Abstract
The prevalence of the bovine torovirus (BToV) and its genetic characterization have been reported in North America, Europe and Japan. Therefore, this study examined the prevalence and genetic diversity of the BToV in a total of 645 diarrheic fecal samples from 629 Korean native beef calf herds using RT-PCR and nested PCR with the primer pairs specific to a part of the BToV membrane (M) gene. Overall, 19 (2.9%) out of 645 diarrheic samples from 19 herds (6.9%) tested positive for BToVs by either RT-PCR or nested PCR. A comparison of the nucleotide (nt) and amino acid (aa) sequences of a part of the BToV M gene (409 bp) among the BToVs showed the Korean BToVs to have comparatively higher sequence homology to the Japanese and Dutch BToVs than to the American and Italian BToVs. Generally, the Korean BToV strains clustered with the Japanese and Dutch BToV strains. However, the American and Italian BToV strains clustered on a separate major branch, suggesting that these are more distantly related to other known BToV strains. These results suggest that the BToV infections are sporadic in diarrheic calves in South Korea, and the Korean BToV strains are more closely related to the Japanese and Dutch BToVs than to the American and Italian BToVs.
Keywords: Bovine torovirus, Prevalence, Genetic diversity, Sporadic
1. Introduction
Toroviruses within the family Coronaviridae are spherical, oval, elongated, or kidney-shaped enveloped viruses that possess a positive-sense single-stranded, polyadenylated RNA genome of approximately 25–30 kb in length (Cornelissen et al., 1997, Horzinek, 1999, Snijder and Horzinek, 1993). Toroviruses have been detected in normal horses as well as in humans, cattle, pigs, and recently in turkeys with diarrhea (Ali and Reynolds, 2000, Beards et al., 1986, Duckmanton et al., 1997, Duckmanton et al., 1998a, Kroneman et al., 1998, Woode, 1994). The first report of toroviruses described the identification of an unclassified virus from diarrheic calves (Woode et al., 1982), which is now known as the Breda virus, or bovine torovirus (BToV). Fecal shedding of the BToVs have been reported in diarrheic calves in several countries including the United States (Hoet et al., 2002, Hoet et al., 2003a, Hoet et al., 2003b, Woode et al., 1982, Woode et al., 1985), Canada (Duckmanton et al., 1998a), Costa Rica (Pérez et al., 1998), Netherlands (Koopmans et al., 1991), Germany (Liebler et al., 1992), Hungary (Matiz et al., 2002), Austria (Haschek et al., 2006), Japan (Ito et al., 2007), and South Africa (Vorster and Gerdes, 1993). However, there are no reports of BToV infections in South Korea. Since the molecular analysis of the BToV has only been carried out in North American, European and Japanese BToV strains (Cornelissen et al., 1997, Draker et al., 2006, Duckmanton et al., 1998b, Gonzálex et al., 2003, Haschek et al., 2006, Hoet et al., 2002, Hoet et al., 2003a, Hoet et al., 2003b, Ito et al., 2007, Smits et al., 2003), it is unclear if the BToVs circulating in other countries have distinct genetic characteristics. This paper reports the detection of BToV shedding in diarrheic calves using RT-PCR and nested PCR, along with the genetic diversity of the BToV strains based on a partial BToV membrane (M) gene.
2. Materials and methods
2.1. Specimens
According to farmers’ request for the treatment of calf diarrhea, local veterinary clinicians visited the cattle farms, and treated diarrheic calves with 2–3 days fasting and the administration of an electrolyte fluid and antibiotics. All the diarrheic calves were aged between 3 and 70 days at the time of sample collection, were fed colostrum from their dams, and housed in the stall. The farm sizes ranged from 5 to 153 animals (average 53). The farms were geographically distributed in Chonnam province, which is the South-Western province in South Korea. In order to diagnose the enteric pathogens, local veterinary clinicians sampled one or two diarrheic fecal specimens from each farm and immediately submitted the sample to the College of Veterinary Medicine, Chonnam National University.
