Abstract
Feline coronaviruses are widespread and come in different flavors. There are two main serotypes both of which occur in two pathotypes, the avirulent enteric viruses and the virulent, usually fatal peritonitis viruses, the latter in turn occurring either in a ‘wet’ or exudative form or in a ‘dry’ or proliferative form. In this paper a concise overview is given of the molecular features of these viruses. Special attention is given to the genetic dynamics of the viruses as these now allow us to begin to understand the origin of the different phenotypes, in particular the genesis of virulence during persistent infection. As discussed, the surprising new insights obtained over the last few years call for a critical reevaluation of strategies for protection.
Keywords: Feline coronavirus, Feline infectious peritonitis, Coronavirus persistence
1. Introduction
The Utrecht group has been accumulating data on feline coronaviruses for more than two decades; Marian C. Horzinek started the work on feline infectious peritonitis in 1975, and with Raoul de Groot, Harry Vennema, Herman Egberink and Bart Haagmans our group has made major contributions on this topic. Arnold Herrewegh’s dissertation, based on five papers, is the last coherent addition to the subject (Herrewegh, 1997).
Coronaviruses are positive-stranded RNA viruses, recently accommodated in the order Nidovirales. The family Coronaviridae includes the genera Coronavirus and Torovirus, whereas the Arteriviridae contains but a single genus (Arterivirus) so far. The viruses have been classified in the order Nidovirales mainly on the basis of two features: their genome organization and their replication strategy (for review, see de Vries et al., 1997).
The organization of the genomes of the corona-, toro-, and arteriviruses is quite similar: they have a large open reading frame at the 5’-end of their genome, which encodes the polymerase (Fig. 1 ). It is synthesized as a precursor protein, which is in part generated by ribosomal frameshifting and which is processed by proteolytic cleavages to generate the functional proteins. Further downstream from the polymerase gene there is a collection of open reading frames, amongst which the genes for the structural proteins are located, as well as genes of largely unknown function. There is some variation in the genetic make-up of coronaviruses, e.g., with respect to the hemagglutinin/esterase (HE) gene, which is lacking in feline viruses. The genes for the structural proteins are obviously present in all viruses, but again there are differences: thus toroviruses lack an E protein, which for coronaviruses is essential in the assembly process.
Fig. 1.
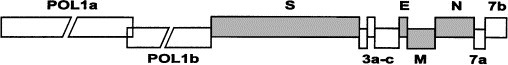
Genome organization of feline coronavirus. The gene for the polymerase polyprotein is indicated (POLla and POLib). The genes for the structural proteins spike (S), envelope (E), membrane (M), and nucleocapsid (N) are shaded. Genes encoding proteins of unknown function are designated by numerals.
The other common feature is the replication strategy, more specifically: the way these viruses make their proteins. The polymerase protein is translated from the genome, the incoming viral RNA. The other open reading frames are translated from subgenomic messenger RNAs that are generated by a specific mechanism of discontinuous transcription. Each mRNA of this collection is responsible for one protein by translation of the 5’-most open reading frame.
Coronaviruses can be grouped into three clusters on the basis of genetic comparisons (Table 1 ). We find the feline coronaviruses in Group 1, together with e.g., porcine transmissible gastroenteritis virus and canine coronavirus. The most prominent member of Group 2 is mouse hepatitis virus, also the coronavirus type species. Infectious bronchitis virus and its many variants constitute Group 3. Electron microscopically these viruses present a characteristic picture, with a ‘corona’ surrounding the enveloped particle, a halo of very typical surface projections. Within the envelope resides the nucleocapsid, which harbors the plus-stranded huge RNA molecule of some 30 kb, the longest mature mRNA known to occur in nature. The RNA is packaged by one type of protein, the nucleocapsid (N) protein, which is surrounded in turn by an envelope containing three membrane proteins. The spike (S) protein, is the most obvious protein in that it forms the above-mentioned ‘corona’. Another envelope constituent is the membrane protein (M), which is most abundant. Finally there is the small envelope protein (E), which has been discovered only recently and which occurs only in very low numbers in virions. In feline coronaviruses the number has not yet been determined but in the related TGEV it was estimated to occur at a rate of about 20 molecules per particle (Godet et al., 1992)
Table 1.
