Highlights
-
•
PEDV ORF3 is a transmembrane protein that localizes at endoplasmic reticulum to induce ER stress response.
-
•
PEDV ORF3 protein has no effect on apoptosis, but induces autophagy dependent on ER stress response.
-
•
This work provides some new findings for the biological function of the PEDV ORF3 protein.
Keywords: Porcine epidemic diarrhea virus, Accessory protein ORF3, Endoplasmic reticulum stress, Apoptosis, Autophagy
Abstract
Porcine epidemic diarrhea virus (PEDV), the causative agent of PED, is an enveloped, positive-stranded RNA virus in the genus Alphacoronavirus, family Coronaviridae, order Nidovirales. PEDV non-structural accessory protein ORF3 is an ion channel related to viral infectivity and pathogenicity. Our previous study showed that PEDV ORF3 has expression characteristic of aggregation in cytoplasm, but its biological function remains elusive. Thus in this study, we initiated the construction of various vectors to express ORF3, and found ORF3 localized in the cytoplasm in the aggregation manner. Subsequently, confocal microscopy analysis showed that the aggregated ORF3 localized in endoplasmic reticulum (ER) to trigger ER stress response via up-regulation of GRP78 protein expression and activation of PERK-eIF2α signaling pathway. In addition, our results showed that PEDV ORF3 could induce the autophagy through inducing conversion of LC3-I to LC3-II, but couldn’t influence the apoptosis. In contrast, conversion of LC3-I/LC3-II could be significantly inhibited by 4-PBA, an ER stress inhibitor, indicating that ORF3-induced autophagy is dependent on ER stress response. This work not only provides some new findings for the biological function of the PEDV ORF3 protein, but also help us for the further understanding the molecular interaction between PEDV ORF3 protein and cells.
1. Introduction
Porcine epidemic diarrhea (PED), as a highly contagious and deadly swine disease, is characterized by vomiting, anorexia, watery diarrhea, severe dehydration, and high mortality rates in neonatal piglets, approximately arriving at 80–100% fatality rate in suckling piglets (Knuchel et al., 1992; Kusanagi et al., 1992). PED is first reported by Pensaert and DeBouck in Belgium and the United Kingdom in the 1970s (Pensaert and de Bouck, 1978; Debouck and Pensaert, 1979), and then spread to multiple swine-producing countries in Europe (Knuchel et al., 1992; Bridgen et al., 1993). Since the 1990s, PED cases have become rare in Europe, but which have been continually reported in many Asian and American countries or regions, including China, Japan, Korea, Philippines, Thailand, Vietnam, Taiwan, United State, Canada, Mexico, and so on (Lee, 2015). Especially, a large number of PED outbreaks have been reported in China in 2010, and a particularly large epidemic PED swept throughout all of North America (Sun et al., 2012; Vlasova et al., 2014). PED has become an increasing problem in many swine-breeding countries, resulting in tremendous economic losses to swine industry worldwide.
Porcine epidemic diarrhea virus (PEDV), the causative agent of PED, is an enveloped, positive-stranded RNA virus in the genus Alphacoronavirus, family Coronaviridae, order Nidovirales (Sato et al., 2018; Yu et al., 2018). The genome of a PEDV is approximately 28 k nt in length and is composed of seven open reading frames (ORFs) arranging in the order 5′-ORF1a/1b-S-ORF3-E-M-N-3′, which encodes four structural proteins and seventeen non-structural proteins (nsp1-nsp16, and ORF3) (Ye et al., 2015; Cheun-Arom et al., 2016). The 5′ two thirds of the genome contains two overlapping ORFs, ORF1a and ORF1b, which encode nsps that are cleaved by viral encoded proteases into 16 nsps (nsp1-16), directing genome replication, transcription, translation and viral polyprotein processing (Huang et al., 2013). The remaining one third of genome encodes four structural proteins responsible for assembly of infectious virus including the glycoprotein (S, 180–220 kDa), membrane (M, 27–32 kDa), envelope (E, 7 kDa), and the nucleocapsid (N, 55–58 kDa) proteins, respectively, and one non-structural accessory protein (ORF3, 27 kDa) (Bridgen et al., 1998).
The PEDV ORF3 gene, located between the S and E genes, has to be found the low sequence conservation through analysis of amino acid sequences and their homologs across the alphacoronavirus genus (Park et al., 2008). The ORF3 protein exhibits a genetic variation among PEDVs. Sequence comparison of virulent and attenuated PEDV strains revealed that some of attenuation-related mutations are located in the ORF3 genes (Chen et al., 2010; Su et al., 2016), suggesting that ORF3 protein exhibits a genetic variation among PEDVs. In addition, ORF3, as the only accessory gene in PEDV, is thought to function as an ion channel and to influence virus production and virulence (Wang et al., 2012). PEDV ORF3 protein can prolong cellular S-phase, facilitate formation of vesicles and thus promote the proliferation of attenuated PEDV rather than virulent PEDV (Ye et al., 2015). Previous study demonstrated that ORF3 and S are co-localized in several cellular compartments in infected cells, and ORF3 may interact with S during PEDV assembly and consequently exert its activity at this step of virus replication (Kaewborisuth et al., 2018). On the contrary, the ORF3 accessory protein has been reported to impede reverse genetics of cell-culture-adapted PEDV, because full-length ORF3, but not the truncated form, can inhibit recovery of reverse-genetics derived PEDV when expressed in trans (Wongthida et al., 2017), and the attenuated PEDV rather than virulent PEDV can grow better in ORF3-expressing Vero cells (Ye et al., 2015). Whereas, much more studies report that infectious PEDV could be rescued from a full-length cDNA clone with intact ORF3. For example, it is reported that the infectious-clone-derived PEDV (icPEDV) replicates as efficiently as the parental virus in cell culture and in pigs, resulting in lethal disease in vivo (Beall et al., 2016). These controversial events altogether suggest that PEDV ORF3 may be a multifunctional protein that is involved in many cellular processes. Whereas, the exact biological function of PEDV ORF3 need to be further defined.
PEDV ORF3 protein consisting of four transmembrane domains (TMDs) is localized in the ER pointing to the possibility that ORF3 might trigger the endoplasmic reticulum (ER) stress associated with either apoptosis or autophagy. However, there are lack of reports about the influence of PEDV ORF3 on the ER stress, apoptosis and autophagy. Up to date, the investigation of ORF3 is hampered by difficulty of detecting protein expression (Kaewborisuth et al., 2018). Thus in present study, we initiated the construction of various recombinant plasmids expressing PEDV ORF3 protein fused with different tags, and found ORF3 localized in the cytoplasm in the aggregation manner. PEDV ORF3 is a transmembrane protein localized in ER, which triggered ER stress via up-regulation of GRP78 and activation of PERK-eIF2α signaling pathway. Furthermore, our results showed that PEDV ORF3 could induce the autophagy through inducing conversion of LC3-I to LC3-II, but couldn’t influence the apoptosis. Nevertheless, conversion from LC3-I to LC3-II could be obviously inhibited by 4-PBA, an ER stress inhibitor, indicating that ORF3-induced autophagy is dependent on ER stress response. To our knowledge, this is the first research about PEDV ORF3 protein functions on ER stress, apoptosis and autophagy simultaneously. These findings have potentially important implications for augments of our understanding of ORF3 biological function.
2. Materials and methods
2.1. Cell lines
African green monkey cells E6 (Vero), human embryonic kidney (HEK) cells (293 T), human cervical carcinoma cells (HeLa) were stored in our laboratory. The established porcine kidney-15 cells (PK15) were kindly provided by prof. Huaji Qiu (Harbin Veterinary Research Institute of Chinese Academy of Agricultural Sciences). These cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin at 37 °C in a 5% CO2 humidified atmosphere.