A total of 645 diarrheic fecal specimens were collected from 629 Korean native beef (Hanwoo) calf herds (aged 3–70 days) between January 2004 and December 2005 during the spring (407 samples/406 herds), summer (107 samples/98 herds), autumn (73 samples/69 herds) and winter (58 samples/56 herds). Upon arrival of the fecal samples, they were immediately examined for common enteric viral, bacterial and protozoan pathogens including the BToV, bovine coronavirus (BCoV), groups A, B and C bovine rotaviruses (BRV A-C), new bovine enteric calicivirus genus known as becovirus, bovine norovirus (BNoV), bovine viral diarrhea virus (BVDV), Salmonella spp., Clostridium spp., Campylobacter spp., shiga-toxin-producing Escherichia coli, Coccidium spp. and Cryptosporidium spp. (Asakura et al., 1998, Park et al., in press-a, Park et al., in press-b, Soulsby, 1982, Timoney et al., 1988). The results of bovine norovirus and bovine coronavirus have been described previously (Park et al., in press-a, Park et al., in press-b). Data for other enteric viruses will be reported elsewhere.
2.2. RNA extraction
The RNA was extracted from a 200 μl starting volume of centrifuged 10% fecal suspensions using the Trizol-LS (Gibco-BRL, Life Tech, Grand Island, USA) procedure. The total RNA recovered was suspended in 50 μl of RNase free water and stored at −80 °C.
2.3. RT-PCR and nested PCR
The primer sets and RT-PCR and nested PCR conditions used for the detection of the BToV, BCoV, BRV A-C, becovirus, BNoV, and BVDV are described elsewhere (Cho et al., 2001, Park et al., 2006, Park et al., in press-a, Park et al., in press-b). As a negative control, the RNA was extracted from the normal feces of colostrum-deprived calf that had been inoculated with 50 Ml sterile PBS, and used for all RT-PCR assays. In order to confirm the absence of cross-contamination from RT-PCR to nested PCR, RT-PCR was performed with RNA extracted from a normal mock-infected calf fecal sample, and nested PCR was then carried out using 5 μl of this RT-PCR product. The amplification products were analyzed using 1.5 or 2% agarose gel electrophoresis and visualized by irradiating the ethidium bromide stained samples with UV.
2.4. DNA sequencing
The nested PCR products for a portion of the M gene (409 bp) were selected, purified and sequenced directly to verify the reaction specificity as well as to obtain the genomic data for phylogenetic analysis. The nested PCR products were purified using a GenClean II kit (BIO 101, Inc., LaJolla, USA) according to the manufacturer's instructions. DNA sequencing was carried out using an automated DNA sequencer (ABI system 3700, Applied Biosystem Inc., Foster City, USA).
2.5. Molecular analysis
The nucleotide (nt) and deduced amino acid (aa) sequences of the partial BToV M gene were compared with those of the other known toroviruses using the DNA Basic module (DNAsis MAX, Alameda, USA) (Table 1 ). Phylogenetic and bootstrap (1000 replicates) analyses based on the nt and aa alignments were constructed using the neighbor-joining method and the unweighted-pair group method with the average linkages of Molecular Evolutionary Genetics Analysis (MEGA, Version 3.1) with a pairwise distance (Kumar et al., 2004). A sequence similarity search was carried out for the bovine calicivirus the RdRp protein using the LALIGN Query program of the GENESTREAM network server at the Institut de Génétque Humaine, Montpellier, France (http://www.eng.uiowa.edu/∼tscheetz/sequence-analysis/examples/LALIGN/lalign-guess.html).
Table 1.
