Abstract
Avian rotaviruses (AvRVs) represent a diverse group of intestinal viruses, which are suspected as the cause of several diseases in poultry with symptoms of diarrhoea, growth retardation or runting and stunting syndrome (RSS). To assess the distribution of AvRVs in chickens and turkeys, we have developed specific PCR protocols. These protocols were applied in two field studies investigating faecal samples or intestinal contents of diseased birds derived from several European countries and Bangladesh. In the first study, samples of 166 chickens and 33 turkeys collected between 2005 and 2008 were tested by PAGE and conventional RT-PCR and AvRVs were detected in 46.2%. In detail, 16.1% and 39.2% were positive for AvRVs of groups A or D, respectively. 11.1% of the samples contained both of them and only four samples (2.0%) contained rotaviruses showing a PAGE pattern typical for groups F and G. In the second study, samples from 375 chickens and 18 turkeys collected between 2009 and 2010 were analyzed using a more sensitive group A-specific and a new group D-specific real-time RT-PCR. In this survey, 85.0% were AvRV-positive, 58.8% for group A AvRVs, 65.9% for group D AvRVs and 38.9% for both of them. Although geographical differences exist, the results generally indicate a very high prevalence of group A and D rotaviruses in chicken and turkey flocks with cases of diarrhoea, growth retardation or RSS. The newly developed diagnostic tools will help to investigate the epidemiology and clinical significance of AvRV infections in poultry.
Keywords: Chicken, Real-time RT-PCR, Rotavirus, Rotavirus group, Turkey
1. Introduction
Viral enteric diseases in poultry, especially in turkeys and chickens, have an important economic impact because of production losses due to poor weight gain. The most important viruses associated with poult enteritis are adenoviruses, astroviruses, turkey coronavirus, enterovirus-like viruses, reovirus, rotavirus (RV) and turkey torovirus (Fitzgerald, 2008, Jones, 2008, Reynolds et al., 1987a, Reynolds and Schultz-Cherry, 2008, Saif, 2008). RV infection is suspected to be a cause of diarrhoea and it has been associated with runting and stunting syndrome (RSS) in chicken or turkey broilers (Otto et al., 2006, Pantin-Jackwood et al., 2007, Reynolds et al., 1987a, Reynolds et al., 1987b, Saif et al., 1985, Spackman et al., 2010, Yason and Schat, 1986, Yason and Schat, 1987). Numerous surveys of virus incidence have shown that poultry flocks are frequently infected with RV (Alfieri et al., 1989, Bellinzoni et al., 1987, Gough et al., 1992, Legrottaglie et al., 1997, McNulty et al., 1978, McNulty et al., 1979, McNulty et al., 1984a, McNulty et al., 1984b, Minamoto et al., 1988, Otto et al., 2006, Pantin-Jackwood et al., 2007, Reynolds et al., 1987b, Reynolds et al., 1987c, Saif et al., 1985, Takase et al., 1990).
Rotaviruses belong to the family Reoviridae and have a non-enveloped viral capsid containing 11 segments of double-stranded RNA (dsRNA) (Estes and Kapikian, 2007). Viral particles consist of three protein layers. The intermediate layer consists of the viral structure protein (VP) 6, which is used for serological grouping of RVs. The electrophoretic migration pattern of the genome segments separated by polyacrylamide gel electrophoresis (PAGE) was also used for a rapid grouping of RVs (Bridger, 1994, Estes and Kapikian, 2007, Johne et al., 2011, Theil et al., 1986, Trojnar et al., 2010). Most of the RV strains detected in humans and animals belong to group A (RV-A). Accordingly, a high percentage of avian RVs (AvRVs) belong to group A, but group D (RV-D) strains have also been frequently detected in avian species. Furthermore, AvRVs of group F and group G have been occasionally found. Commonly, electron microscopy, PAGE, or enzyme-linked immunosorbent assays (ELISA) are used to detect AvRVs (Bellinzoni et al., 1987, Elschner et al., 2002, Legrottaglie et al., 1997, McNulty et al., 1979, Otto et al., 2006, Theil et al., 1986, Todd and McNulty, 1986). More recently, reverse transcription-polymerase chain reaction (RT-PCR) protocols have been employed (Day et al., 2007, Pantin-Jackwood et al., 2007). However, until now the examination of poultry samples by the latter method was restricted to detection of AvRVs of group A, because sequence data of other AvRV groups were lacking.
The aim of this study was to assess the distribution of AvRVs of the different RV groups in poultry flocks with intestinal diseases. Due to the availability of the genomic sequences of group A and D AvRV strains (Otto et al., 2006, Schumann et al., 2009, Trojnar et al., 2009, Trojnar et al., 2010), PCR-based methods could be developed for detection of both virus groups. Two studies were performed with samples derived from a variety of European countries and Bangladesh. In the first study, PAGE and conventional RT-PCR were applied to AvRV RNA detection. The PAGE is almost as sensitive as electron microscopy and enables identification of non-AvRV-A or–D strains (Otto et al., 2006, Johne et al., 2011). In the second study, more sensitive real-time RT-PCR assays were used for AvRV-A and -D detection.
2. Materials and methods
2.1. Samples, virus strains and RNA extraction
In total, faecal samples and intestinal contents were obtained from 541 chickens from 83 flocks, and intestinal contents were collected from 51 turkeys from 21 flocks. Details on the origin of the samples from study A and study B are given in Table 1, Table 2 , respectively. The samples of study A were collected between 2005 and 2008 and analyzed by PAGE and AvRV-A-specific conventional RT-PCR. The samples from study B were collected between 2009 and 2010 and analyzed by AvRV-A- and AvRV-D-specific real-time RT-PCRs.