2.2. Reagents and antibodies
Thapsigargin (Tg) (T9033), 4-phenylbutyrate (4-PBA) (STBG8666) and rapamycin (V900930) were purchased from Sigma-Aldrich (Darmstadt, German). TRIzol reagent (101002) was purchased from Thermo Fisher Scientific (Shanghai, China). Lipofectamine™2000 (11668019) was purchased from Invitrogen (Shanghai, China). Reverse transcriptase M-MLV (RNase H-) (2641) and Ex Taq polymerase (R060) were purchased from Takara (Dalian, China). Rabbit anti-PDI monoclonal antibody (AP233), rabbit anti-LC3B polyclonal antibody (AL221), rabbit anti-GRP78 monoclonal antibody (AF0171), rabbit anti-PERK monoclonal antibody (AP328), rabbit anti-phosphorylated eIF2α monoclonal antibody (AF1237) and Annexin V-FITC apoptosis detection kit (C1062S) were purchased from Beyotime (Shanghai, China). Rabbit anti-eIF2α antibody (9722) and rabbit anti-pPERK antibody (9451) was purchased from Cell Signaling Technology (Boston, USA). Rabbit anti-GFP monoclonal antibody (ab1218) was purchased from Abcam (Cambridge, MA, USA). Mouse anti-Myc monoclonal antibody (19C2) and mouse anti-Flag monoclonal antibody (M2008) were purchased from Abmart (Shanghai, China). Mouse anti-β-actin monoclonal antibody (ZM-0001), mouse anti-α-tubulin monoclonal antibody (ZS-80350), secondary antibodies conjugated to horseradish peroxidase (HRP) (goat anti-mouse (ZB-2305) and goat anti-rabbit (ZB-2301)) used for Western blot and secondary antibodies conjugated FITC (goat anti-rabbit (ZF-0311) and goat anti-mouse (ZF-0312)) and TRITC (goat anti-rabbit [ZF-0316] and goat anti-mouse [ZF-0313]) used for immunofluorescence were purchased from Zhongshan Biotech (Beijing, China).
2.3. Plasmids and transfection
The eukaryotic expression vectors pEGFP-N1, pCMV-Myc, and pcDNA3.0-Flag were stored in our laboratory. Plasmid pEGFP-LC3, indicator of autophagy, was kindly provided by prof. Xiaojun Wang (Harbin Veterinary Research Institute of Chinese Academy of Agricultural Sciences). PEDV genome was extracted from virus stock with TRIzol reagent, and was used as template to amplify the PEDV orf3 gene fragment using RT-PCR method according to our experimental methods (Zhang et al., 2018). Plasmid of the PEDV ORF3 fused with GFP tag was designed by cloning the open reading frame (ORF) of orf3 into the Xho I and Bam HI sites of the pEGFP-N1 vector. To construct plasmid expressing the PEDV ORF3 fused with Myc tag, fragment of orf3 was cloned into the EcoR I and Kpn I sites of pCMV-Myc vector. Amplicons containing Flag tag fused to ORF3 was inserted cloned into the Bam HI and Xho I sites of pcDNA3.0-Flag. These sequence-specific primers for amplification of orf3 gene and construction of recombinant plasmids were list in Table 1 .
Table 1.
Primer sequences used for RT-PCR amplification and construction of corresponding plasmids.
| Primer name | Orientation | Primer sequence (5’-3’) | Application |
|---|---|---|---|
| ORF3 | Forward | ATGTTTCTTGGACTTTTTCAATAC | RT-PCR |
| Reverse | TCATTCACTAATTGTAGCATACTC | RT-PCR | |
| GFP-ORF3 | Forward | CCCTCGAGATGTTTCTTGGACTTTTTCAATACA | cloning |
| Reverse | CGGGATCCCGTTCACTAATTGTAGCATACTCGTCT | cloning | |
| Myc-ORF3 | Forward | GCGGATCCATGTTTCTTGGACTTTTTCAATACA | cloning |
| Reverse | CGCTCGAGTCATTCACTAATTGTAGCATACTCG | cloning | |
| Flag-ORF3 | Forward | CGGAATTCGGATGTTTCTTGGACTTTTTCAATACA | cloning |
| Reverse | GGGGTACCTCATTCACTAATTGTAGCATACTCG | cloning |
The underlined letters indicate the restriction enzyme digestion sites.
For transient transfections, cells were transfected with plasmid DNA using Lipofectamine™2000 according to the manufacturer’s protocols. Briefly, cells were plated in 24-well culture dishes with complete medium and allowed to grow for 24 h to 80% confluency. A mixture of Opti-MEM medium (Gibco) and Lipofectamine™2000 was incubated for 5 min at room temperature and was then incubated with corresponding plasmids for 20 min at room temperature to allow complex formation. This mixture including plasmids was then added to each well. Four to six hours after transfection, the medium was changed with fresh medium, and analyses were performed at the indicated times after transfection (Zhang et al., 2015).
2.4. Western blot analysis
Western blot analysis was performed in according to our previous methods with minor modification (Zhang et al., 2014a,b). Any of Vero, 293 T, HeLa and PK15 cells were washed two times with PBS and disrupted with lysis buffer (50 mM Tris−HCl, pH 8.0, 150 mM NaCl and 1% Triton X-100, supplemented with 1 tablet of Complete-Mini Protease Inhibitor Cocktail (Dhand et al., 1994) per 50 ml buffer). Cell lysates were centrifugated at 12, 000 g for 10 min to harvest supernatants. Protein assays were performed on all supernatant using the Bradford method (Lau et al., 2008). Samples, each containing 25–30 μg of protein equivalent, were separated in 12% gradient SDS-PAGE and transferred to Hybond-P PVDF membranes (GE, USA). Membranes were blocked for 2 h with 5% non-fat dry milk solution in Tris-buffered saline containing 0.5% Tween-20 (TBST). The membranes were then blotted with the required specific primary antibodies, followed by incubation with the corresponding secondary antibodies conjugated with HRP. Blots were developed with the Western Lightning Chemiluminescence Kit (NC4109), following the manufacturer’s instruction (Pierce, USA).
2.5. Confocal microscopy analysis
Confocal microscopy analysis was performed in accordance with previous described method (Zhang et al., 2014a,b; Zhang et al., 2016). Briefly, any kind of cells were cultured to sub-confluency on 9-mm glass coverslips (Thomas Scientific) in 48-well plates and were transfected with each corresponding plasmid in figure legends. At the indicated time points, the cells were prepared for microscopy in the following steps: fixed with 4% paraformaldehyde for 15 min, quenched with 100 mM glycine for 15 min, permeabilized with 0.1% Triton X-100 for 15 min, blocked with 10% goat serum plus 1% BSA in PBS for 2 h, washed three times with PBS and either incubated with diluted primary monoclonal antibodies followed by secondary antibodies conjugated TRITC or neglection of the antibodies staining for the cells transfected with pEGFP-N1-ORF3 or pEGFP-N1. Then the cells were washed extensively with PBS before being incubated for 10 min in mounting medium with 0.1% DAPI to stain the nuclei. Cells were examined and images were captured using 100 × objectives with the confocal microscope (Leica SP8, German). The images were refined and figures were generated using Adobe Photoshop software (Adobe Systems, San Jose, CA).
2.6. Flow cytometry
The percentages of apoptotic cells were determined by flow cytofluorimetric analysis on the basis of our previous reported methods (Zhang et al., 2014a,b). In brief, 293 T cells were transfected with corresponding plasmids indicated in figure legend. After 6–12 h post transfection, aliquots of 2 × 105 cells were harvested, fixed with 70% cold ethanol at −20 °C for 2 h, and then incubated with 10 μL Annexin V-FITC (Molecular Probes) and 2 μL propidiumiodide (PI) stocks for 15 min at 37 °C. The Annexin V-FITC fluorescence and PI fluorescence were measured with BD FACSCalibur (Becton Dickinson). Two populations of cells were separated, one consisting of the living cells characterized by bright fluorescence labeling and the second being apoptotic cells with a characteristic distinct pattern of increased fluorescence intensity. A minimum of 30, 000 cells were analyzed for each sample. Data were analyzed with Cellquest software (Becton Dickinson).