Genbank Accession numbers of the reference strains of bovine, equine and porcine toroviruses used in nucleotide and amino acid sequence comparisons, and phylogenetic analysis
| Host | Virus | Designation | Origin |
Accession number | References | |
|---|---|---|---|---|---|---|
| Country | Year | |||||
| Bovine | BToV | BRV(Breda) | USA | 1982 | AY427798 | Draker et al. (2006) |
| B6 | Italy | 1990 | AJ575374 | Smits et al. (2003) | ||
| B145 | The Netherlands | 1998 | AJ575375 | Smits et al. (2003) | ||
| B150 | The Netherlands | 1998 | AJ575376 | Smits et al. (2003) | ||
| B155 | The Netherlands | 1998 | AJ575377 | Smits et al. (2003) | ||
| K-567 | Japan | 2004 | AB270905 | Ito et al. (2007) | ||
| K-637 | Japan | 2004 | AB270908 | Ito et al. (2007) | ||
| K-645 | Japan | 2005 | AB270911 | Ito et al. (2007) | ||
| K-674 | Japan | 2005 | AB270913 | Ito et al. (2007) | ||
| K-676 | Japan | 2005 | AB270915 | Ito et al. (2007) | ||
| K-683 | Japan | 2005 | AB270917 | Ito et al. (2007) | ||
| K34 | South Korea | 2004 | DQ778041 | This study | ||
| K49 | South Korea | 2004 | DQ778042 | This study | ||
| K96 | South Korea | 2004 | DQ778043 | This study | ||
| K110 | South Korea | 2004 | DQ778044 | This study | ||
| K119 | South Korea | 2004 | DQ778045 | This study | ||
| K375 | South Korea | 2004 | DQ778046 | This study | ||
| K490 | South Korea | 2005 | DQ778047 | This study | ||
| K501 | South Korea | 2005 | DQ778048 | This study | ||
| K536 | South Korea | 2005 | DQ778049 | This study | ||
| K537 | South Korea | 2005 | DQ778050 | This study | ||
| K540 | South Korea | 2005 | DQ778051 | This study | ||
| K546 | South Korea | 2005 | DQ778052 | This study | ||
| K577 | South Korea | 2005 | DQ778053 | This study | ||
| Equine | EqTV | BEV(Berne) | Switzerland | 1972 | X52506 | Snijder et al. (1990) |
| Porcine | PToV | P4 | Italy | 1990 | AJ575369 | Smits et al. (2003) |
| P9 | Italy | 1996 | AJ575370 | Smits et al. (2003) | ||
| P10 | Italy | 1999 | AJ575371 | Smits et al. (2003) | ||
| P-MAR | The Netherlands | 1995 | AJ575368 | Smits et al. (2003) | ||
3. Results and discussion
A one-step RT-PCR assay, which targets a 603 bp fragment of the M gene in the BToV, detected three positive fecal samples from three herds. The nested PCR assay, which targeted a 409 bp fragment of the M gene, detected 19 positive fecal samples (2.9%) from 19 herds (6.9%). The calculated percentage of BToV infections in the diarrheic calves in South Korea was comparatively low compared with that reported in other countries, where the reported fecal prevalence of the BToV in calf diarrhea was 5% in Lower Saxony, Germany (Liebler et al., 1992), 6.5% in Japan (Ito et al., 2007), 9.7% in the USA (Hoet et al., 2003a), and 36.4% in southern Ontario, Canada (Duckmanton et al., 1998a). This suggests that BToV infections are sporadic in diarrheic calves in South Korea.
Of the 19 BToV-positive fecal specimens from the 19 calf herds, 4 fecal samples from 4 herds tested positive to the BToV alone, while the other 15 BToV-positive fecal samples from the 15 herds also tested positive to other enteric pathogens (Table 2 ). This suggests that other bovine enteric pathogens play an important role in the clinical and pathological presentation of this disease. An increase in the severity of the disease after a natural co-infection with other bovine enteric pathogens has been reported (Duckmanton et al., 1998a, Hoet et al., 2003a).
Table 2.