Division of coronavirusesa
| Group 1 |
Group 2 |
Group 3 |
| CCV | BCV | IBV |
| FIPV | HCV OC43 | |
| HCV | HEV | |
| PEDV | MHV | |
| TGEV 229E | TCV |
Explanation of acronyms: bovine coronavirus: BCV; canine coronavirus: CCV; feline infectious peritonitis virus: FIPV; human coronavirus 229E: HCV 229E; human coronavirus 0C43: HCV 0C43; hemagglutinating encephalomyelitis virus: HEV; infectious bronchitis virus: IBV; mouse hepatitis virus: MHV; porcine epidemic diarrhea virus: PEDV; transmissible gastroenteritis virus: TGEV; turkey coronavirus: TCV.
Coronaviruses do not bud from the plasma membrane but are assembled within the cell, at intracellular membranes. These have been identified as the membranes from the intermediate compartment, a complex situated between the endoplasmic reticulum and the Golgi apparatus. This is how it happens: the nucleocapsid, the ribonucleoprotein structure composed of the RNA and the nucleocapsid protein is synthesized in the cytoplasm. It subsequently encounters the membrane proteins which after their synthesis in the endoplasmic reticulum have traveled to the intermediate compartment. In this compartment the membrane proteins accumulate and interact with the nucleocapsid. Subsequent budding gives rise to viral particles which are moved to and through the Golgi complex by vesicular transport, undergoing various modifications while on their way. From the Golgi complex they are transported (again by vesicles) to the plasma membrane and exocytosed.
Many properties of the membrane proteins are known. The S-protein is a homotrimer of a glycoprotein of some 180 kDa. The M-protein is a triple-spanning membrane protein – a typical Nidovirales protein found also in toro- and arteriviruses in that configuration. Finally there is the largely hydrophobic 10 kDa E-protein. As we have shown, only the last approximately 25 residues of this polypeptide are exposed at the cytoplasmic side of cellular membranes; these residues end up in the interior of the virus particle after assembly.
We have extensively studied the coronaviral envelope proteins, amongst others by coexpressing their genes in different combinations. Using the vaccinia virus/T7 polymerase expression system with plasmids coding for the respective proteins we asked the question what would emerge from these cells. When we coexpressed all three membrane proteins, we encountered particles in the medium, which were morphologically indistinguishale from coronavirions (Vennema et al., 1996). We have done the experiment for mouse hepatitis virus and repeated it with feline coronaviruses, with the same result the particles produced by coexpression are indistinguishable from authentic virions. The surprise was great when we studied the various combinations and found that, with only the M and E envelope proteins particles were still formed, while nothing happened when the proteins were expressed separately. This finding means that while the M protein alone is unable to assemble coexpression of the E protein allows the formation of a curvature in the membrane, and eventually budding of vesicles with virion size. If the S protein is present, it is co-incorporated. We know that the S protein interacts with the M protein whereby it is apparently drawn into these particles. The conclusion of this experiment is that the assembly of the coronavirus envelope is independent of the nucleocapsid, the M and E proteins being essential, while the spike protein is also dispensable. These findings are interesting not only for fundamental studies on virus assembly, but also for applied research.
2. Feline coronaviruses
Feline coronaviruses can be grouped on the basis of different criteria, one of them being virulence for the host. This has led to the definition of biotypes, or pathotypes as we may want to call them. We have on the one hand avirulent strains which cause usually mild or even subclinical enteric infections. They are mostly referred to as feline enteric coronaviruses (FECV). On the other hand we have the virulent strains which cause feline infectious peritonitis (FIP), and we then talk about FIP viruses (FIPV). For a general review, see de Groot and Horzinek (1995).