3. Results
3.1. Construction of pEGFP-N1-ORF3 recombinant plasmid and confirmation of the GFP-ORF3 fusion protein expression
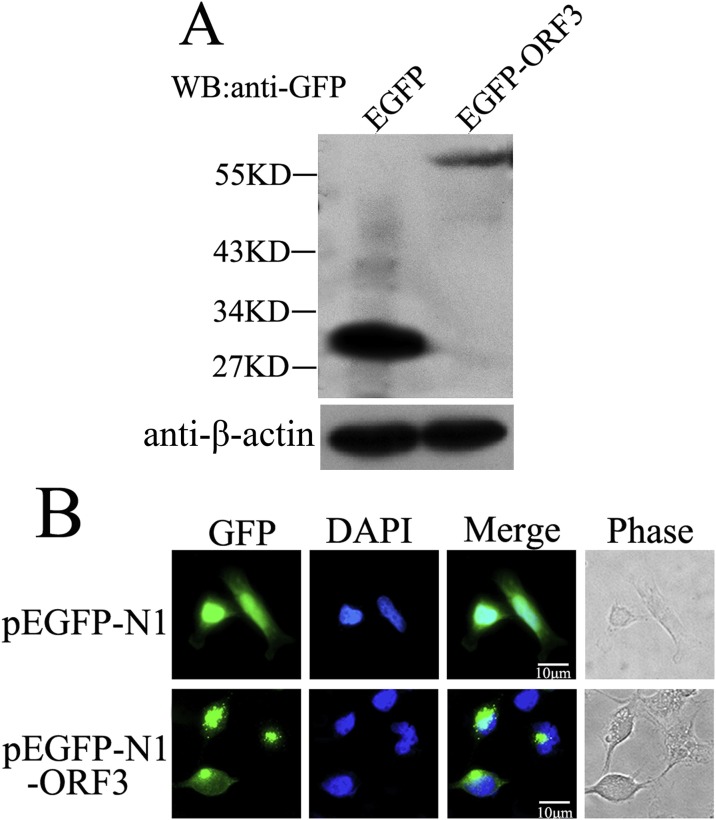
In order to readily visualize dynamic expression of ORF3 protein in cells, the orf3 gene was amplified from PEDV virus genome and then inserted into pEGFP-N1 vector to result into recombinant plasmid pEGFP-N1-ORF3. This plasmid was transiently transfected into HeLa cells, and the expression of GFP-ORF3 was detected in the lysates of transfected cells by Western blot probed with mouse anti-GFP monoclonal antibody. The β-actin was measured to serve as internal protein-loading control. The results showed that protein bands with molecular mass of approximately 27 kDa and 54 kDa were observed in the HeLa cells transfected with pEGFP-N1 and pEGFP-N1-ORF3, respectively (Fig. 1 A), which meet the agreement with the predicted size of fusion protein. According to the amount of β-actin, although the expression of fusion protein GFP-ORF3 was comparatively lower than control GFP, the amount of GFP-ORF3 was sufficiently for next experiments. In the other hand, the HeLa cells transfected with indicated plasmids were fixed and permeabilized, and the intracellular locations of GFP-ORF3 was detected by confocal microscopy. As showed in Fig. 1B, strong punctate accumulation of GFP fluorescence in the cytosol was observed in the cells expressing GFP-ORF3, and the fusion protein completely located in cytoplasm with the aggregation manner, while the control GFP distributed throughout whole pEGFP-N1-transfected cells with diffusing manner.
Fig. 1.
Detection of the GFP-ORF3 fusion protein expression. (A) HeLa cells were transiently transfected with either pEGFP-N1 or PEGFP-N1-ORF3 plasmids for 24 h. Cell lysates were harvested and subjected to Western Blot probed with the mouse monoclonal anti-GFP antibody followed by the goat-anti-mouse-HRP antibody. β-actin was detected to serve as internal loading control. (B) HeLa cells transfected with indicated plasmids for 24 h were fixed, permeabilized and then detected using confocal immunofluorescence. Nuclear material was stained with DAPI (blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.2. PEDV ORF3 protein expression characteristic
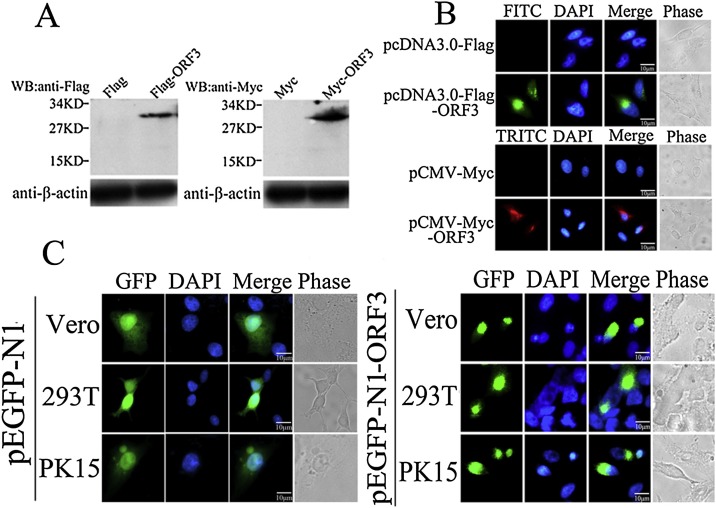
Although the PEDV ORF3 protein fused with GFP tag express in the cytoplasm correctly, we can’t confirm the protein aggregation is caused by ORF3 protein, due to the GFP tag has the same molecular mass size with ORF3 protein. To test whether the GFP tag affects the ORF3 expression and subsequent aggregation, two other smaller tags Flag and Myc were fused with ORF3 protein, and then their expression manner in HeLa cells were investigated. The results of Western Blot probed with either mouse anti-Flag or -Myc monoclonal antibodies showed that both Flag-ORF3 and Myc-ORF3 fusion proteins migrated to an expected molecular mass sizes at 27 kDa in cells transfected with indicated recombinant plasmids (Fig. 2 A). After fixed and permeabilized, the transfected HeLa cells were stained with mouse anti-Flag or -Myc monoclonal antibodies and FITC- or TRITC-labeled goat-anti-mouse IgG, and the aggregation of fusion ORF3 was detected by confocal microscopy. The results showed that both ORF3 proteins fused with either Flag or Myc tags were concentrated in the nucleate of cytoplasm similar with the GFP-ORF3 protein (Fig. 2B), indicating that expression tag has no effects on the aggregation of ORF3 protein. To exclude the possibility of ORF3 aggregation related with cell types, the GFP-ORF3 was expressed in different cell lines (Vero, 293 T and PK15) and examined using a GFP filter of confocal immunofluorescence. The results showed that compared with the controls, each ORF3 proteins concentrated and aggregated to form nucleate in all of cytoplasms, but the control GFP distributed diffusely in whole cell (Fig. 2C). These above data suggest that PEDV ORF3 protein expression characteristic of aggregation has nothing to do with the fusion tags no matter what cell lines used for its expression.
Fig. 2.
