Highlights
-
•
Equine arteritis virus lacking PLP2 DUB activity is replication competent in vivo.
-
•
EAV lacking PLP2 DUB activity protects against challenge with KY84 virus.
-
•
An enhanced immune response was not detected using this experimental set-up.
Keywords: Deubiquitinase, Interferon, Vaccine, EAV, PRRSV, Arterivirus
Abstract
Arteriviruses are a family of positive-stranded RNA viruses that includes the prototypic equine arteritis virus (EAV) and porcine reproductive and respiratory syndrome virus (PRRSV). Although several vaccines against these viruses are commercially available there is room for improvement, especially in the case of PRRSV. The ability of arteriviruses to counteract the immune response is thought to decrease the efficacy of the current modified live virus vaccines. We have recently shown that the deubiquitinase (DUB) activity of EAV papain-like protease 2 (PLP2) is important for the inhibition of innate immune activation during infection. A vaccine virus lacking PLP2 DUB activity may therefore be more immunogenic and provide improved protection against subsequent challenge than its DUB-competent counterpart. To test this hypothesis, twenty Shetland mares were randomly assigned to one of three groups. Two groups were vaccinated, either with DUB-positive (n = 9) or DUB-negative (n = 9) recombinant EAV. The third group (n = 2) was not vaccinated. All horses were subsequently challenged with the virulent KY84 strain of EAV. Both vaccine viruses proved to be replication competent in vivo. In addition, the DUB-negative virus provided a similar degree of protection against clinical disease as its DUB-positive parental counterpart. Owing to the already high level of protection provided by the parental virus, a possible improvement due to inactivation of PLP2 DUB activity could not be detected under these experimental conditions. Taken together, the data obtained in this study warrant further in vivo investigations into the potential of using DUB-mutant viruses for the improvement of arterivirus vaccines.
1. Introduction
Arteriviruses are a family of animal viruses that includes the prototypic equine arteritis virus (EAV) and porcine reproductive and respiratory syndrome virus (PRRSV). Whereas outbreaks of equine viral arteritis (EVA) are only occasionally reported, infections with PRRSV pose a major threat to swine-farming industries worldwide. Especially the emergence of highly virulent strains of PRRSV in China since 2006 has been a major concern (Zhou and Yang, 2010). Although current vaccine-based control strategies for EAV are considered adequate, some concerns regarding efficacy (i.e. limited cross protection) and safety (i.e. shedding of vaccine virus) provide room for improvement (Balasuriya et al., 2013, McCollum et al., 1987). In contrast, whereas currently available vaccines against PRRSV generally provide protection against clinical disease, they do not consistently prevent replication of field strains and shedding of infectious virus, and especially the limited protection against heterologous field strains is a major issue (Kimman et al., 2009). The design of (novel) PRRSV vaccines that provide improved protection against both homologous and heterologous field strains is therefore of significant importance.
It has been suggested that the immune-evasive capabilities of PRRSV play an important role in reducing vaccine efficacy (Kimman et al., 2009, Wang et al., 2013b). One of the arterivirus protein domains that have been suggested to be involved in downregulating the innate immune response is papain-like protease 2 (PLP2) (Frias-Staheli et al., 2007, Sun et al., 2010, van Kasteren et al., 2012), which functions both as a deubiquitinase (DUB) and plays an essential role in the autoproteolytic maturation of the viral replicase polyproteins. Importantly, we have recently been able to show that the DUB activity of EAV PLP2 inhibits the innate immune response in infected cells, via the structure-based design of a viable mutant virus lacking this activity (van Kasteren et al., 2013). The aim of the current study was to determine in vivo whether this DUB-negative virus provides better protection against subsequent challenge infection than its parental DUB-competent counterpart. Since to date the separation of DUB and polyprotein processing functions of arterivirus PLP2 has only been successful for EAV, we have used this virus for the present animal study.
2. Methods
2.1. Cells and viruses
BHK-21 cells were cultured in Glasgow minimum essential medium (Lonza) supplemented with 5% foetal bovine serum (FBS), 10% tryptose phosphate broth, and 10 mM Hepes (pH 7.4). Primary equine lung fibroblasts (ELFs) were cultured in minimum essential medium (Lonza) supplemented with 10% FBS and grown on collagen-coated plastics for a maximum of 10 passages. Vero cells were cultured in proprietary cell culture medium (MSD Animal Health) supplemented with 1% FBS. All culture media contained 100 U/mL of penicillin and 100 mg/mL of streptomycin or neomycin.
The cloning and production of the DUB-competent (in our previous publication referred to as “wild-type”) and DUB-negative (T312A/I313V/I353R, amino acid numbering based on polyprotein) viruses (strain Bucyrus; EAN551) that were used for vaccination were described previously (van Kasteren et al., 2013). Both viruses have been thoroughly characterized in cell culture experiments, revealing no differences in replication kinetics, yet a strongly decreased DUB activity and an enhanced induction of interferon beta mRNA expression (a hallmark of innate immune activation) of the mutant virus compared to its parental counterpart (van Kasteren et al., 2013). Viral titres were determined by standard plaque assay on ELFs. For experimental challenge, we used the virulent Kentucky 1984 (KY84) strain of EAV, which has been previously described (Zhang et al., 2012).