Another distinction is made on the basis of serology, which has allowed the definition of two serotypes. The epidemiology of coronavirus infections can be summarized as follows. All viruses are transmitted by the oro-faecal route, the viruses being widespread in all cat populations known. If you look in catteries or community shelters where cats are in crowded situations, seropositivity is high; but even many single-cat households appear to be seropositive. On the other hand FIP is a rare consequence of the infection, occurring in only 1–5 % of the infected animals, mainly in kittens, but sometimes also in old cats. The signs of the infection leading to FIP are chronic undulating fever, anorexia, general malaise, ocular and neurological disorders. Very characteristic is the abdominal extension which is caused by the formation of ascitic fluid in the ‘wet’ or exudative form of FIP; there is also a ‘dry’ or proliferative form where only little if any exudate is found.
The pathogenesis of FIP is clearly immune-mediated. The key pathogenic event appears to be the infection of cells of the monocyte/macrophage lineage. As shown recently by Bart Haagmans there is bystander apoptosis of activated T-cells mediated by some soluble factor, while the T-cells are not replicating the virus (Haagmans et al., 1996). Also antibody enhancement is a very well-known phenomenon, with early death as a consequence under experimental conditions. The enhanced infection of macrophages is probably mediated by antibodies complexed with virions and taken up via a Fc receptor.
What is the origin of the coronaviruses causing FIP? This question has kept us busy for many years. What is the relation of FIPV with FECV, so to say. How do FIP-inducing coronaviruses arise? A quote from a recent paper of Harry Vennema (Vennema et al., 1998) is appropriate here: FIPV is derived by mutation from endemic enteric coronaviruses and the paper provides strong indications for that. There were already indications from the work of Arnold Herrewegh and others before, but this paper most convincingly shows that FIPV is a variant, a mutant of FECV arising in an infected animal. Vennema and coworkers came to this conclusion on the basis of extensive sequence analyses and sequence comparisons. They compared sequences of FIPV viruses from different geographic areas with those of FECV viruses from these same areas and concluded that the viruses isolated from FIP cases are most similar to the coronaviruses from the area where this particular case of FIP occurred. This important conclusion also has implications e.g., for protection.
Another pathogenetic indication obtained was that virulence, the sudden appearance of an FIPV biotype, appears to correlate with deletions in the 3c and 7b genes. What the 3c gene actually does is unknown; in essence the same is true for the 7b gene product, though we know that this is a small glycoprotein that is released into the medium from infected cells in culture. We have suggested that it might have a signalling function, acting as a ‘virokine’.
3. Serotypes
The two feline coronavirus serotypes can be distinguished on the basis of their relationship to canine coronavirus. CCV specific sera neutralize type II viruses but fail to neutralize, or neutralize only poorly those of type I. The viruses also have different in vitro characteristics: type II strains grow well in tissue culture, type I strains grow poorly (Pedersen et al., 1984). As a consequence more is known about type II viruses than about type I viruses. In the field, however, type I strains are predominant; in Europe for instance, type II strains have hardly been found, if ever, and also in the US they have been detected only incidentally. In Japan they are reported to constitute 10–20% of the viruses.
What is the origin of the serotype II, what is the molecular basis for the serotypes? It has become clear from sequence comparisons by Vennema et al. (1995), Motokawa et al. (1996) and Herrewegh et al. (1998) that the type II strains are the result of double recombination events between FECV type I strains and CCV (see abstract from Harry Vennema in this issue). Recombinations have been found to occur somewhere in the E and the M gene, the second cross-over point being found in ORF 1. Since in the various cases of type II viruses analyzed the same picture has been found (where the spike protein of CCV has been acquired by a type I virus) it is understandable why type II viruses are CCV related serologically. The fact that several genetically different FECV type I/CCV recombinant strains have been recognized indicates that they have been independently generated, and this double recombination is apparently not a rare event.