Determination of PEDV ORF3 protein expression characteristic. (A) HeLa cells were transiently transfected with indicated plasmids for 24 h, respectively. Cell lysates were harvested and subjected to Western Blot probed with either mouse monoclonal anti-Flag (left panel) or anti-Myc (right panel) antibodies followed by the goat-anti-mouse-HRP antibody. β-actin was detected to serve as internal loading control. (B) HeLa cells were transiently transfected with indicated plasmids for 24 h, respectively. Cells were fixed, permeabilized and then probed with mouse monoclonal anti-Flag (upper panel) or anti-Myc (lower panel) antibodies, followed by FITC-labeled goat-anti-mouse (green) or TRITC-labeled goat-anti-mouse (red) antibodies with confocal microscope analysis. Nuclei were identified by DAPI staining. (C) Three types of cells, including Vero, 293 T and PK15, were either transfected with pEGFP-N1 or pEGFP-N1-ORF3 for 24 h, respectively. Cells were fixed, permeabilized, and directly examined using a GFP filter of confocal immunofluorescence. Nuclei were identified by DAPI staining. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.3. PEDV ORF3 is a transmembrane protein that localizes at ER
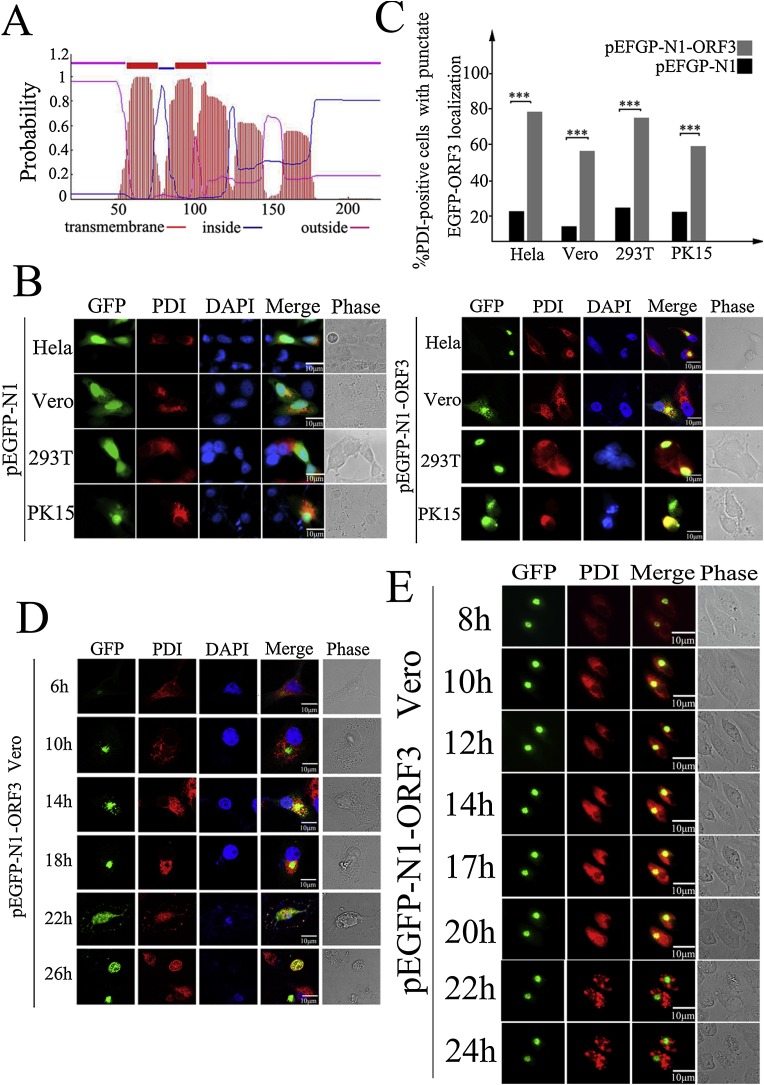
Different from GFP protein that is distributed throughout the nucleus and cytoplasm, GFP-ORF3 fusion protein only localizes as aggregate in the cytoplasm, but its exact subcellular localization site is not determined. Thus we next performed the bioinformatic analysis to determine whether the ORF3 protein is present in specific cytosolic sites. PEDV orf3 gene encodes an ion channel protein and regulates virus production (Wang et al., 2012), which promote us to predict its transmembrane domain using the software TMHMM program (Sonnhammer et al., 1998). The results revealed that ORF3 is secondary transmembrane protein containing four putative TMDs (Fig. 3 A). Subcellular localization of ORF3 was also predicted in silico by WoLFPSORT program (https://www.genscript.com/wolf-psort.html), and results revealed that plas: 27, mito: 3, nucl: 1, cyto: 1, indicating that the protein is likely to be associated with membranes system. To address the possibility of ORF3 localization, different cell lines (HeLa, Vero, 293 T and PK15) expressing either control GFP or GFP-ORF3 were stained with rabbit anti-PDI monoclonal antibody and TRITC-labeled anti-rabbit IgG. confocal immunofluorescence. Colocalization analysis results showed that control GFP distributed diffusely in whole cells and no colocalization with ER was observed, but the colocalization of ORF3 with the ER-associated protein PDI in various cell lines with no statistical difference was confirmed (Fig. 3B). Furthermore, we performed quantification analysis through counting the percentage of PDI-positive cells with punctate of GFP or GFP-ORF3, and the results indicated that more than 50%–80% of GFP-ORF3 expressing cells exhibited colocalization between ORF3 and PDI, which was significantly higher than that in GFP control cells with lower 20% punctate (Fig. 3C). In contrast to normal ER distributing at perinuclear boundary uniformly, ER aggregated to form punctate bodies or pattern in cells expressing the GFP-ORF3, indicating that ORF3 associates with ER presumably affecting the cell survival. Then we performed confocal immunofluorescence staining of ER-associated protein PDI in Vero cells transiently expressing GFP-ORF3 to analyze the effects of ORF3-ER interaction on the cells growth. The results showed that ER distributed at perinuclear boundary uniformly before 6 h post pEGFP-ORF3 transfection (Fig. 3D). As the protein expression amount increased at 10 to 14 h, ORF3 recruited the ER shifting to one side of cytoplasm and forming the punctate bodies to colocalize with each other. Intriguingly, when we further performed the immunofluorescence analysis of their colocalization, whereas, we observed that ORF3 protein aggregated to a granules and was packaged or wrapped by ER at 18 h. After 22 h, the aggregated ORF3 became to diffusion manner, and ER was broken leading to the cell death eventually. To test the observation more directly, we examined a time course of colocalization of ORF3 and PDI by live cell imaging of GFP-ORF3-expressing Vero cells. We found that as the transfection times prolonging, expressed ORF3 aggregated to form a granule and recruited the ER to interplay each other, lastly resulting into cell disruption at 22 h post transfection, which supported the similar results with the above observation (Fig. 3E). These data confirm that PEDV ORF3 is a transmembrane protein that localizes at ER and associates with each other to affect the cell fate.
Fig. 3.
PEDV ORF3 is a predicted transmembrane protein that localizes in and associates with ER to affect cell fate. (A) Schematic representation of the predicted transmembrane topology (TMHMM (transmembrane helices in proteins) Server version. 2.0) of PEDV ORF3 protein. (B) Four types of cells were either transfected with pEGFP-N1 or pEGFP-N1-ORF3 for 12 h, respectively. Cells were fixed, permeabilized and then probed with rabbit monoclonal anti-PDI antibody, followed by TRITC-labeled goat-anti-rabbit antibody (red) for confocal immunofluorescence detecting the colocalization of PEDV3 and ER. Nuclei were stained with DAPI. (C) Quantitative analysis of colocalization of either GFP or ORF3 with ER in different cells from (B). The percentages of the cells with colocalization were determined by counting more than 100 cells for each experiment. The results of three independent experiments were shown as mean ± standard deviation (SD). * P < 0.05; ** P < 0.01; *** P < 0.001. (D) Vero cells were transfected with pEGFP-N1-ORF3 for 6, 10, 14, 18, 22, and 24 h, respectively. At the indicated time points, ORF3-expressing cells were stained with rabbit monoclonal anti-PDI antibody, followed by TRITC-labeled goat-anti-rabbit antibody. Colocalization between PEDV ORF3 and ER, and cell fate were analyzed by confocal microscope. Nuclei were stained with DAPI (blue). (E) Vero cells were transfected with pEGFP-N1-ORF3, then the colocalization and dynamic change of interplay between ORF3 and ER along with time-lapse was determined by live-cell laser confocal imaging system. Elapsed time is indicated at h post transfection. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.4. PEDV ORF3 protein causes ER stress response via up-regulation of GRP78 and activation of PERK-eIF2α signaling pathway
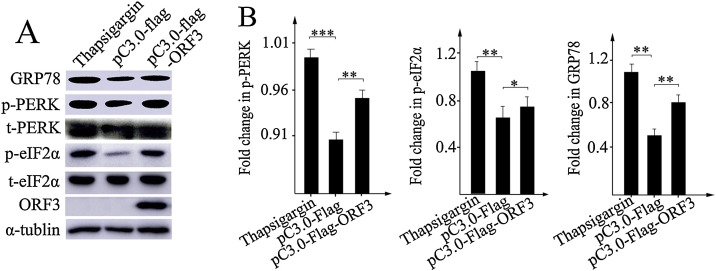
During many Coronaviridae replication, substantial amounts of viral transmembrane glycoproteins are produced to induce ER stress response (Minakshi et al., 2009; Xu et al., 2013a,b). Since the PEDV ORF3 protein is also a transmembrane protein that localizes to the ER, we speculate that it has potential to induce ER stress response. In order to examine the role of ORF3 as a potential ER stress inducer, we transfected 293 T cells with either pcDNA3.0-Flag-ORF3 or pcDNA3.0-Flag, and set the TG-treated cells as positive control. Whole-cell lysates prepared from these harvested cells were analyzed by Western blot using the indicated antibodies in figure. The results showed that either expression of ORF3 or Tg treatment caused significant elevation of the amount of GRP78 (Fig. 4 A), which encodes crucial ER chaperones that are markers of ER stress, indicating that PEDV ORF3 protein exactly induces the ER stress response. As expected, the ORF3 also gave a similar increase of phosphorylated form of p-PERK and p-eIF2α, while the levels of total PERK and eIF2α remained unchanged. Densitometry scans conducted using ImageJ software showed that ORF3 significantly induced more than 1–2 folds of GRP78, p-PERK and p-eIF2α expressions (Fig. 4B), when compared with pcDNA3.0-Flag-transfected control. These results suggest that ORF3 is responsible for ER stress response through activation PERK-eIF2α signaling pathway.
