084/jem.92.5.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armesto M., Cavanagh D., Britton P. The replicase gene of avian coronavirus infectious bronchitis virus is a determinant of pathogenicity. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley K., Keep S.M., Armesto M., Britton P. Identification of a noncanonically transcribed subgenomic mRNA of infectious bronchitis virus and other gammacoronaviruses. J. Virol. 2013;87:2128–2136. doi: 10.1128/JVI.02967-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlenga G., Cook J.K., Gelb J., Jr., de Wit J.J. Development and use of the H strain of avian infectious bronchitis virus from the Netherlands as a vaccine: a review. Avian Pathol. 2004;33:550–557. doi: 10.1080/03079450400013154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M.E., Brown T.D., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus IBV glycopolypeptides: size of their polypeptide moieties and nature of their oligosaccharides. J. Gen. Virol. 1983;64:1187–1191. doi: 10.1099/0022-1317-64-5-1187. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Gelb J., Jr. Infectious bronchitis. In: Saif Y.M., Fadly A.M., Glisson J.R., McDougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. 12th edn. Wiley-Blackwell Publishing; Iowa: 2008. pp. 117–135. [Google Scholar]
- Cavanagh D., Davis P.J., Cook J.K., Li D., Kant A., Koch G. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992;21:33–43. doi: 10.1080/03079459208418816. [DOI] [PubMed] [Google Scholar]
- Chen L., Zhang T., Han Z., Liang S., Xu Y., Xu Q., Chen Y., Zhao Y., Shao Y., Li H., Wang K., Kong X., Liu S. Molecular and antigenic characteristics of Massachusetts genotype infectious bronchitis coronavirus in China. Vet. Microbiol. 2015;181:241–251. doi: 10.1016/j.vetmic.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Jiang L., Zhao W., Liu L., Zhao Y., Shao Y., Li H., Han Z., Liu S. Identification and molecular characterization of a novel serotype infectious bronchitis virus (GI-28) in China. Vet. Microbiol. 2017;198:108–115. doi: 10.1016/j.vetmic.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J.J., Nieuwenhuisen-van Wilgen J., Hoogkamer A., van de Sande H., Zuidam G.J., Fabri T.H. Induction of cystic oviducts and protection against early challenge with infectious bronchitis virus serotype D388 (genotype QX) by maternally derived antibodies and by early vaccination. Avian Pathol. 2011;40:463–471. doi: 10.1080/03079457.2011.599060. [DOI] [PubMed] [Google Scholar]
- Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez M.F., Martin A.M., Owoade A.A., Olatoye I.O., Alkali B.R., Maikano I., Snoeck C.J., Sausy A., Cordioli P., Muller C.P. Characterization of a new genotype and serotype of infectious bronchitis virus in Western Africa. J. Gen. Virol. 2009;90:2679–2685. doi: 10.1099/vir.0.012476-0. [DOI] [PubMed] [Google Scholar]
- Gao M., Wang Q., Zhao W., Chen Y., Zhang T., Han Z., Xu Q., Kong X., Liu S. Serotype, antigenicity, and pathogenicity of a naturally recombinant TW I genotype infectious bronchitis coronavirus in China. Vet. Microbiol. 2016;15:1–8. doi: 10.1016/j.vetmic.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb J.Jr, Keeler C.L.Jr, Nix W.A., Rosenberger J.K., Cloud S.S. Antigenic and S-1 genomic characterization of the Delaware variant serotype of infectious bronchitis virus. Avian Dis. 1997;41:661–669. [PubMed] [Google Scholar]
- Han Z., Zhao W., Chen Y., Xu Q., Sun J., Zhang T., Zhao Y., Liang S., Gao M., Wang Q., Kong X., Liu S. Genetic, antigenic, and pathogenic characteristics of avian infectious bronchitis viruses genotypically related to 793/B in China. Vet. Microbiol. 2017;203:125–135. doi: 10.1016/j.vetmic.2017.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Gao M., Chen Y., Zhao W., Sun J., Zhao Y., Liu S. Genetics, antigenicity and virulence properties of three infectious bronchitis viruses isolated from a single tracheal sample in a chicken with respiratory problems. Virus Res. 2018;257:82–93. doi: 10.1016/j.virusres.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Boynton T.O., Hilt D.A., McKinley E.T., Kissinger J.C., Paterson A.H., Robertson J., Lemke C., McCall A.W., Williams S.M., Jackwood J.W., Byrd L.A. Emergence of a group 3 coronavirus through recombination. Virology. 2010;398:98–108. doi: 10.1016/j.virol.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zhao W., Han Z., Chen Y., Zhao Y., Sun J., Li H., Shao Y., Liu L., Liu S. Genome characterization, antigenicity and pathogenicity of a novel infectious bronchitis virus type isolated from south China. Infect. Genet. Evol. 2017;54:437–446. doi: 10.1016/j.meegid.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant A., Koch G., van Roozelaar D.J., Kusters J.G., Poelwijk F.A., van der Zeijst B.A. Location of antigenic sites defined by neutralizing monoclonal antibodies on the S1 avian infectious bronchitis virus glycopolypeptide. J. Gen. Virol. 1992;73:591–596. doi: 10.1099/0022-1317-73-3-591. [DOI] [PubMed] [Google Scholar]