Summary of the viral enteric pathogens detected in the diarrheic fecal samples from calves
| Sample name | County | Sampling month | Age (days) | Fecal condition | BToV PCR results |
Other enteric pathogens detecteda | |
|---|---|---|---|---|---|---|---|
| RT-PCR | Nested PCR | ||||||
| K34 | Muan | April/2004 | 8 | Diarrhea | − | + | BRV A |
| K49 | Muan | May/2004 | 14 | Diarrhea | − | + | BRV A |
| K96 | Muan | June/2004 | 70 | Diarrhea | + | + | BCoV, BRV A plus BRV C |
| K110 | Muan | June/2004 | 3 | Diarrhea | − | + | BVD |
| K119 | Muan | June/2004 | 65 | Diarrhea | + | + | BCoV |
| K122 | Hampyeong | June/2004 | 14 | Diarrhea | − | + | BCoV |
| K124 | Sinan | June/2004 | 18 | Diarrhea | − | + | BNoV |
| K125 | Muan | June/2004 | 20 | Diarrhea | − | + | BCoV plus BRV A |
| K127 | Sinan | June/2004 | 10 | Diarrhea | − | + | BCoV plus Beco |
| K375 | Muan | November/2004 | 50 | Diarrhea | + | + | BNoV |
| K490 | Muan | January/2005 | 60 | Diarrhea | − | + | BNoV plus BVD |
| K501 | Muan | January/2005 | 7 | Diarrhea | − | + | − |
| K536 | Muan | January/2005 | 40 | Diarrhea | − | + | − |
| K537 | Muan | January/2005 | 10 | Diarrhea | − | + | BCoV |
| K540 | Muan | January/2005 | 30 | Diarrhea | − | + | − |
| K546 | Muan | February/2005 | 5 | Diarrhea | − | + | BCoV |
| K577 | Muan | February/2005 | 8 | Diarrhea | − | + | − |
| K726 | Muan | April/2005 | 7 | Diarrhea | − | + | BCoV, BVD, BRV A plus BRV C |
| K728 | Muan | April/2005 | 7 | Diarrhea | − | + | BCoV plus BVD |
BToV: bovine torovirus; BCoV: bovine coronavirus; BRV A–C: group A, B, C bovine rotaviruses; Beco: becovirus; BNoV: bovine norovirus.
Seasonally, BToV infections were less prevalent in the diarrheic fecal samples of calves obtained in the spring than in the other seasons (Table 2); 4 (1.0%) out of 407 fecal samples tested positive in spring; 1 (1.4%) out of 73 fecal samples tested positive in autumn; 2 (3.4%) out of 58 fecal samples tested positive in winter; and 7 (6.5%) out of 107 fecal samples tested positive in summer. This result is very similar to that reported by Hoet et al. (2003a), who showed that two peaks of BToV shedding were observed, one during the winter months and the other during the summer season.
Thirteen nested PCR products were selected, purified and sequenced directly, and their nt and aa sequences were compared with those of other known toroviruses (Table 1). Table 3 shows the sequence relationships of the nt and aa of the partial M gene among the torovirus strains. Interestingly, a comparison of the nt and aa sequences among the BToVs revealed all the Korean BToVs to share 100% nt and aa identity to each other, except for the K96 strain (99.0% nt and 98.8% aa identity). In order to eliminate cross contamination, the negative control for RT-PCR and nested PCR was used when the field diarrheic fecal samples were analyzed by RT-PCR and nested PCR. However, there was no positive reaction detected in this negative control. Therefore, this identical nt and aa sequence homology of the M gene among the Korean BToVs might be due to the fact that the positive fecal samples used for DNA sequencing originated from the same county, Muan, even though they had been sampled from different farms at different times (Table 2, Table 3). The Korean BToVs shared higher nt and aa sequence homology to the Japanese BToVs (98.5–99.5% nt and 98.5–100% aa identity) and the Dutch BToVs (98.3–99.3% nt and 99.3–100% aa identity) than the Italian (96.6% nt and 99.3–100% aa identity) and American (96.6% nt and 98.5–99.3% aa identity) BToVs. In addition, the BToVs including the American, European, Japanese and Korean BToVs had a lower nt and aa identity to the equine torovirus strain, Berne (43.2–44.9% nt and 9.8–10.5% aa identity), than to porcine toroviruses (78.2–81.2% nt and 88.9–91.1% aa identity). From these results, the Korean BToVs were more closely related to the Japanese and Dutch BToVs than to the American and Italian BToVs. In addition, the overall nt and aa sequence homology of the BToV M gene in this study is higher than of the spike (S) protein gene (Ito et al., 2007, Koopmans and Horzinek, 1994). The relative conservation of the M protein of all BToVs suggests that the structural constraints on this protein are stern, resulting in more limited evolution of this protein than the S protein (Koopmans and Horzinek, 1994). Therefore, future studies will be needed to generate more information on the genetic diversity by including other parts of the genome particularly the S gene as well as including the fecal specimens from other regions in South Korea.
Table 3.