Fig. 4.
PEDV ORF3 up-regulates the level of GRP78 expression and induces the phosphorylation of PERK and eIF2α. (A) The 293 T cells were either transfected with indicated plasmids or treated with Tg used as for positive control. At the indicated time point, cell lysates were harvested and subjected to Western Blot analysis using the corresponding primary antibodies and second antibodies conjugated with HRP. Cellular α-tubulin was employed as an internal loading control protein. (B) Densitometry scans were conducted using ImageJ software (NIH). Densitometry of the GRP78, phospho-PERK and phospho-eIF2α bands normalized to controls are presented as fold change ± SEM compared with the controls. * P < 0.05; ** P < 0.01; *** P < 0.001.
3.5. PEDV ORF3 protein has no effect on apoptosis
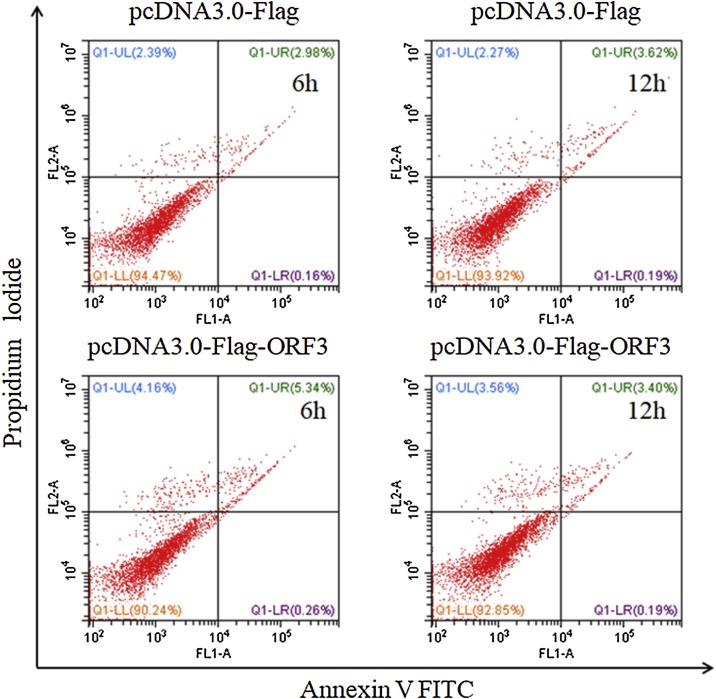
Prolonged ER stress is known to activate related signaling to trigger ER stress-dependent apoptotic cell death (Tabas and Ron, 2011). It has also been demonstrated that Coronavirus proteins induce apoptosis in vitro and in vivo (An et al., 1999; DeDiego et al., 2011). Our results so far suggest that ORF3 might induce the apoptosis via triggering the ER stress. To test the possibility, we transfected 293 T cells with pcNDA3.0-Flag-ORF3 and pcDNA3.0-Flag plasmids for the indicated times, respectively. After cells were fixed and stained by Annexin V-FITC and PI, we performed flow cytometry analysis to determine whether ORF3 expression affect the apoptosis. The results showed no statistical difference of FITC fluorescence intensity in cells expressed ORF3 proteins and control cells (Fig. 5 ). Even though the transfection time prolonged to 12 h, we couldn’t detect the increased fluorescence signal in all of cells. The percentage of apoptotic cells remained stable during the whole process of ORF3 expression indicating PEDV ORF3 can’t influence the apoptosis.
Fig. 5.
PEDV ORF3 has no effect on the apoptosis. The 293 T cells were transfected with either pcDNA3.0-Flag or pcDNA3.0-Flag-ORF3 for 6 h and 12 h. Cells were harvested, fixed with 70% cold ethanol and stained with Annexin V-FITC and PI, followed by examination with flow cytometry analysis. The percentages of apoptotic cells corresponding to the fluorescenceintensity for each of the sample are indicated.
3.6. PEDV ORF3 protein-induced autophagy is dependent on ER stress response
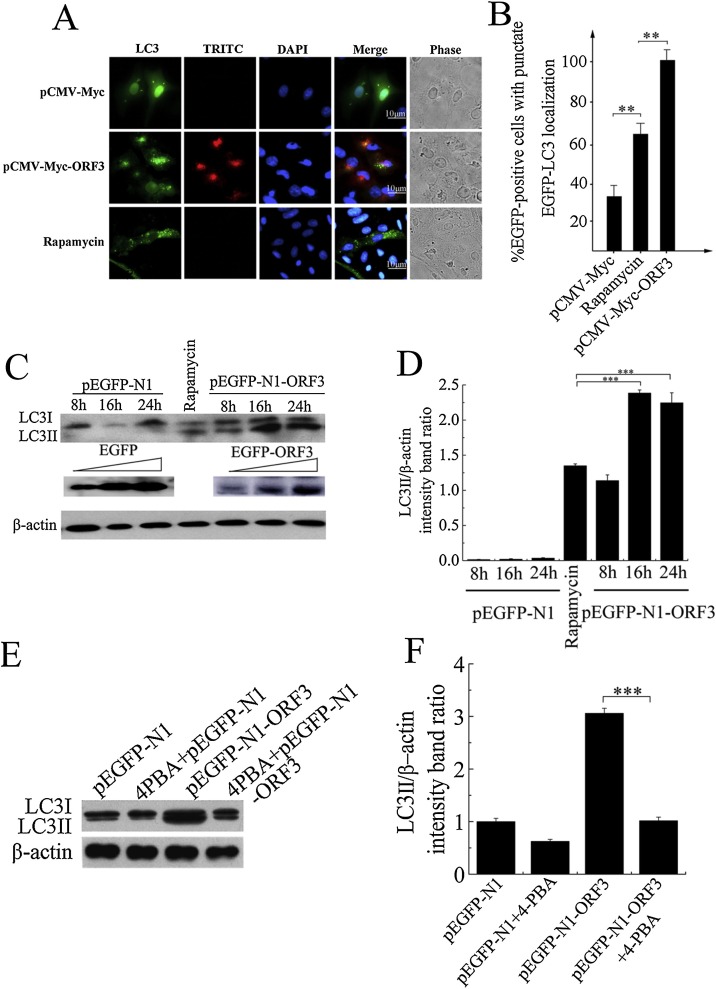
ER stress response is closely associated with a number of major signaling pathways, such as apoptosis and autophagy (Ogata et al., 2006). Prolonged ER stress can trigger apoptosis, but unfolded protein response (UPR) can also facilitate the resolution of ER stress and induce the autophagy (Wei et al., 2008). Coronavirus proteins can generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate to trigger autophagy (Cottam et al., 2011). Now that PEDV ORF3 protein can’t affect the apoptosis, we next investigate the effect of ORF3 on autophagy induction. Vero cells were transfected with pEGFP-LC3 to expression autophagy marker LC3, and then cotransfected with either pCMV-Myc-ORF3 or pCMV-Myc plasmids. Cells treated with rapamycin, an autophagy inducer of mammalian cells, were set as the positive control. The confocal microscopy analysis showed that LC3 generated diffuse signals indicating even distribution throughout in control cells, but the LC3 fluorescence redistributed to punctate vesicles indicative of autophagosome in ORF3-expressing cells (Fig. 6 A). Rapamycin also caused redistribution of GFP-LC3 to punctate structures in cytoplasm indicative of autophagosomes similar with cells expressed ORF3 protein. Quantitative analysis of the cells containing the punctate vesicles showed that cells expressing ORF3 resulted into more than 80% of autophagy, which was significantly higher than both rapamycin and negative control (Fig. 6B). To further confirm the role of ORF3 protein in autophagy formation, we transfected Vero cells with either pEGFP-N1 or pEGFP-N1-ORF3, and examined the autophagy induction by analyzing conversion from LC3-I to LC3-II using Western Blot. After 16 h post transfection, we observed a significant increase of LC3-II protein in ORF3-transfected cells, and the ratio of LC3-II to LC3-I was much higher in ORF3-transfected cells when compared to the control (Fig. 6C). Densitometry scans were conducted to represent the fold change of LC3-II expression, and the results showed the expression level of LC3-II in ORF3-transfected cells increased 1–2.5 folds (Fig. 6D). In contrast, conversion of LC3-I/LC3-II in 293 T cells could be inhibited by 4-PBA, an ER stress inhibitor, indicating ER stress response contributes to ORF3-induced autophagy (Fig. 6E). In addition, the results of densitometry analysis showed 3 folds increasing of LC3-II expression in ORF3-expressed cells, but the 4-PBA counteracted the amount of LC3-II to normal expression level (Fig. 6F). These results suggest PEDV ORF3 plays an important role in autophagy induction, which is dependent on ER stress response.