Table 1.
Origin data of chickens and turkeys which were included in Study A.
| Serial no. | Year | Country | Host | Age | Number of animals | Number of flocks | Clinical signs | % Positive for AvRV-A | % Positive for AvRV-D | % Positive for AvRV-A + AvRV-D |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2005 | Scotland | Chicken | Unknown | 16 | 4 | RSS | 12.5 | 18.8 | 6.3 |
| 2 | 2006 | Scotland | Chicken | Unknown | 3 | 1 | RSS | 0.0 | 0.0 | 0.0 |
| 3 | 2006 | Sweden | Chicken | 3–23 days | 28 | 5 | Growth retardation | 3.6 | 0.0 | 0.0 |
| 4 | 2006 | Germany | Chicken | 5–28 days | 61 | 23 | RSS | 9.8 | 26.2 | 3.3 |
| 5 | 2006 | UK | Chicken | 7–18 days | 30 | 6 | RSS | 6.7 | 23.3 | 3.3 |
| 6 | 2006 | Germany | Turkey | 2 days to 9 weeks | 20 | 6 | Diarrhoea | 0.0 | 70.0 | 0.0 |
| 7 | 2007 | Sweden | Chicken | 9–21 days | 6 | 3 | Growth retardation | 100.0 | 100.0 | 100.0 |
| 8 | 2007 | Germany | Chicken | 7–21 days | 13 | 5 | RSS | 46.2 | 92.3 | 38.5 |
| 9 | 2007 | Poland | Chicken | 18 days | 4 | 1 | RSS | 0.0 | 0.0 | 0.0 |
| 10 | 2007 | Germany | Turkey | 2–21 days | 13 | 7 | Diarrhoea | 5.9 | 38.4 | 7.7 |
| 11 | 2008 | Germany | Chicken | 10–23 days | 5 | 4 | RSS | 0.0 | 20.0 | 0.0 |
| Total | – | – | – | – | 199 | 65 | – | 16.1 | 39.2 | 11.1 |
Table 2.
Origin data of chickens and turkeys which were included in Study B.
| Serial No. | Year | Country | Host | Age | Number of animals | Number of flocks | Clinical signs | % Positive for AvRV-A | % Positive for AvRV-D | % Positive for AvRv-A +AVRV-D |
|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 2009 | Italy | Chicken | 3–11 days | 41 | 2 | RSS | 26.8 | 17.1 | 14.6 |
| 13 | 2009 | Germany | Chicken | 15 days | 243 | 13 | RSS | 61.7 | 91.4 | 52.7 |
| 14 | 2009 | Germany | Chicken | 15 days | 10 | 2 | None | 0 .0 | 40.0 | 0.0 |
| 15 | 2010 | Bangladesh | Chicken | 15 days | 30 | 3 | RSS | 76.7 | 3.3 | 3.3 |
| 16 | 2010 | Germany | Turkey | 3–12 weeks | 14 | 6 | Diarrhoea | 64.3 | 7.1 | 7.1 |
| 17 | 2010 | Germany | Chicken | 9–21 days | 7 | 4 | RSS | 85.8 | 14.3 | 14.3 |
| 18 | 2010 | Netherlands | Chicken | 1–35 days | 44 | 7 | RSS, diarrhoea | 72.8 | 61.3 | 36.7 |
| 19 | 2010 | Netherlands | Turkey | 6 weeks | 4 | 2 | Diarrhoea | 0.0 | 0.0 | 0.0 |
| Total | – | – | – | – | 393 | 39 | – | 58.8 | 65.9 | 38.9 |
An intestinal sample containing strain RVD/Chicken-wt/DEU/05V0049/2005/GXP[X] (Trojnar et al., 2010, designation according to Matthijnssens et al., 2011) was used as a positive control for AvRV-D. The chicken AvRV-A strain RVA/Chicken-tc/DEU/02V0002G3/2002/G19P[30] and the turkey AvRV-A strain RVA/Turkey-tc/DEU/03V0001E10/2003/G22P[35], both propagated in MA104 cells (Schumann et al., 2009), were used as positive controls for AvRV-A.
The faecal samples and intestinal contents were initially suspended at 1:5 (v:v) dilution in phosphate-buffered saline (PBS) pH 7.4, homogenised for 30 s in an ultrasonic water bath at level 4 (UST 20, K.-W. Meinhardt Ultraschalltechnik, Leipzig, Germany) and then clarified at 3345 × g for 20 min. The supernatants were collected and stored at −20 °C until analysis. Viral RNA was extracted from 140 μl of faecal supernatants or cell culture supernatants using QIAamp Viral RNA Kit® (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. The viral RNA was stored at −80 °C until further analysis.
2.2. Polyacrylamide gel electrophoresis
Extracted viral RNAs were analyzed for their pattern using PAGE as described by Otto et al. (1999). Gels were dried in a GelAirDryer (Bio-Rad Laboratories, Munich, Germany) and scanned on an Epson GT-15000 (Epson Deutschland GmbH, Meerbusch, Germany).
2.3. Conventional reverse transcription-polymerase chain reaction
Primer sequences targeting the VP6-encoding genes of AvRV-A (Otto et al., 2006, Schumann et al., 2009, Trojnar et al., 2009) were selected on a basis of an alignment of available sequences. Primers ARVA6-1F and ARVA6-1R were used, which amplify a 493 bp PCR product. All primer sequences and the sizes of the expected amplicons are listed in Table 3 .