Sequence relationships of the nucleotide and amino acid of the partial membrane gene among torovirus strains
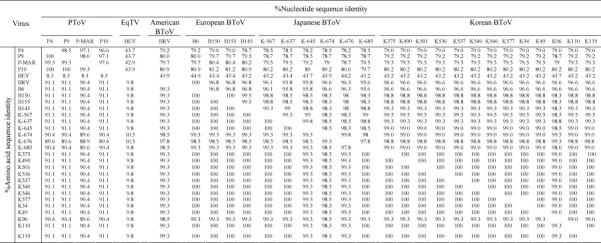
Generally, phylogenetic data of nt sequence of the partial BToV M gene revealed that the Korean BToV strains were closely related to the Japanese and Dutch BToV strains (Fig. 1 ). Of the Korean BToV strains, all Korean BToV strains clustered together with the exception of the K96 strain, which clustered on a separate branch with the Japanese BToV strains, K-674 and K-676. Other Japanese and Dutch BToVs made separate branches. In addition, the American Breda and the Italian B6 strains clustered on a separate major branch, suggesting these were the most distantly related to the other known BToV strains. Phylogenetic analysis of the aa sequence did not show any significant classification of the BToVs because the homology of the aa sequence among the BToV strains was higher than that of the nt sequence (data not shown). This data supports the above mentioned hypothesis that the Korean BToVs were more closely related to the Japanese and Dutch BToVs than to the American and Italian BToVs. South Korea imports live cows mainly from countries in North American and Oceania, not from Japan and European countries. Hence, the similarity in phylogenetic data on the Korean BToV with the Japanese and Dutch BToVs is quite surprising. Therefore, continuous monitoring and intensive studies will be needed for a more detailed characterization of the BToV with other genes.
Fig. 1.
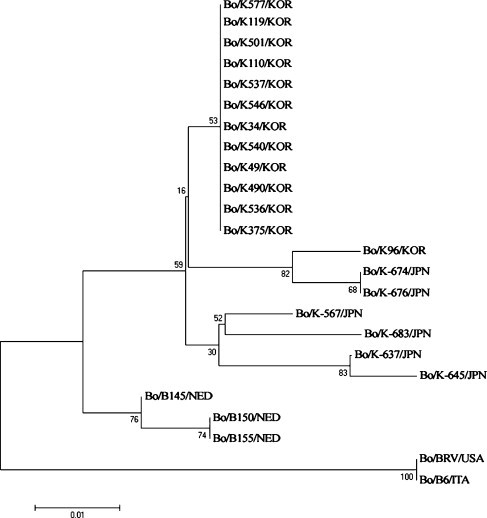
A phylogenetic tree of the nucleotide sequence of the M gene of the bovine torovirus strains was made using the neighbor-joining method of Molecular Evolutionary Genetics Analysis (Kumar et al., 2004). The names of the viruses used are listed in Table 1.
In conclusion, BToV infections are sporadic in diarrheic calves in South Korea. Based on the partial sequence of the BToV M gene, the Korean BToVs were more closely related to the Japanese and Dutch BToVs than to the American and Italian BToVs. To our knowledge, this is the first report of the detection of BToV shedding and its genetic diversity in diarrheic calves in South Korea.
Acknowledgments
This study was supported by grant No. RTI05-01-01 from the Regional Technology Innovation Program of the Ministry of Commerce, Industry and Energy (MOCIE), Republic of Korea. The authors acknowledge a graduate fellowship provided by the Korean Ministry of Education and Human Resources Development through the Brain Korea 21 project.