Fig. 6.
PEDV ORF3 protein induces the autophagy that is dependent on ER stress response. (A) Vero cells were transfected with pEGFP-LC3, and then either transfected with pCMV-Myc-ORF3 or treated with rapamycin used for positive control, as well as pCMV-Myc-transfected cells used for negative control. At 12 h post transfection, cells were fixed, permeabilized and then probed with mouse monoclonal anti-Myc antibody, followed by TRITC-labeled goat-anti-mouse antibody (red). Nuclei were stained with DAPI (blue), and the GFP-LC3 punctate structures were visualized under confocal microscope. (B) Average numbers of LC3-positive vesicles per cell were quantified from three biological replicates. * P < 0.05; ** P < 0.01; *** P < 0.001. (C) Vero cells were transfected with pEGFP-LC3, and then either transfected with pEGFP-N1 or pEGFP-N1-ORF3 for 8, 16 and 24 h, and cell lysates were harvested and subjected to Western Blot probed with the mouse monoclonal anti-LC3 and anti-GFP antibodies, followed by the goat-anti-mouse-HRP antibody. Cellular β-actin was detected as the control protein. (D) Densitometry scans of the LC3-II brand of Western blot from (C). * P < 0.05; ** P < 0.01; *** P < 0.001. (E) The 293T cells were either treated with 4-PBA or not for 4 h, then transfected with pEGFP-N1-ORF3 or pEGFP-N1 for 24 h. Western Blot was performed for testing the endogenous LC3-I/LC3-II conversion with the mouse monoclonal anti-LC3 antibody followed by the goat-anti-mouse-HRP antibody. Cellular β-actin was employed as an internal loading control protein. (F) Densitometry scans of the LC3-II brand of Western blot from (E). * P < 0.05; ** P < 0.01; *** P < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
4. Discussion
As we all know, many reports focus on the differentiation of the orf3 gene used for genetic diversity and molecular epidemiology studies of PEDV (Park et al., 2008). Recently, the accessory orf3 gene has received a large amount of attention in the context of PEDV virulence which can be reduced by altering the orf3 gene (Park et al., 2007, 2012). However up to date, little is known about the function of orf3 gene compared to other Coronaviruses. Therefore, it is need to investigate the novel function of PEDV ORF3 for elucidating the mechanism by which this protein regulates PEDV life cycle. In this study, we constructed three different eukaryotic expression vectors expressing the PEDV ORF3 protein fused with the EGFP, Myc and Flag tags, respectively, which were used for analysis of its subcellular localization and biological function. Our results showed that PEDV ORF3 as transmembrane protein located at perinuclear area forming the punctate bodies, and plenty of PEDV ORF3 protein colocalized with ER to causes ER stress response via up-regulation of GRP78 and activation of PERK-eIF2α signaling pathway. PEDV ORF3 protein induced the autophagy through promoting conversion of LC3-I to LC3-II, but had no effect on apoptosis. Whereas, conversion from LC3-I to LC3-II could be notably inhibited by 4-PBA, an ER stress inhibitor, indicating that ER stress response contributes to ORF3-induced autophagy and ORF3 protein plays multiple roles in PEDV life cycle.
Similar with some other accessory proteins of Coronaviruses, such as severe acute respiratory syndrome-associated coronavirus (SARS-CoV) 3a protein (Chan et al., 2009), and human pathogenic coronavirus NL63 (hCoV-NL63) ORF3 protein (Muller et al., 2010), the PEDV ORF3 protein has been also testified to encode an ion channel protein that regulates virus production, assembly and release. The ion-channel function is dependent on TMDs, because the ion channel activity is reduced in ORF3 protein of attenuated typical PEDV while its amino acid substitution or deletion (Wang et al., 2012). Besides, it is reported that the ORF3 gene is removed and replaced with a red fluorescent protein (RFP) gene to generate icPEDV-ORF3-RFP molecular clone, which replicates efficiently in vitro and in vivo, is efficiently transmitted among pigs, and produces lethal disease outcomes (Beall et al., 2016). In our study, we firstly constructed different eukaryotic expression vectors to expression ORF3 protein, and found ORF3 protein distributed in perinuclear cytoplasm, and then shifted for aggregation and concentrated in the nucleate of cytoplasm to form punctate patter. This distribution manner of PEDV ORF3 is similar with one study that ORF3 proteins localizes in the cytoplasm in punctate manner (Ye et al., 2015), but distinguishes from the other report of uniform distribution in cytoplasm diffusely (Wongthida et al., 2017; Kaewborisuth et al., 2018). We speculate that the localization and distribution manner of PEDV ORF3 might be dependent on the virus strain-type-specific differences.
Furthermore, we found that PEDV ORF3 is secondary transmembrane protein containing four putative TMDs through prediction analysis, which is in accordance with previous report that proposed a structure model of the ORF3 protein consisting of four TM domains and forming a tetrameric assembly (Wang et al., 2012). PEDV ORF3 protein expression characteristic of aggregation has nothing to do with the fusion tags no matter what cell lines used for its expression, indicating that ORF3 might induce accumulation of misfolded or unfolded proteins within the ER. At the same time, we performed subcellular localization analysis of PEDV ORF3 and revealed the protein likely to locate in the endoplasmic reticulum, which was confirmed by next confocal immunofluorescence analysis that showed the colocalization of ORF3 with the ER. It is noteworthy that as the amount of ORF3 expression increased, ORF3 dissociated from ER to come into being diffusion manner, and ER was broken leading to the cell death eventually, suggesting that ORF3 localizes at ER and associates with each other to affect the cell survival or death, which might be dependent on ER stress response induced by the accumulation of misfolded or unfolded proteins within the ER to initiates a number of cellular responses to restore ER homeostasis. Whereas, whether the PEDV ORF3 TMDs induce ER stress response and subsequently cause the protein aggregation in perinuclear boundary area remain to be further elucidated.