Table 3.
Oligonucleotide primers and TaqMan probes used in RT-PCR and real-time RT-PCR amplification of parts of the VP6 gene of avian group A and group D rotavirus.
| Specificity (mode) | Name | Sequences (5′ → 3′) | Position (nt)a | Amplicon size |
|---|---|---|---|---|
| AvRV-A (RT-PCR) | ARVA6-1F | CAC CAC GAC TTA TGC AGA GA | 709–728 | 493 bp |
| ARVA6-1R | CTC CGA ATG GAT GCT ACT GT | 1201–1182 | ||
| AvRV-A (chicken strains) (real-time RT-PCR) | ARVA6-9F | GAG CAA CTA TTG ATT ACT TCA TTG A | 268–292 | 114 bp |
| ARVA6-9R | AAA GTT GCC TTA RTG CAT TAG A | 381–360 | ||
| ARVA6probe3 | AGG AGC TAT TCC ATT ACG TTG AGA TTC | 330–356 | ||
| AvRV-A (turkey strains) (real-time RT-PCR) | ARVA6-10F | CAT TTG ACT TTG GAA CAC TCG G | 202–223 | 80 bp |
| ARVA6-10R | GTC AAT TGT TGT TCT TGC ATT CTC | 281–258 | ||
| ARVA6probe4 | ACG ACC TTA CTW AAC TTG GAC GCG A | 225–247 | ||
| AvRV-D (RT-PCR and real-time RT-PCR) | ARVD6-1F | GCG ACA ACT GAG ACA ACT G | 1008–1026 | 186 bp |
| ARVD6-1R | GGA AGC AGT TGT CAT CAA C | 1193–1175 | ||
| ARVD6probe1 | TTG CAT ATT AGA TTG TCT CGC TGG TGT ATA | 1142–1171 |
The RT-PCR was conducted as an one-step RT-PCR using the Ready-to-Go (RTG) RT-PCR-Beads (GE Healthcare, Munich, Germany). Briefly, 1 μl of each of the primers (100 pmol/μl) was added to 10 μl of extracted RNA, denaturated for 5 min at 98 °C, and cooled down on ice. One RTG bead, dissolved with 38 μl DNase and RNase-free water was given to the RNA-primer mix. Amplifications were performed in a Mini Cycler™ (MJ Research Inc., Waltham, US) or Mastercycler (Eppendorf, Hamburg, Germany), and the incubation steps consisted of 42 °C for 30 min and 94 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 54 °C for 1 min, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. The PCR products were examined by electrophoresis in a 2% agarose gel, stained with ethidium bromide and visualized by ultraviolet transillumination.
2.4. Real-time RT-PCR
Primers and probes for the real-time RT-PCR assays (Table 3) targeting highly conserved regions of the VP6-encoding genes of AvRV-A or AvRV-D strains were designed using an alignment of published sequences (Otto et al., 2006, Schumann et al., 2009, Trojnar et al., 2009, Trojnar et al., 2010). Probes were labelled at the 5′ end with 5′-carboxyfluorescein (FAM) and at the 3′ end with the blackhole quencher BBQ (TIB Molbiol, Berlin, Germany). Primers ARVA6-9F and ARVA6-9R were used in combination with probe ARVA6probe3 for detection of AvRV-A in study B. As it became later evident that this real-time RT-PCR assay detects strains derived from turkey with only a low sensitivity, another assay using primers ARVA6-10F and ARVA6-10R together with probe ARVA6probe4 was established for more sensitive detection of those strains. For AvRV-D detection, primers ARVD6-1F and ARVD6-1R were used together with probe ARVD6probe1.
All real-time RT-PCR assays were carried out using several commercial one-step RT-PCR kits. The RT-PCR master mixes were prepared following the manufacturer's protocols (2× QuantiTect Probe RT-PCR Master Mix, Qiagen; Brilliant II QRT-PCR Low ROX Master Mix, Agilent Technologies, Waldbronn, Germany; EXPRESS One-Step SuperScript qRT-PCR Universal; Invitrogen, Karlsruhe, Germany) including 0.5 μl of TaqMan probes (5 pmol/μl). Thereafter, 2 μl of extracted RNA were mixed in a separate tube with 0.5 μl of each primer (10 pmol/μl) and subjected to 5 min of incubation at 95 °C to allow the denaturation of the rotaviral dsRNA. The mixture was placed immediately on ice, and then the RT-PCR master mix was added. The tubes were briefly centrifuged before being loaded into the cycler (ABI Prism 7000, Applied Biosystems Applera Deutschland GmbH, Darmstadt, Germany; Mx 3000P, Stratagene, Agilent Technologies, Santa Clara, US). After reverse transcription and initial denaturation steps according to the instructions of the one-step RT-PCR manufacturer, the conditions for the PCR were 45 cycles of 15 s at 95 °C and 60 s at 60 °C. Fluorescence was measured after the annealing step starting with cycle 3.
2.5. Transmission electron microscopy (TEM)
The samples were investigated by TEM using the negative staining method. Briefly, supernatants of the samples were applied to polioform-carbon-coated, 400-mesh copper grids (Plano GmbH, Wetzlar, Germany), stained with 2% of aqueous uranyl acetate solution and examined by TEM (JEM-1010 JEOL, Tokyo, Japan) at 80 kV accelerated voltage.