4. Virus persistence and coronavirus evolution
For many years there have been indications that feline coronaviruses can persist in their natural host. Epidemiological studies have indicated that and there is of course Niels Pedersen’s classical experiment where he infected a cat with a sublethal dose of FIPV, put the cat in isolation, waited for 4 months and then superinfected it with FeLV. The immunosuppression caused by the retrovirus made the coronavirus reappear, it was activated to replicate, and eventually the disease picture of FIP was seen (Pedersen and Floyd, 1985). More recently, using our sensitive RT-PCR methods (Herrewegh et al., 1995a, Herrewegh et al., 1995b), we designed studies in a closed cat facility in Hannover, Germany, which confirmed and extended these observations. This was the laboratory cat facility of the Medical School, and its history was well documented. It had been FECV-free for many years when, in 1991, feline coronavirus has been inadvertently introduced and from that time on there were yearly cases of FTP; a total of some 5% of the cats succumbed to the disease. When we came to analyze the situation, no FIP cases had been observed over the last 3 years. Irrespective of this fact, the virus was really still around: of the 42 cats studied, 29 were seropositive by immunofluorescence; 85% of these cats were RT-PCR positive in faeces and/or serum and/or plasma. After 3 months we re-examined five RT-PCR positives and found that four of them were still positive. (Herrewegh et al., 1997). These data strongly suggested virus persistence in this cattery. The question was then asked whether the continuous presence of the virus was due to recurrent infections between animals or to its extended presence in individual cats. Consequently, two PCR-positive cats were placed in strict isolation and their faeces collected. After 4 months, one cat (#H324) was sacrificed, and the collected samples were tested by RT-PCR. The results shown in Fig. 2 clearly show that this animal had remained RT-PCR positive for the entire observation period. The long bars in Fig. 2 indicate that the samples were positive by direct PCR, the short bars indicate that a nested PCR was required to show a positive result. In essence the same was true for the cat that had been maintained in isolation (cat #H419). We saw that the infection waned and some samples were negative even using a nested PCR. Later on positive results were recorded for up to 7 months. This finding demonstrates that the virus can really persist in isolated cats for a long time.
Fig. 2.
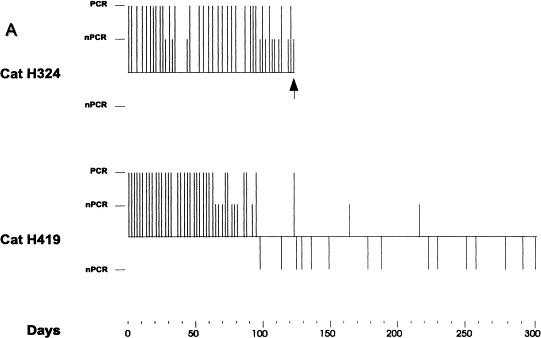
Chronic shedding of feline coronavirus as monitored by RT-PCR on feces. Amplification reactions were targeted to the 3’ nontranslated region of the viral genome. Cats H324 and H419 were placed in isolation at Day 0. The numbers of days that the cats were in isolation is on the horizontal axis. The RT-PCR results are expressed by bars above (i.e. positive in RT-PCR) or below (i.e. negative) the horizontal axis. Long and short bars represent single and nested RT-PCR.
From cat #H324, the animal that had been sacrificed, a number of tissues were examined for the presence of virus in different tissues. By PCR, viral (genomic) RNA was found in many organs. We then employed a PCR designed to detect only mRNA, to check for replication occurring in these tissues, and there oniy the lower part of the gastro-intestinal tract was found positive. Immunohistochemistry performed by Bart Haagmans evidenced single, scattered antigen-positive cells in various areas in the ileum, close to Peyer’s patches. The conclusion from these experiments is that feline coronaviruses may persist in the lower intestinal tract where the virus continues to replicate at low levels.
We used the samples collected in this cattery also for sequencing, to learn more about the evolution of the viruses, about their genetic changes. The 7b gene is quite a stable gene (Herrewegh et al., 1995c) and was taken for comparison. We found that feline coronaviruses in this colony formed a distinct dade (Herrewegh et al., 1997). When we compared them with viruses from other sources and with laboratory strains, they were quite distinct. The viruses from the Hannover cattery form one dade, as do viruses originating from another cattery. Most probably the viruses in the Hannover cattery originated from a single founder infection. The most interesting finding came when we compared the viruses in their 5’ end of the S gene, known to be highly variable. It appeared that each cat harbors its own distinct feline coronavirus quasispecies. Conceivably, the persisting virus confers to its host resistance against superinfection by the closely related feline coronaviruses, which were infecting the other cats. This is an important piece of information when we think about protection, especially the fact that there is resistance against superinfection.