The genomes of Coronaviridae are well known to code for a reasonable amount of ion channels, referred as viroporins, which usually contain TMDs and play important roles in pathogenesis, virus assembly and release. For example, E proteins of MHV (Madan et al., 2005), SARS-CoV (Liao et al., 2006), HCoV-229E (Wilson et al., 2004) and IBV (Ye and Hogue, 2007) possess ion channel activity. PEDV E protein has also been found to localize in the ER and the nucleus and it can cause ER stress response (Xu et al., 2013a,b), suggesting other PEDV proteins containing TMDs, such as ORF3, have the similar potential to cause ER stress. In this study, our results showed that PEDV ORF3 protein was able to induce ER stress response, as indicated by the significant up-regulation of the molecule chaperone GRP78, a typical marker of ER stress. ER stress can activate multiple cell-signaling pathways, such as UPR, to regulate gene expression at both transcriptional and translational levels, and to increase the capacity of the ER machinery to fold proteins correctly and activate the degradation of aberrant proteins (Grootjans et al., 2016). We found that as one of three major arms of UPR, PERK pathway was activated by PEDV ORF3, resulting in the elevated phosphorylation of eIF2α, suggesting that ORF3 causes ER stress response to subsequently inhibit translation and protein input into the ER.
It is well known that UPR can adjust the biosynthetic burden and capacity of the ER to maintain homeostasis. While the damage to the ER is severe or persistent, the ER stress triggers apoptosis (Jheng et al., 2014). Apoptosis, cell cycle change and the association of each other are common cellular responses to many Coronaviridae infections. Previous study reported that ORF3 prolong the S phase, which may associate with the virus replication (Ye et al., 2015). It has also been demonstrated that Coronavirus infection induces apoptosis and arrests cell cycle (Chen et al., 2004), which is caspase-dependent but p53-independent manner (Li et al., 2007). Nevertheless, PEDV ORF3 was found to have no influence on apoptosis in our study. Coronavirus infection triggers autophagy, and the autophagosome membranes have been shown to be derived from the ER (Cottam et al., 2011). The decision between apoptosis and autophagy in cells under ER stress may be intricately regulated by multiple mechanisms (Castillo et al., 2011). These events promote us to investigate the other signaling pathways associated with ER stress, for instance autophagy induced by PEDV ORF3 protein. Our observations indicated that autophagy marker LC3 redistributed to punctate vesicles in cytoplasm indicative of autophagosome in ORF3-expressing cells. We also observed the conversion from LC3-I to LC3-II, because the ratio of LC3-II to LC3-I was much higher in ORF3-transfected cells, indicating PEDV ORF3 plays an important role in autophagy induction via increasing conversion of LC3-I to LC3-II. Moreover, when 4-PBA, an ER stress inhibitor, was used to block the ER stress response, the result of conversion from LC3-I to LC3-II could be significantly inhibited, suggesting that PEDV ORF3 protein-induced autophagy is dependent on ER stress response.
In conclusion, PEDV ORF3 is found to be a transmembrane protein, which can localize in ER to trigger ER stress through increasing expression level of GRP78 and activating PERK-eIF2α signaling pathway. In addition, our results showed that PEDV ORF3 can induce the autophagy through facilitating conversion of LC3-I to LC3-II, but it can’t influence the apoptosis. However, conversion of LC3-I to LC3-II can be significantly inhibited by 4-PBA, an ER stress inhibitor, indicating that ORF3-induced autophagy is dependent on ER stress response. Therefore, our study not only provides some new findings for the biological function of the PEDV ORF3 protein, but also accelerates the understanding the molecular interaction between PEDV ORF3 protein and cells.
Declaration of Competing Interest
There is no conflict of interest among the contributors of this paper.
Acknowledgements
This work is supported by State Key Laboratory of Veterinary Biotechnology Foundation, Harbin Veterinary Research Institute of Chinese Academy of Agricultural Sciences (SKLVBF201913), National Natural Science Foundation of China (NSFC, Grant No. 31570159 and 31200121), Program for Young Scholars with Creative Talents in HeiLongJiang BaYi Agricultural University (CXRC2016-12), Doctor’s Research Foundation, HeiLongJiang BaYi Agricultural University (XDB2015-16 and XDB2015-18), Postdoctoral Scientific Research Start-up Foundation of HeiLongJiang Province (LBH-Q17135), Joint Guidance Project of Natural Science Foundation of HeiLongJiang Province (LH2019C046), Key Project of Cultivation Foundation, HeiLongJiang BaYi Agricultural University (XA2017-02), and Earmarked Fund for China Agriculture Research System (No. CARS35). We are grateful to Prof. Songdong Meng (Professor of Institute of Microbiology, Chinese Academy of Sciences) and Prof. Paul Chu (Guest Professor of Institute of Microbiology, Chinese Academy of Sciences) for critical reading of the manuscript. We also thank Dr. Tingting Li, Dr. Li Huang and Prof. Changjiang Weng (Harbin Veterinary Research Institute of Chinese Academy of Agricultural Sciences) for assistance with confocal microscope experiment.
Contributor Information
Hua Zhang, Email: byndhzh@163.com.
Hongwei Cao, Email: byndcaohongwei@163.com.
References
- An S., Chen C.J., Yu X., Leibowitz J.L., Makino S. Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J. Virol. 1999;73:7853–7859. doi: 10.1128/jvi.73.9.7853-7859.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall A., Yount B., Lin C.M., Hou Y., Wang Q., Saif L., Baric R. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. MBio. 2016;7:e01451–15. doi: 10.1128/mBio.01451-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgen A., Duarte M., Tobler K., Laude H., Ackermann M. Sequence determination of the nucleocapsid protein gene of the porcine epidemic diarrhoea virus confirms that this virus is a coronavirus related to human coronavirus 229E and porcine transmissible gastroenteritis virus. J. Gen. Virol. 1993;74(Pt 9):1795–1804. doi: 10.1099/0022-1317-74-9-1795. [DOI] [PubMed] [Google Scholar]
- Bridgen A., Kocherhans R., Tobler K., Carvajal A., Ackermann M. Further analysis of the genome of porcine epidemic diarrhoea virus. Adv. Exp. Med. Biol. 1998;440:781–786. doi: 10.1007/978-1-4615-5331-1_101. [DOI] [PubMed] [Google Scholar]
- Castillo K., Rojas-Rivera D., Lisbona F., Caballero B., Nassif M., Court F.A., Schuck S., Ibar C., Walter P., Sierralta J. BAX inhibitor-1 regulates autophagy by controlling the IRE1α branch of the unfolded protein response. EMBO J. 2011;30:4465–4478. doi: 10.1038/emboj.2011.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.M., Tsoi H., Chan W.M., Zhai S., Wong C.O., Yao X., Chan W.Y., Tsui S.K., Chan H.Y. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009;41:2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Sugiyama K., Kubo H., Huang C., Makino S. Murine coronavirus nonstructural protein p28 arrests cell cycle in G0/G1 phase. J. Virol. 2004;78:10410–10419. doi: 10.1128/JVI.78.19.10410-10419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang C., Shi H., Qiu H., Liu S., Chen X., Zhang Z., Feng L. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010;155:1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheun-Arom T., Temeeyasen G., Tripipat T., Kaewprommal P., Piriyapongsa J., Sukrong S., Chongcharoen W., Tantituvanont A., Nilubol D. Full-length genome analysis of two genetically distinct variants of porcine epidemic diarrhea virus in Thailand. Infect. Genet. Evol. 2016;44:114–121. doi: 10.1016/j.meegid.2016.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouck P., Pensaert M. Experimental infection of pigs with Belgian isolates of the porcine rotavirus. Zentralblatt Veterinarmedizin Reihe B. 1979;26:517–526. doi: 10.1111/j.1439-0450.1979.tb00845.x. [DOI] [PubMed] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Jiménez-Guarde˜no J.M., Regla-Nava J.A., Álvarez E., Oliveros J.C., Zhao J., Fett C., Perlman S., Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhand R., Hiles I., Panayotou G., Roche S., Fry M.J., Gout I., Totty N.F., Truong O., Vicendo P., Yonezawa K., Kasuga M., Courtneidge S.A., Waterfield M.D. PI3-kinase is a dual specificity enzyme: autoregulation by an intrinsic protein-serine kinase activity. EMBO J. 1994;13:522–533. doi: 10.1002/j.1460-2075.1994.tb06290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootjans J., Kaser A., Kaufman R.J., Blumberg R.S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016;16:469–484. doi: 10.1038/nri.