2.6. Assessment of (real-time) RT-PCR sensitivity
The performance of the real-time RT-PCR assays was assessed using dilution series of in vitro-transcribed RNAs. Briefly, RT-PCR products were amplified from AvRV-A strain RVA/Chicken-tc/DEU/02V0002G3/2002/G19P[30] with primers AVRVA6-9F and AVRVA6-9R (Table 3) and from AvRV-D strain RVD/chicken-wt/DEU/05V0049/2005/GXP[X] with primers AVRVD6-1F and AVRVD6-1R (Table 3), respectively, and subsequently cloned into vector pCR4-Topo (Invitrogen). After confirmation of the cloned sequences by DNA sequencing, the insert together with a flanking T7-promotor was amplified by PCR with primers M13F and M13R (Invitrogen) and purified using the QIAquick Gel Extraction Kit (Qiagen). The DNA fragment was transcribed into RNA using the MEGAscript T7 Kit (Ambion, Applied Biosystems, Darmstadt, Germany) and the remaining DNA was removed by digestion with Turbo DNAse (Applied Biosystems). After purification using the High Pure RNA Isolation Kit (Roche, Basel, Switzerland), the RNA concentration was measured photometrically using a NanoDrop device (Analytik Jena AG, Jena, Germany). Ten-fold dilutions of RNA containing 1 × 10−1 to 1 × 1013 molecules were tested with the real-time RT-PCR protocols as described above and the R 2 values, PCR efficiency and minimum number of detected copies per reaction were calculated from the established standard curves.
In order to compare the sensitivities of the different PCR assays (conventional and real-time), preparations containing infectious rotavirus particles were used. The cell culture adapted AvRV-A strains from chicken and turkey were used for infectivity titrations by preparing serial 10-fold dilutions of supernatants from infected MA-104 cells (Schumann et al., 2009). Aliquots of each dilution were inoculated onto four wells containing a confluent MA-104 cell monolayer. The presence of a cytopathic effect was observed daily and the 50% tissue culture infective dose (TCID50) was estimated according to Spearman and Kärber (Mayr et al., 1974) at day 4 post inoculation. Thereafter, the RNA was isolated from the undiluted cell culture supernatants using the QIAamp Viral RNA Kit® (Qiagen) and serial 10-fold RNA dilutions were prepared in duplicate. The preparations were subsequently tested in parallel using the different real-time RT-PCR protocols and the detection limits were correlated with the TCID50 of the corresponding dilutions. To compare the sensitivity of AvRV-D-specific real-time and conventional PCR protocols, a similar dilution series was prepared from RNA of a faecal sample containing AvRV-D strain and the dilutions were tested in parallel by the assays.
3. Results
3.1. Study A
Conventional molecular assays (PAGE and RT-PCR) were established for the detection of AvRV-A and AvRV-D. Using RNA of the AvRV-A strain RVA/Chicken-tc/DEU/02V0002G3/2002/G19P[30], a 493 bp amplicon was produced by the AvRV-A-specific RT-PCR.
Between 2005 and 2008, faecal samples or intestinal contents were collected from 166 chickens and 33 turkeys originating from several commercial flocks of five different European countries (Table 1). All of the samples were tested using the PAGE and the RT-PCR assay. Results are presented in Table 1. AvRVs were detected in 92 samples (46.2%). In detail, 7 and 56 samples (3.5 and 28.2%) contained only AvRV-A or AvRV-D, respectively, and 22 samples (11.1%) contained both. The detection rates according to the different avian species were 18.7% AvRV-A in chicken, 3.0% AvRV-A in turkey, 35.5% AvRV-D in chicken and 57.6% AvRV-D in turkey. All of the samples showing electropherotypes typical for AvRV-A by PAGE were also tested positive using the RT-PCR assay. However, some of the samples, which were negative in PAGE showed positive RT-PCR results indicating a higher sensitivity of the RT-PCR assay.
By PAGE analysis, four chicken samples revealed the presence of RVs distinct from AvRV groups A and D (Fig. 1 ). The samples 06V0034 and 06V0044 originated from Germany and Sweden, respectively, showed a dsRNA migration pattern with a faster migrating segment 5 as compared to AvRV-A or AvRV-D and very closely migrating segments 6 and 7 (Fig. 1C). These dsRNA migration patterns have been described as RV-F (Saif et al., 1994, Johne et al., 2011). The samples 05V0497 and 05V0499, both originated from the UK, produced a dsRNA profile characterized by very closely migrating segments 5 and 6 and very closely migrating segments 9 and 10 (Fig. 1D). These dsRNA migration patterns are typical for RV-G (Johne et al., 2011). The presence of RV in these four samples could be confirmed by electron microscopy (Fig. 2 ). No conspicuous differences in particle morphology could be observed between AvRV-A and AvRV of other groups as already described by Johne et al., 2011.
Fig. 1.

Analysis of the genome profiles of avian rotavirus (AvRV) dsRNA after polyacrylamide gel electrophoresis followed by silver staining. RNA segments are indicated by numbers according to their mobility, an additional segment indicated by arrow could point to a mixed virus infection. (A) AvRV-A strain 05V0498, (B) AvRV-D strain 06V0065, and (C) AvRV-F strain 06V0044 and (D) AvRV-G strain 05V0499.
Fig. 2.