References
- Ali A., Reynolds D.L. Characterization of the stunting syndrome agent: relatedness to known viruses. Avian Dis. 2000;44:45–50. [PubMed] [Google Scholar]
- Asakura H., Makino S., Shirahata T., Tsukamoto T., Kurazono H., Ikeda T., Takeshi K. Detection and genetical characterization of Shiga toxin-producing Escherichia coli from wild deer. Microbiol. Immunol. 1998;42:815–822. doi: 10.1111/j.1348-0421.1998.tb02356.x. [DOI] [PubMed] [Google Scholar]
- Beards G.M., Brown D.W.G., Green J., Flewett T.H. Preliminary characterization of torovirus-like particles of humans: comparison with Berne virus of horses and Breda virus of calves. J. Med. Virol. 1986;20:67–78. doi: 10.1002/jmv.1890200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K.O., Hasoksuz M., Nielsen P.R., Chang K.O., Lathrop S., Saif L.J. Cross-protection studies between respiratory and calf diarrhea and winter dysentery coronavirus strains in calves and RT-PCR and nested PCR for their detection. Arch. Virol. 2001;146:2401–2419. doi: 10.1007/s007050170011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen L.A., Wierda C.M., van der Meer F.J., Herrewegh A.A., Horzinek M.C., Egberink H.F., de Groot R.J. Hemagglutinin-esterase, a novel structural protein of torovirus. J. Virol. 1997;71:5277–5286. doi: 10.1128/jvi.71.7.5277-5286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R., Roper R.L., Petric M., Tellier R. The complete sequence of the bovine torovirus genome. Virus Res. 2006;115:56–68. doi: 10.1016/j.virusres.2005.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckmanton L.M., Carman S., Nagy E., Petric M. Detection of bovine torovirus in fecal specimens of calves with diarrhea from Ontario farms. J. Clin. Microbiol. 1998;36:1266–1270. doi: 10.1128/jcm.36.5.1266-1270.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckmanton L.M., Tellier R., Liu P., Petric M. Bovine torovirus: sequencing of the structural genes and expression of the nucleocapsid protein of Breda virus. Virus Res. 1998;58:83–96. doi: 10.1016/S0168-1702(98)00104-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckmanton L.M., Luan B., Devenish J., Tellier R., Petric M. Characterization of torovirus from human fecal specimens. Virology. 1997;239:158–168. doi: 10.1006/viro.1997.8879. [DOI] [PubMed] [Google Scholar]
- Gonzálex J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschek B., Klein D., Benetka V., Herrera C., Sommerfeld-Stur I., Vilcek Š., Moestl K., Baumgartner W. Detection of bovine torovirus in neonatal calf diarrhea in lower Austria and Styria (Austria) J. Vet. Med. B. 2006;53:160–165. doi: 10.1111/j.1439-0450.2006.00936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoet A.E., Nielsen P.R., Hasoksuz M., Thomas C., Wittum T.E., Saif L.J. Detection of bovine torovirus and other enteric pathogens in feces from diarrhea cases in cattle. J. Vet. Diagn. Invest. 2003;15:205–212. doi: 10.1177/104063870301500301. [DOI] [PubMed] [Google Scholar]
- Hoet A.E., Smiley J., Thomas C., Nielsen P.R., Wittum T.E., Saif L.J. Association of enteric shedding of bovine torovirus (Breda virus) and other enteropathogens with diarrhea in veal calves. Am. J. Vet. Res. 2003;64:485–490. doi: 10.2460/ajvr.2003.64.485. [DOI] [PubMed] [Google Scholar]
- Hoet A.E., Cho K.O., Chang K.O., Loerch S.C., Wittum T.E., Saif L.J. Enteric and nasal shedding of bovine torovirus (Breda virus) in feedlot cattle. Am. J. Vet. Res. 2002;63:342–348. doi: 10.2460/ajvr.2002.63.342. [DOI] [PubMed] [Google Scholar]
- Horzinek M.C. Molecular evolution of corona- and toroviruses. Adv. Exp. Med. Biol. 1999;473:61–72. doi: 10.1007/978-1-4615-4143-1_5. [DOI] [PubMed] [Google Scholar]
- Ito T., Okada N., Fukuyama S. Epidemiological analysis of bovine torovirus in Japan. Virus Res. 2007;126:32–37. doi: 10.1016/j.virusres.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M., Horzinek M.C. Toroviruses of animals and humans: a review. Adv. Virus Res. 1994;43:233–273. doi: 10.1016/S0065-3527(08)60050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M., van Wuijckhuise-Sjouke L., Schukken Y.H., Cremers H., Horzinek M.C. Association of diarrhea in cattle with torovirus infections on farms. Am. J. Vet. Res. 1991;52:1769–1773. [PubMed] [Google Scholar]