We have seen that the feline coronaviruses are harbored in the lower intestinal tract. The question remains whether they are restricted to the enteric epithelium or whether they can cross it. The idea was that ‘feline enteric coronaviruses’ are indeed restricted in tropism, while ‘FIP viruses’ would cross the epithelium, infect macrophages and go systemically. Epithelial cells are specialized cells; they possess an apical membrane facing e.g. the gut lumen, and a basolateral membrane keeping contact with the neighboring cells and facing e.g., the submucosa; ‘tight junctions’ form a barrier between both the compartments and make sure that different proteins and lipids can be kept separately in them. These cells are specialized in the sorting of proteins and lipids, in the transport of immunoglobulins by transcytosis etc. To study their functions, these cells are grown on filter membranes, whereby the basolateral and apical membranes are separately accessible, e.g., for a virus infection. When the monolayer is complete, there is no transport of medium from the upper part into the lower part of the filter device. Using this system, John Rossen from our group studied the infection and release behavior of FIPV, FECV, CCV, MHV, and TGEV. We knew from earlier experiments that TGEV is secreted into the apical medium, and by inference into the gut lumen if one considers the intestinal tract, with progression of the infection through apical virus uptake. In contrast, MHV is released into the basolateral medium, and in the mouse, this virus causes a systemic infection, giving rise to hepatitis amongst other symptoms. Thus, its basolateral release would be in line with the systemic infection (Rossen et al., 1995a, Rossen et al., 1995b, Rossen et al., 1996).
When examining FIPV, FECV, and CCV we found virus release into the basolateral medium in all cases, which would mean that the direction of release is toward the blood stream. So we will probably need to modify our preconception that these viruses are restricted to the intestine. It needs to be examined, of course, if the same holds true for the natural situation.
5. Vaccination and protection
There have been many attempts to protect cats against FIP, using various approaches. The result of all these studies is that generally there is no protection when an antibody response to the spike protein is induced there is rather an enhancement of the infection, with an ‘early death’ phenomenon. All these attempts although fruitless in the practical sense have gathered a body of information that allows a number of better focused questions to be asked. One important question is: do we really want to protect against the FIPV infection? Since FIP-inducing coronavirus mutants are generated within the persistently infected animal the objective would rather be to prevent FECV infetion and persistence. When FIPV is generated in an animal and causes disease, it is probably shed to some extent, but other cats usually do not get infected. So the transmission is either not very efficient, or most of these cats are resistant to superinfection due to an infection with a related coronavirus variant. Usually one sees catteries with one or two isolated cases but the virulent virus obviously does not spread very efficiently. So who is the enemy? The enemy is FECV. What we should want to prevent is the establishment of an FECV infection, because it will eventually be the source of an FIPV strain. What we would want to achieve is to protect cats against an infection with FECV. It is highly unlikely that any vaccination strategy will result in sterile immunity. It is unlikely that any vaccine will prevent an infection to occur. There will probably always be a few cells that become infected. What one would want to achieve then is to have an immune system in place that can either terminate or contain that infection immediately. Replication of the infecting FECV must be kept to a minimum, such that the viral load and the eplication determinants for the stochastic appearance of FIPV mutants are as low as possible. What are the correlates of protection? Cellular immunity is probably most important, and there are many indications that this is where the solution must be sought. The vaccinological objective can be summed up in one sentence: Novel vaccines aimed at protection against FIP should induce a vigorous T-cell response, above all against type I viruses.
References
- de Groot, R.J., M.C. Horzinek, 1995. Feline infectious peritonitis. In: Siddell, S.G. (Ed.), The Coronaviridae. Plenum Press, New York, pp. 293–309.
- de Vries A.A.F., Horzinek M.C., Rottier P.J.M., de Groot R.J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Seminars in Virology. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet M., L’Haridon R., Vautherot J.-F., Laude H. TGEV coronavirus ORF 4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188:666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., Egberink H.F., Horzinek M.C. Apoptosis and T-cell depletion during feline infectious peritonitis. J. Virol. 1996;70:8977–8983. doi: 10.1128/jvi.70.12.8977-8983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh, A.A.P.M., 1997. Molecular genetics, persistence and evolution of FCoV. Thesis (ISBN 90-393-1972-3), Utrecht University, Utrecht.