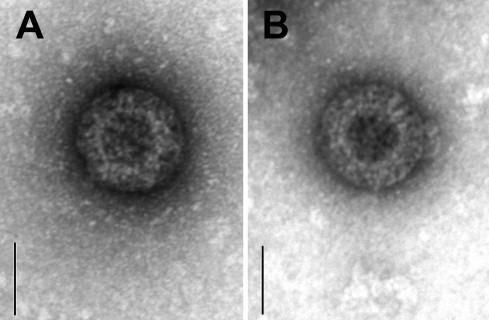
Identification of avian rotavirus particles by negative staining electron microscopy. (A) group F rotavirus 06V0034 and (B) group G rotavirus 05V0497. The bar corresponds to 50 nm.
3.2. Study B
Real-time RT-PCR assays were established for the detection of AvRV-A and AvRV-D. Testing of RNA from reference strains of AvRV-A and AvRV-D as well as from selected AvRV-A- or AvRV-D-positive chicken samples derived from study A yielded concordant results in the real-time RT-PCR assays (data not shown). Performance and sensitivity of the real-time RT-PCR assays were tested using dilutions series of in vitro-transcribed RNAs, which contained sequences of AvRV-A strain RVA/Chicken-tc/DEU/02V0002G3/2002/G19P[30] or AvRV-D strain RVD/chicken-wt/DEU/05V0049/2005/GXP[X]. By this, the AvRV-A-specific assay showed an R 2 value of 0.99 and a PCR efficiency of 84.0%. The minimum number of detected copies of this assay was 2.65 × 102 per reaction. The AvRV-D-specific assay showed an R 2 value of 0.99 and a PCR efficiency of 81.5%; the minimum number of detected copies was 3.38 × 103 per reaction
From 2009 and 2010 faecal samples or intestinal contents were collected from 375 chickens and 18 turkeys that originated from several commercial flocks in three different European countries and Bangladesh (Table 2). All of the samples were examined by real-time RT-PCR for detection of AvRV-A and AvRV-D. With this method, 337 samples (85.8%) showed a positive result for AvRV. In detail, 231 samples (58.8%) were positive for AvRV-A and 259 samples (65.9%) were positive for AvRV-D. AvRVs of both groups could be detected in 152 samples (38.9%).
The samples from two commercial flocks (Serial No. 14 in Table 2) originated from broiler chickens which did not show clinical signs of diarrhoea or RSS, also revealed AvRV-Ds in 4 of these 10 samples; however, the resulting ct-values of 34.06, 38.13, 39.22 and 43.70 were relatively high indicating a low amount of virus within the samples.
To compare the sensitivity of the conventional RT-PCR and the real-time RT-PCR assays using infectious AvRV-A as starting material, RNA dilution series were prepared from cell culture supernatants containing the chicken AvRV-A strain RVA/Chicken-tc/DEU/02V0002G3/2002/G19P[30] and the turkey AvRV-A strain RVA/Turkey-tc/DEU/03V0001E10/2003/G22P[35]. The infectivity titres of the preparations of the chicken and turkey AvRV-A strains were 105.75 and 106.00 TCID50/ml, respectively. For the conventional AvRV-A-specific RT-PCR, the lower limit of detection for the chicken AvRV-A strain was 103.75 TCID50/ml, whereas the turkey AvRV-A strain was not detected by this assay. For the AvRV-A-specific real-time RT-PCR assay (with primer set ARVA6-9F/R as used in Study B), the lower limit of detection for the chicken AvRV-A strain was 101,25 TCID50/ml, but only 104 TCID50/ml for the turkey AvRV-A strain. Using another set of primers and probes (ARVA6-10F/R, Table 3), designed on the basis of RV sequences derived from turkeys, the lower limit of detection for this strain was 1 TCID50/ml. For AvRV-D, a serial dilution of a faecal sample containing AvRV-D strain RVD/chicken-wt/DEU/05V0049/2005/GXP[X] was tested either by real-time RT-PCR as described or by a conventional RT-PCR using primers ARVD-1F/R followed by electrophoretic analysis of the PCR product. In both assays, the conventional as well as the real-time RT-PCR, detected this strain until a dilution of 10−2.
4. Discussion
The occurrence of RVs in many avian species, such as ducks, pheasants, chickens, turkeys, pigeons and wild birds, has been reported previously from many countries worldwide (Bellinzoni et al., 1987, Day et al., 2007, Gough et al., 1992, McNulty et al., 1979, Minamoto et al., 1988, Pantin-Jackwood et al., 2007, Reynolds et al., 1987c, Takase et al., 1986, Takehara et al., 1991, Theil et al., 1986, Todd and McNulty, 1986). In these studies, a range of rotavirus detection systems with different specificity and sensitivity was applied leading to high variations in the estimation of the prevalence of AvRV in poultry. Only a few RT-PCR protocols are available for detection of AvRVs and all of them exclusively detect AvRV-A (Day et al., 2007, Elschner et al., 2002, Pantin-Jackwood et al., 2007). Therefore, the main objective of this study was the identification of the most prevalent AvRV groups and the subsequent development of RT-PCR methods for their detection. As it became evident, that AvRV-D occurs with high very frequency in chickens and turkeys, sensitive real-time RT-PCR assays for detection of both, AvRV-A and AvRV-D have been successfully developed. To our knowledge, this is the first study applying such assays to assess of the distribution of AvRV in diseased chicken and turkey.