- Kottier S.A., Cavanagh D., Britton P. Experimental evidence of recombination in coronavirus infectious bronchitis virus. Virology. 1995;213:569–580. doi: 10.1006/viro.1995.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladman B.S., Loupos A.B., Gelb J., Jr. Infectious bronchitis virus S1 gene sequence comparison is a better predictor of challenge of immunity in chickens than serotyping by virus neutralization. Avian Pathol. 2006;35:127–133. doi: 10.1080/03079450600597865. [DOI] [PubMed] [Google Scholar]
- Lee C.W., Hilt D.A., Jackwood M.W. Typing of field isolates of infectious bronchitis virus based on the sequence of the hypervariable region in the S1 gene. J. Vet. Diagn. Invest. 2003;15:344–348. doi: 10.1177/104063870301500407. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhang Q., Chen J., Han Z., Liu X., Feng L., Shao Y., Rong J., Kong X., Tong G. Genetic diversity of avian infectious bronchitis coronavirus strains isolated in China between 1995 and 2004. Arch. Virol. 2006;151:1133–1148. doi: 10.1007/s00705-005-0695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Han Z., Chen J., Liu X., Shao Y., Kong X., Tong G., Rong J. S1 gene sequence heterogeneity of a pathogenic infectious bronchitis virus strain and its embryo-passaged, attenuated derivatives. Avian Pathol. 2007;36:231–244. doi: 10.1080/03079450701338730. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhang X., Gong L., Yan B., Li C., Han Z., Shao Y., Li H., Kong X. Altered pathogenicity, immunogenicity, tissue tropism and 3’-7 kb region sequence of an avian infectious bronchitis coronavirus strain after serial passage in embryos. Vaccine. 2009;27:4630–4640. doi: 10.1016/j.vaccine.2009.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ma H., Xu Q., Sun N., Han Z., Sun C., Guo H., Shao Y., Kong X., Liu S. Characterization of a recombinant coronavirus infectious bronchitis virus with distinct S1 subunits of spike and nucleocapsid genes and a 3’ untranslated region. Vet. Microbiol. 2013;162:429–436. doi: 10.1016/j.vetmic.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manswr B., Ball C., Forrester A., Chantrey J., Ganapathy K. Evaluation of full S1 gene sequencing of classical and variant infectious bronchitis viruses extracted from allantoic fluid and FTA cards. Avian Pathol. 2018;47:418–426. doi: 10.1080/03079457.2018.1471196. [DOI] [PubMed] [Google Scholar]
- Moreno A., Franzo G., Massi P., Tosi G., Blanco A., Antilles N., Biarnes M., Majó N., Nofrarías M., Dolz R., Lelli D., Sozzi E., Lavazza A., Cecchinato M. A novel variant of the infectious bronchitis virus resulting from recombination events in Italy and Spain. Avian Pathol. 2017;46:28–35. doi: 10.1080/03079457.2016.1200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesters H.G., Lenstra J.A., Spaan W.J., Zijderveld A.J., Bleumink-Pluym N.M., Hong F., van Scharrenburg G.J., Horzinek M.C., van der Zeijst B.A. The peplomer protein sequence of the M41 strain of coronavirus IBV and its comparison with Beaudette strains. Virus Res. 1986;5:253–263. doi: 10.1016/0168-1702(86)90022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Schalk A.F., Hawn M.C. An apparently new respiratory disease of baby chicks. J. Am. Vet. Med. Assoc. 1931;78:413–422. [Google Scholar]
- Schikora B.M., Shih L.M., Hietala S.K. Genetic diversity of avian infectious bronchitis virus California variants isolated between 1988 and 2001 based on the S1 subunit of the spike glycoprotein. Arch. Virol. 2003;148:115–136. doi: 10.1007/s00705-002-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q., Wang C., Keirs R.W. Genetic relationships of infectious bronchitis virus isolates from Mississippi broilers. Avian Dis. 2000;44:66–73. [PubMed] [Google Scholar]
- Simon-Loriere E., Holmes E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011;9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Lavazza A., Matiz K., Horzinek M.C., Koopmans M.P., de Groot R.J. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J. Virol. 2003;77:9567–9577. doi: 10.1128/JVI.77.17.9567-9577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G., Monne I. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect. Genet. Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.H., Huang Y.C. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch. Virol. 2000;145:291–300. doi: 10.1007/s007050050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Han Z., Wang Q., Zhang T., Gao M., Zhao Y., Shao Y., Li H., Kong X., Liu S. Emergence of novel nephropathogenic infectious bronchitis viruses currently circulating in Chinese chicken flocks. Avian Pathol. 2016;45:54–65. doi: 10.1080/03079457.2015.1118435. [DOI] [PubMed] [Google Scholar]
- Xu L., Han Z., Jiang L., Sun J., Zhao Y., Liu S. Genetic diversity of avian infectious bronchitis virus in China in recent years. Infect. Genet. Evol. 2018;66:82–94. doi: 10.1016/j.meegid.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Jiang Y., Low S., Wang Z., Nam S.J., Liu W., Kwangac J. Characterization of three infectious bronchitis virus isolates from China associated with proventriculus in vaccinated chickens. Avian Dis. 2001;45:416–424. [PubMed] [Google Scholar]
- Zhang L., Wu C., Zhang Z., He Y., Li H., Qin L., Wei T., Mo M., Wei P. Sequencing and serologic identification of S1 genes of infectious bronchitis viruses isolated during 2012–2013 in Guangxi Province, China. Bing Du Xue Bao. 2016;32:62–69. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.