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. 2013;4:e00737–13. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jheng J.R., Ho J.Y., Horng J.T. ER stress, autophagy, and RNA viruses. Front. Microbiol. 2014;5:388–390. doi: 10.3389/fmicb.2014.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewborisuth C., He Q.G., Jongkaewwattana A. The accessory protein ORF3 contributes to porcine epidemic diarrhea virus replication by direct binding to the spike protein. Viruses. 2018;10:399. doi: 10.3390/v10080399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuchel M., Ackermann M., Muller H.K., Kihm U. An ELISA for detection of antibodies against porcine epidemic diarrhoea virus (PEDV) based on the specific solubility of the viral surface glycoprotein. Vet. Microbiol. 1992;32:117–134. doi: 10.1016/0378-1135(92)90100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusanagi K., Kuwahara H., Katoh T., Nunoya T., Ishikawa Y., Samejima T., Tajima M. Isolation and serial propagation of porcine epidemic diarrhea virus in cell cultures and partial characterization of the isolate. J. Vet. Med. Sci. 1992;54:313–318. doi: 10.1292/jvms.54.313. [DOI] [PubMed] [Google Scholar]
- Lau C., Wang X., Song L., North M., Wiehier S., Proud D., Chow C.W. Syk associates with clathrin and mediates phosphatidylinositol 3-kinase activation during human rhinovirus internalization. J. Immunol. 2008;180:870–880. doi: 10.4049/jimmunol.180.2.870. [DOI] [PubMed] [Google Scholar]
- Lee C.H. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.Q., Tam J.P., Liu D.X. Cell cycle arrest and apoptosis induced by thecoronavirus infectious bronchitis virus in the absence of p53. Virology. 2007;365:435–445. doi: 10.1016/j.virol.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Yuan Q., Torres J., Tam J.P., Liu D.X. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology. 2006;349:264–275. doi: 10.1016/j.virol.2006.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V., Garcia Mde J., Sanz M.A., Carrasco L. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 2005;579:3607–3612. doi: 10.1016/j.febslet.2005.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4:e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M.A., van der Hoek L., Voss D., Bader O., Lehmann D., Schulz A.R., Kallies S., Suliman T., Niedrig M. Human coronavirus NL63 open reading frame 3 encodes a virion-incorporated N-glycosylated membrane protein. Virol. J. 2010;7:6. doi: 10.1186/1743-422X-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., Shiosaka S., Hammarback J.A., Urano F., Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Kim H.K., Song D.S., An D.J., Park B.K. Complete genome sequences of a Korean virulent porcine epidemic diarrhea virus and its attenuated counterpart. J. Virol. 2012;86:5964. doi: 10.1128/JVI.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Moon H.J., Luo Y.Z., Kim H.K., Kim E.M., Yang J.S., Song D.S., Kang B.K., Lee C.S., Park B.K. Cloning and further sequence analysis of the ORF3 gene of wild-and attenuated-type porcine epidemic diarrhea viruses. Virus Genes. 2008;36:95–104. doi: 10.1007/s11262-007-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Song D.S., Ha G.W., Park B.K. Cloning and further sequence analysis of the spike gene of attenuated porcine epidemic diarrhea virus DR13. Virus Genes. 2007;35:55–64. doi: 10.1007/s11262-006-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Oroku K., Ohshima Y., Furuya Y., Sasakawa C. Efficacy of genogroup 1 based porcine epidemic diarrhea live vaccine against genogroup 2 field strain in Japan. Virol. J. 2018;15:28. doi: 10.1186/s12985-018-0940-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Su Y., Liu Y., Chen Y., Zhao B., Ji P., Xing G., Jiang D., Liu C., Song Y., Wang G., Li D., Deng R., Zhang G. Detection and phylogenetic analysis of porcine epidemic diarrhea virus in central China based on the ORF3 gene and the S1 gene. Virol. J. 2016;13:192. doi: 10.1186/s12985-016-0646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R.Q., Cai R.J., Chen Y.Q., Liang P.S., Chen D.K., Song C.X. Outbreak of porcine epidemic diarrhea in suckling piglets. China. Emerg. Infect. Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I., Ron D. Integrating the mechanisms of apoptosis induced by endo-plasmic reticulum stress. Nat. Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova A.N., Marthaler D., Wang Q., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Lu W., Chen J., Xie S., Shi H., Hsu H., Yu W., Xu K., Bian C., Fischer W.B., Schwarz W., Feng L., Sun B. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586:384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Sinha S., Levine B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy. 2008;4:949–951. doi: 10.4161/auto.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., McKinlay C., Gage P., Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330:322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongthida P., Liwnaree B., Wanasen N., Narkpuk J., Jongkaewwattana A. The role of ORF3 accessory protein in replication of cell-adapted porcine epidemic diarrhea virus (PEDV) Arch. Virol. 2017;162:2553–2563. doi: 10.1007/s00705-017-3390-5. [DOI] [PubMed] [Google Scholar]
- Xu X.G., Zhang H.L., Zhang Q., Huang Y., Dong J., Liang Y.B., Liu H.J., Tong D.W. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.J., Zhang H.L., Zhang Q., Dong J., Liang Y.B., Huang Y., Liu H.J., Tong D.W. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013;10:26. doi: 10.1186/1743-422X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S., Li Z., Chen F., Li W., Guo X., Hu H., He Q. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Genes. 2015;51:385–392. doi: 10.1007/s11262-015-1257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Hogue B.G. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J. Virol. 2007;81:3597–3607. doi: 10.1128/JVI.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Chai X., Cheng Y., Xing G., Liao A., Du L., Wang Y., Lei J., Gu J., Zhou J. Molecular characteristics of the spike gene of porcine epidemic diarrhoea virus strains in Eastern China in 2016. Virus Res. 2018;247:47–54. doi: 10.1016/j.virusres.2018.01.013. [DOI] [PubMed] [Google Scholar]
- Zhang H., Chen N., Li P.F., Pan Z.Y., Ding Y., Zou D.H., Li L.Y., Xiao L.J., Shen B.L., Liu S.X., Cao H.W., Cui Y.D. The nuclear protein Sam68 is recruited to the cytoplasmic stress granules during enterovirus 71 infection. Microb. Pathogenesis. 2016;96:58–66. doi: 10.1016/j.micpath.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Zhang H., Li F.Q., Pan Z.Y., Wu Z.J., Wang Y.H., Cui Y.D. Activation of PI3K/Akt pathway limits JNK-mediated apoptosis during EV71 infection. Virus Res. 2014;192:74–84. doi: 10.1016/j.virusres.2014.07.026. [DOI] [PubMed] [Google Scholar]
- Zhang H., Song L., Cong H.L., Tien P. Nuclear protein Sam68 interacts with the enterovirus 71 internal ribosome entry site and positively regulates viral protein translation. J. Virol. 2015;89:10031–10043. doi: 10.1128/JVI.01677-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.J., Mao H.Y., Yan J.Y., Zhang L., Sun Y., Wang X.Y., Chen Y., Lu Y.Y., Chen E.F., Lv H.K., Gong L.M., Li Z., Gao J., Xu C.P., Feng Y., Ge Q., Xu B.X., Xu F., Yang Z.N., Zhao G.Q., Han J.K., Guus K., Li H., Shu Y.L., Chen Z.P., Xia S.C. Isolation and characterization of H7N9 avian influenza A virus from humans with respiratory diseases in Zhejiang, China. Virus Res. 2014;189:158–164. doi: 10.1016/j.virusres.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Zhang Y.T., Yao L.L., Xu X., Han H.S., Li P.F., Zou D.H., Li X.Z., Zheng L., Cheng L.X., Shen Y.J., Wang X.H., Wu X.N., Xu J.X., Song B.F., Xu S.Y., Zhang H., Cao H.W. Enterovirus 71 inhibits cytoplasmic stress granule formation during the late stage of infection. Virus Res. 2018;255:55–67. doi: 10.1016/j.virusres.2018.07.006. [DOI] [PubMed] [Google Scholar]