The detailed analysis of the results of study A indicated that two groups of AvRVs dominated in chickens and turkeys with diarrhoea, growth retardation and/or RSS: AvRV-A and AvRV-D (16.1 and 39.2%). As reported in several previous studies, AvRV-D was the most commonly found rotavirus in turkeys (McNulty and Reynolds, 2008, Reynolds et al., 1987a, Reynolds et al., 1987b, Theil et al., 1986). Co-infections with AvRV-D and astrovirus in turkeys have also been frequently detected (Pantin-Jackwood et al., 2007). In contrast, very little information is available on the occurrence of AvRV-D in chicken. Here, we show that AvRV-D is also frequently present in chicken, representing the most abundant AvRV group in many of the investigated chicken flocks. In contrast, only 4 chicken samples contained AvRV particles with electropherotypes typically for AvRV-F and AvRV-G groups. Prototype viruses of these RV groups, the A4 and 555 isolates, were first observed in gut contents of chicken in Northern Ireland (McNulty et al., 1984a, Ramig et al., 2005). Based on our results, AvRV-F and AvRV-G seem to represent rare rotavirus groups in chickens, at least in the timeframe and the geographical regions investigated in this study. It should be taken into account, however, that identification of RV groups by PAGE is difficult as genome rearrangements may occur in rotaviruses (Desselberger, 1996). In addition, the sensitivity of PAGE is generally low. The availability of genomic sequences of AvRV-F and AvRV-G (Johne et al., 2011) may enable the future development of RT-PCR methods for detection of these RV groups, which could be more sensitive and specific than PAGE.
Due to the dominating presence of AvRV-A and -D, Study B was performed to estimate the prevalence of these RV groups using more sensitive real-time RT-PCRs. As a result, the detection rates of both AvRV groups were very high, with 65.9% of the samples positive for AvRV-D and 58.8% for AvRV-A. Although differences between the flocks of different geographical origin exist, these high detection rates were found in all countries tested. As only 2 out of the analyzed 30 flocks did not contain birds showing diarrhoea, growth retardation or RSS, the clinical significance of the rotaviruses detected cannot be inferred from this study. However, the relatively low AvRV incidence combined with the low virus amount detected in the samples from these clinically healthy flocks may give a first indication for involvement of these viruses in disease development.
The detection rate of AvRVs was much lower in Study A as compared to Study B, which is likely due to the different sensitivities of the applied RT-PCR assays. Indeed, the real-time RT-PCR assay for detection of the AvRV-A turned out to be about 1000-fold more sensitive than the conventional RT-PCR assay as assessed by using an infectious AvRV-A strain from chicken. This difference in sensitivity might be due to the use of different primer binding sequences in the two assays. In contrast, the real-time RT-PCR assay for detection of AvRV-D showed the same sensitivity as compared to the respective conventional RT-PCR protocol as assessed by using a defined AvRV-D-containing intestinal sample. This might be explained by the fact, that both assays used the same primer pair. In concordance with this result, the relative low sensitivity of the AvRV-D-specific real-time RT-PCR was confirmed by using in vitro-transcribed RNA. Therefore, a further optimization of this assay might be useful in future.
Although both the AvRV-A-specific assays, the conventional and the real-time RT-PCR, yielded positive results in field samples of turkeys, they failed to detect sensitively a tissue-culture-derived AvRV strain originally isolated from a turkey. A recent sequence analysis of AvRV-A strains showed that isolates from chickens and turkeys cluster into two different phylogenetic lineages (Schumann et al., 2009). These sequence differences were also observed, when the VP6-encoding genome segment was compared, which is the target used for the RT-PCR assays. Therefore, it might be necessary to use separate assays for sensitive detection of chicken and turkey RV-A strains. The additional real-time RT-PCR protocol developed here with modified primers and probes for detection of the turkey strains showed a high sensitivity with the cell-culture-derived turkey virus and may be applied in future field studies.
In conclusion, this study shows that AvRV-A and -D are highly prevalent in diseased chicken and turkey originating from commercial flocks in Europe and Bangladesh. Further investigations are needed to determine the etiological role of the viruses detected. The real-time RT-PCR protocols developed here are useful tools, which should be optimized for further application in larger field studies or as quantitative methods in experimental investigations in order to answer these questions.
Conflict of interest
None of the authors of this paper has a financial or personal relationships with other people or organisations that could inappropriately influence or bias the content of the paper.
Acknowledgements
We would like to thank Petra Sippach for excellent technical assistance. We are grateful to Josef Bachmeier, Eva Berndtson, Gerhard Glünder, Silvia Jodas, Ulrich Löhren, Christian Prins, Fausto Ruffini, Dietmar Schmitt, Otto van Tuijl, Gert-Jan Zuidam, colleaques and farmers for accessing samples of chickens and turkeys. We thank Olfert Landt (TIB Molbiol, Berlin, Germany) for modifying the primers and probes and Wolfram Maginot for taking the photos. We wish to thank Prof. Ian N. Clarke, University of Southampton, Molecular Microbiology Group, Division of Infection, Inflammation and Immunity, for reviewing the English language in the manuscript. This investigation was supported in part by the Deutsche Forschungsgemeinschaft, projects JO369/4-1 (Eva Trojnar) and SFB852 (Patrycja Machnowska).