- Herrewegh A.A.P.M., Mahler M., Hedrich H.J., Haagmans B.L., Egberink H.F., Horzinek M.C., Rottier P.J.M., de Groot R.J. Persistence and evolution of feline coronavirus in a closed cat-breeding colony. Virology. 1997;234:349–363. doi: 10.1006/viro.1997.8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., de Groot R.J., Cepica A., Egberink H.F., Horzinek M.C., Rottier P.J.M. Detection of feline coronavirus RNA in feces, tissues and body fluids of naturally infected cats by reverse transcriptase PCR. J. Clin. Microbiol. 1995;33:684–689. doi: 10.1128/jcm.33.3.684-689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., Egberink H.F., Horzinek M.C., Rottier P.J.M., de Groot R.J. Polymerase chain reaction (PCR) for the diagnosis of naturally occurring feline coronavirus infections. Feline Practice. 1995;23:56–60. [Google Scholar]
- Herrewegh A.A.P.M., Vennema H., Horzinek M.C., Rottier P.J.M., de Groot R.J. The molecular genetics of feline coronaviruses: comparative sequence analysis of the ORF7a/7b transcription unit of different biotypes. Virology. 1995;212:622–631. doi: 10.1006/viro.1995.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., Smeenk I., Horzinek M.C., Rottier P.J.M., de Groot R.J. Feline coronavirus type II strains FECV 79-1683 and FIPV 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J.Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motokawa K., Hohdatsu T., Hashimoto H., Koyama H. Comparison of the amino acid sequence and phylogenetic analysis of the peplomer integral membrane and nucleocapsid proteins of feline canine and porcine coronaviruses. Microbiol. Immunol. 1996;40:425–433. doi: 10.1111/j.1348-0421.1996.tb01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., J.W. Black, J.F. Boyle, J.F. Evermann, A.J. McKeiman, R.L. Ott, 1984. Pathogenic differences between various feline coronavirus isolates. In: Rottier, P.J.M., van der Zeijst, B.A.M., Spaan, W.J.M., Horzinek, M.C. (Eds), Molecular Biology and Pathogenesis of Coronaviruses. Plenum Press, New York, pp. 365–380.
- Pedersen N.C., Floyd K. Experimental studies with three new strains of feline infectious peritonitis virus: FIPV-UCD2, FIPV-UCD4, and FIPV-UCD4. Compend.Contin. Educ. Pract. Vet. 1985;7:1001–1011. [Google Scholar]
- Rossen J.W.A., Bekker C.P.J., Strous G.J.A.M., Horzinek M.C., Dveksler G.S., Holmes K.V., Rottier P.J.M. A murine and a porcine coronavirus are released from opposite surfaces of the same epithelial cells. Virology. 1996;224:345–351. doi: 10.1006/viro.1996.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen J.W.A., Horzinek M.C., Rottier P.J.M. Coronavirus infection of polarized epithelial cells. Trends in Microbiol. 1995;3:486–490. doi: 10.1016/S0966-842X(00)89018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen J.W.A., Voorhout W.F., Horzinek M.C., van der Ende A., Strous G.J.A.M., Rottier P.J.M. MHV-A59 enters polarized murine epithelial cells through the apical surface but is released basolaterally. Virology. 1995;210:54–66. doi: 10.1006/viro.1995.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G.-.J., Rossen J.W.A., Voorhout W.F., Horzinek M.C., Opstelten D.-J.E., Rottier P.J.M. Nucleocapsid-independent assembly of coronavirus-like particles by coexpression of viral envelope proteins. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Poland A., Floyd Hawkins K., Pedersen N.C. A comparison of the genomes of FECVs and FIPVs and what they tell us about the relationships between feline coronaviruses and their evolution. Feline Pract. 1995;23:40–44. [Google Scholar]