- Kroneman A., Cornelissen L.A., Horzinek M.C., de Groot R.J., Egberink H.F. Identification and characterization of a porcine torovirus. J. Virol. 1998;72:3507–3511. doi: 10.1128/jvi.72.5.3507-3511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Masatoshi N. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Liebler E.M., Kluver S., Pohlenz J., Koopmans M. The significance of bredavirus as a diarrhea agent in calf herds in Lower Saxony. Dtsch. Tierarztl. Wochenschr. 1992;99:195–200. [PubMed] [Google Scholar]
- Matiz K., Kecskemeti S., Kiss I., Adam Z., Tanyi J., Nagy B. Torovirus detection in faecal specimens of calves and pigs in Hungary: short communication. Acta Vet. Hung. 2002;50:293–296. doi: 10.1556/AVet.50.2002.3.5. [DOI] [PubMed] [Google Scholar]
- Park, S.I., Jeong, C., Kim, H.H., Park, S.H., Park, S.J., Hyun, B.H., Yang, D.K., Kim, S.K., Kang, M.I., Cho, K.O. Molecular epidemiology of bovine noroviruses in South Korea. Vet. Microbiol., in press-a. [DOI] [PMC free article] [PubMed]
- Park, S.J., Lim, G.K., Park, S.I., Kim, H.H., Cho, K.O. Detection and molecular characterization of calf diarrhea bovine coronaviruses circulating in South Korea during 2004–2005. Zoonoses Public Health, in press-b. [DOI] [PubMed]
- Park S.J., Jeong C., Yoon S.S., Choy H.E., Saif L.J., Park S.H., Kim Y.J., Jeong J.H., Park S.I., Kim H.H., Lee B.J., Cho H.S., Kim S.K., Kang M.I., Cho K.O. Detection and characterization of bovine coronaviruses in fecal specimens of adult cattle with diarrhea during the warmer seasons. J. Clin. Microbiol. 2006;44:3178–3188. doi: 10.1128/JCM.02667-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez E., Kummeling A., Janssen M.M., Jimenez C., Alvarado R., Caballero M., Donado P., Dwinger R.H. Infectious agents associated with diarrhea of calves in the canton of Tilaran Costa Rica. Pre. Vet. Med. 1998;33:195–205. doi: 10.1016/S0167-5877(97)00038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Lavazza A., Matiz K., Horzinek M.C., Koopmans M.P., de Groot R.J. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J. Virol. 2003;77:9567–9577. doi: 10.1128/JVI.77.17.9567-9577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Horzinek M.C. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. J. Gen. Virol. 1993;74:2305–2316. doi: 10.1099/0022-1317-74-11-2305. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Den Boon J.A., Spaan W.J., Weiss M., Horzinek M.C. Primary structure and post-translational processing of the Berne virus peplomer protein. Virology. 1990;178:355–363. doi: 10.1016/0042-6822(90)90332-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulsby E.J.L. seventh ed. Lea and Febiger; Philadelphia: 1982. Helminths Arthropods and Protozoa of Domesticated Animals. [Google Scholar]
- Timoney J.F., Gillespie J.H., Scott F.W., Barlough J.E. eighth ed. Cornell University Press; Ithaca: 1988. Hagan and Bruner's Microbiology and Infectious Diseases of Domestic Animals. [Google Scholar]
- Vorster J.H., Gerdes G.H. Breda virus-like particles in calves in South Africa. J. S. Afr. Vet. Assoc. 1993;64:58. [PubMed] [Google Scholar]
- Woode G.N. The toroviruses: bovine (Breda virus) and equine (Berne virus) and the torovirus-like agents of humans and animals. In: Kapikian A.Z., editor. Viral Infections of the Gastrointestinal Tract. Marcel Dekker; New York: 1994. pp. 581–602. [Google Scholar]
- Woode G.N., Saif L.J., Quesada M., Winand N.J., Pohlenz J.F., Gourley N.K. Comparative studies on three isolates of Breda virus of calves. Am. J. Vet. Res. 1985;46:1003–1010. [PubMed] [Google Scholar]
- Woode G.N., Reed D.E., Runnels P.L., Herrig M.A., Hill H.T. Studies with an unclassified virus isolated from diarrheic calves. Vet. Microbiol. 1982;7:221–240. doi: 10.1016/0378-1135(82)90036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