References
- Alfieri A.F., Resende M., Resende J.S., Alfieri A.A. Atypical rotavirus infections among broiler chickens in Brazil. Arq. Bras. Med. Vet. Zoot. 1989;41:81–82. [Google Scholar]
- Bridger J.C. Non-group rotaviruses. In: Kapikian A.Z., editor. Viral Infections of the Gastrointestinal Tract. 2nd ed. Marcel Dekker Inc.; New York/Basel/Hong Kong: 1994. pp. 369–407. [Google Scholar]
- Bellinzoni R., Mattion N., Vallejos L., La Torre J.L., Scodeller A. Atypical rotavirus in chickens in Argentina. Res. Vet. Sci. 1987;43:130–131. [PubMed] [Google Scholar]
- Day J.M., Spackman E., Pantin-Jackwood M. A multiplex RT-PCR for the differential identification of turkey astrovirus type 1, turkey astrovirus type 2, chicken astrovirus, avian nephritis virus, and rotavirus. Avian Dis. 2007;51:681–684. doi: 10.1637/0005-2086(2007)51[681:AMRTFT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Desselberger U. Genome rearrangements of rotaviruses. Adv. Virus Res. 1996;46:69–95. doi: 10.1016/s0065-3527(08)60070-6. [DOI] [PubMed] [Google Scholar]
- Elschner M., Prudlo J., Hotzel H., Otto P., Sachse K. Nested reverse transcriptase-polymerase chain reaction for the detection of group A rotaviruses. J. Vet. Med. B. 2002;49:77–81. doi: 10.1046/j.1439-0450.2002.00510.x. [DOI] [PubMed] [Google Scholar]
- Estes M.K., Kapikian A.Z. Rotaviruses. In: Knipe D.M., Howley P.M., Griffin M.A., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Fields Virology. 5th ed. Lippincott, Williams and Wilkins; Philadelphia, PA: 2007. pp. 1917–19704. [Google Scholar]
- Fitzgerald S.D. Adenovirus infections. In: Saif Y.M., Fadley A.M., Glission J.R., Mc Dougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. 12th ed. Blackwell Publishing Professional; Ames, IA, USA: 2008. pp. 251–252. [Google Scholar]
- Gough R.E., Cox W.J., Devoy J. Isolation and identification of rotavirus from racing pigeons. Vet. Rec. 1992;130:273. doi: 10.1136/vr.130.13.273. [DOI] [PubMed] [Google Scholar]
- Jones C. Reovirus infections. In: Saif Y.M., Fadley A.M., Glission J.R., Mc Dougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. 12th ed. Blackwell Publishing Professional; Ames, IA, USA: 2008. pp. 309–328. [Google Scholar]
- Johne R., Otto P., Roth B., Löhren U., Belnap D., Reetz J., Trojnar E. Sequence analysis of the VP6-encoding genome segment of avian group F and G rotaviruses. Virology. 2011;412:384–391. doi: 10.1016/j.virol.2011.01.031. [DOI] [PubMed] [Google Scholar]
- Legrottaglie R., Rizzi V., Agrimi P. Isolation and identification of avian rotavirus from pheasant chicks with signs of clinical enteritis. Comp. Immunol. Microbiol. Infect. Dis. 1997;20:205–210. doi: 10.1016/S0147-9571(97)00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr, A., Bachmann, P.A., Bibrack, B., Wittmann, G., 1974. 3. Quantitative Bestimmung der Virusinfektiösität (Virustitration). In: Virologische Arbeitsmethoden. 1. Aufl., Gustav Fischer Verlag Jena, S. 35–39.
- Matthijnssens J., Ciarlet M., McDonald S.M., Attoui H., Bányai K., Brister J.R., Buesa J., Esona M.D., Estes M.K., Gentsch J.R., Iturriza-Gómara M., Johne R., Kirkwood C.D., Martella V., Mertens P.P.C., Nakagomi O., Parreño V., Rahman M., Ruggeri F.M., Saif L.J., Santos N., Steyer A., Taniguchi K., Patton J.T., Desselberger U., van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch. Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty M.S., Allan G.M., Stuart J.C. Rotavirus infection in avian species. Vet. Rec. 1978;103:319–320. doi: 10.1136/vr.103.14.319-a. [DOI] [PubMed] [Google Scholar]
- McNulty M.S., Curran W.L., Todd D., McFerran J.B. Detection of viruses in avian faeces by direct electron microscopy. Avian Pathol. 1979;8:239–247. doi: 10.1080/03079457908418349. [DOI] [PubMed] [Google Scholar]
- McNulty M.S., Todd D., Allan G.M., Mc Ferran J.B., Greene J.A. Epidemiology of rotavirus infection in broiler chickens: recognition of four serogroups. Arch. Virol. 1984;81:113–121. doi: 10.1007/BF01309301. [DOI] [PubMed] [Google Scholar]
- McNulty M.S., Allan G.M., Mc Ferran J.B. Prevalence of antibody to conventional and atypical rotaviruses in chickens. Vet. Rec. 1984;114:219. doi: 10.1136/vr.114.9.219. [DOI] [PubMed] [Google Scholar]
- McNulty M.S., Reynolds D.L. Rotavirus infections. In: Saif Y.M., Fadley A.M., Glission J.R., Mc Dougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. 12th ed. Blackwell Publishing Professional; Ames, IA, USA: 2008. pp. 338–350. [Google Scholar]
- Minamoto N., Oki K., Tomita M., Kinjo T., Suzuki Y. Isolation and characterization of rotavirus from feral pigeon in mammalian cell cultures. Epidemiol. Infect. 1988;100:481–492. doi: 10.1017/s0950268800067212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto P., Schulze P., Herbst W. Demonstration of group C rotaviruses in fecal samples of diarrheic dogs from Germany. Arch. Virol. 1999;144:2467–2473. doi: 10.1007/s007050050659. [DOI] [PubMed] [Google Scholar]
- Otto P., Liebler-Tenorio E.M., Elschner M., Reetz J., Löhren U., Diller R. Detection of rotaviruses and intestinal lesions in broiler chicks from flocks with runting and stunting syndrome (RSS) Avian Dis. 2006;50:411–418. doi: 10.1637/7511-020106R.1. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood M.J., Spackman E., Day J.M., Rives D. Periodic monitoring of commercial turkeys for enteric viruses indicates continuous presence of astrovirus and rotavirus on the farms. Avian Dis. 2007;51:674–680. doi: 10.1637/0005-2086(2007)51[674:PMOCTF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Ramig R.F., Ciarlet M., Mertens P.P.C., Dermody T.S. Genus Rotavirus. In: Fauquet C.M., Mayo C.M., Maniloff M.A., J. Desselberger U., Ball L.A., editors. Virus Taxonomy, 8th report. Academic Press; San Diego: 2005. pp. 484–496. [Google Scholar]
- Reynolds D.L., Saif Y.M., Theil K.W. A survey of enteric viruses of turkey poults. Avian Dis. 1987;31:89–98. [PubMed] [Google Scholar]
- Reynolds D.L., Saif Y.M., Theil K.W. Enteric viral infections of turkey poults: incidence of infection. Avian Dis. 1987;31:272–276. [PubMed] [Google Scholar]
- Reynolds D.L., Theil K.W., Saif Y.M. Demonstration of rotavirus and rotavirus-like virus from the intestinal contents of diarrheic pheasant chicks. Avian Dis. 1987;31:376–379. [PubMed] [Google Scholar]
- Reynolds D.L., Schultz-Cherry S.L. Astrovirus infections. In: Saif Y.M., Fadley A.M., Glission J.R., Mc Dougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. 12th ed. Blackwell Publishing Professional; Ames, IA, USA: 2008. pp. 351–355. [Google Scholar]
- Saif L.J., Saif Y.M., Theil K.W. Enteric viruses in diarrheic turkey poults. Avian Dis. 1985;29:798–811. [PubMed] [Google Scholar]
- Saif L.J., Rosen B.I., Parwani A.V. Animal Rotaviruses. In: Kapikian A.Z., editor. Viral Infections of the Gastrointestinal Tract. 2nd ed. Marcel Dekker Inc; New York/Basel/Hong Kong: 1994. pp. 279–367. [Google Scholar]
- Saif Y.M. Viral enteric infections. Introduction. In: Saif Y.M., Fadley A.M., Glission J.R., Mc Dougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. 12th ed. Blackwell Publishing Professional; Ames, IA, USA: 2008. p. 329. [Google Scholar]
- Schumann T., Hotzel H., Otto P., Johne R. Evidence of interspecies transmission and reassortment among avian group A rotaviruses. Virology. 2009;386:334–343. doi: 10.1016/j.virol.2009.01.040. [DOI] [PubMed] [Google Scholar]
- Spackman E., Day J.M., Pantin-Jackwood M.J. Astrovirus, reovirus, and rotavirus concomitant infection causes decreased weight gain in broad-breasted white poults. Avian Dis. 2010;54:16–21. doi: 10.1637/8986-070909-Reg.1. [DOI] [PubMed] [Google Scholar]
- Takehara K., Kiuchi H., Kuwahara M., Yanagisawa F., Mizukami M., Matsuda H., Yoshimura M. Identification and characterization of a plaque forming avian rotavirus isolated from a wild bird in Japan. J. Vet. Sci. 1991;53:479–486. doi: 10.1292/jvms.53.479. [DOI] [PubMed] [Google Scholar]
- Takase K., Nonaka F., Sakaguchi M., Yamada S. Cytopathic avian rotavirus isolated from duck faeces in chicken kidney cell cultures. Avian Pathol. 1986;15:719–730. doi: 10.1080/03079458608436334. [DOI] [PubMed] [Google Scholar]
- Takase K., Uchimura T., Katsuki N., Yamamoto M.A. Survey of chickens sera for antibody to atypical avian rotavirus of duck origin in Japan. Jpn. J. Vet. Sci. 1990;52:1319–1321. doi: 10.1292/jvms1939.52.1319. [DOI] [PubMed] [Google Scholar]
- Theil K.W., Reynolds D.L., Saif Y.M. Comparison of immune electron microscopy and genome electropherotyping techniques for detection of turkey rotaviruses and rotaviruslike viruses in intestinal contents. J. Clin. Microbiol. 1986;23:695–699. doi: 10.1128/jcm.23.4.695-699.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd D., McNulty M.S. Electrophoretic variation of avian rotavirus RNA in polyacrylamid gels. Avian Pathol. 1986;15:149–159. doi: 10.1080/03079458608436274. [DOI] [PubMed] [Google Scholar]
- Trojnar E., Otto P., Johne R. The first complete genome sequence of a chicken group A rotavirus indicates independent evolution of mammalian and avian strains. Virology. 2009;386:325–333. doi: 10.1016/j.virol.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Trojnar E., Otto P., Roth B., Reetz J., Johne R. The genome segments of a group D rotavirus possess group A-like conserved termini but encode group-specific proteins. J. Virol. 2010;84:10254–10265. doi: 10.1128/JVI.00332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yason C.V., Schat K.A. Experimental infection of specific-pathogen-free chickens with avian rotaviruses. Avian Dis. 1986;30:551–556. [PubMed] [Google Scholar]
- Yason C.V., Schat K.A. Pathogenesis of rotavirus infection in various age groups of chickens and turkeys: clinical signs and virology. Am. J. Vet. Res. 1987;48:977–983. [PubMed] [Google Scholar]