To confirm the use of the correct virus for vaccination, the presence or absence of PLP2 mutations was established, both before and after vaccination. Before vaccination, viral RNA was isolated from the produced virus stocks using the QIAamp viral RNA mini kit (Qiagen) and converted to cDNA using RevertAid H Minus reverse transcriptase (RT) (Fermentas) and random hexamer primers. The PLP2-encoding region was subsequently PCR amplified using Pfu DNA polymerase (Fermentas) and sequenced. After vaccination, viral RNA present in the blood of four horses from each of the vaccinated groups at 4 days post vaccination was subjected to sequencing. This was done essentially as described above with the exception that the RT reaction on whole blood total RNA (see below) was performed using a primer that recognizes the EAV genome and thus specifically converts the limited amount of viral RNA present in the samples to cDNA. Primer sequences are available upon request.
2.2. Experimental vaccination and challenge of horses
The experiment was performed in accordance with European Community guidelines and national laws on animal experiments. The design of the experiment was approved by the MSD Animal Health's Committee on the Ethics of Animal Experiments (Dierexperimentencommissie), which is required by national legislation to include both MSD Animal Health employees and independent members, prior to the start of the trial (Permit Number: EXP 12.059). All efforts were made to minimize animal discomfort.
Twenty female Shetland horses (Equus ferus caballus; average age 7.5 ± 5.1 years) that tested negative for EAV-neutralizing antibodies (antibody titres were determined as described previously, Zhang et al., 2012) before the start of the experiment were randomly assigned to one of three treatment groups. After a one-week acclimatization period, horses in Group 1 (n = 9) and Group 2 (n = 9) received an intramuscular (cervical muscle) vaccination of 1 ml phosphate-buffered saline containing 1 × 107 plaque-forming units (PFU) of parental or PLP2 DUB-negative EAV, respectively. Horses in Group 3 (n = 2) were not vaccinated and were included in the study one week before challenge. At 34 days post vaccination (dpv), all horses were challenged by intranasal inoculation with 1 × 105 PFU of EAV KY84 in a total volume of 5 ml phosphate-buffered saline.
Given the fact that the viruses used for vaccination qualify as genetically modified organisms (GMO), vaccinated horses were kept in vBSL3 containment during the entire experiment. All horses were housed in groups, but Group 1 horses were kept separate from Group 2 horses to prevent any cross-contamination. Horses from Group 3 were divided among the two stables upon inclusion, without having direct contact with Group 1 or 2 horses. Water was provided ad libitum and standard feeding procedures were applied.
The general health status of the animals was checked by a veterinarian before vaccination as well as before challenge, and daily by animal care-takers during the entire course of the experiment. In addition, clinical signs were recorded daily from 0 to 14 days post challenge (dpc) and scored according to Table 1 . Rectal temperatures were taken daily from 0 to 14, and at 21 and 28 dpv, and daily from 0 to 14, and at 21 and 27 dpc. Blood samples for serum and total RNA isolation were taken every other day between 0 and 14, and at 21 and 28 dpv, and every other day between 0 and 14, and at 21 and 27 dpc. Animals were euthanized according to standard procedures at 61 dpv (27 dpc). For a schematic overview of the experimental set-up see Fig. 1 .
Table 1.
Clinical signs scoring table.
| Clinical sign | Score/day | Total score [average per animal/day] |
||
|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | ||
| Depressed, normal appetite | 1 | 1 [0.03] | ||
| Depressed, reduced appetite | 2 | 12 [0.40] | ||
| Hives, legs (mild) | 1 | 2 [0.07] | ||
| Hives, legs (marked) | 2 | 4 [0.13] | ||
| Hives, neck | 1 | 5 [0.17] | ||
| Mucopurulent eye discharge | 2 | 6 [0.04] | 22 [0.73] | |
| Muscle tremors | 2 | 2 [0.07] | ||
| Nasal serous discharge | 1 | 38 [0.28] | 40 [0.30] | 6 [0.20] |
| Petechial haemorrhages | 1 | 1 [0.03] | ||
| Red oral mucosa | 1 | 2 [0.01] | 4 [0.13] | |
| Reduced appetite | 1 | 1 [0.03] | ||
| Stiffness | 1 | 1 [0.03] | ||
| Temperature 38.5–39.0 °C | 1 | 1 [0.03] | ||
| Temperature 39.1–39.5 °C | 2 | 6 [0.20] | ||
| Temperature 39.6–40.0 °C | 3 | 9 [0.30] | ||
| Temperature > 40 °C | 4 | 24 [0.80] | ||
| Total | 38 [0.28] | 48 [0.36] | 101 [3.4] | |
Fig. 1.
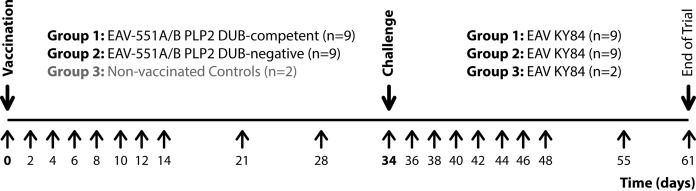
Schematic representation of the animal trial. Twenty female Shetland horses were randomly assigned to one of three groups. At the start of the experiment, horses from Groups 1 and 2 (n = 9 each) were vaccinated with PLP2 DUB-competent or DUB-negative EAV, respectively. Horses in Group 3 (n = 2) were not vaccinated. At 34 dpv, all horses were challenged with moderately virulent EAV KY84. The experiment ended at 61 dpv. Blood samples for serum and total RNA isolation were taken at the indicated days (lower arrows).
2.3. Virus neutralization assay
Blood for serum neutralizing antibody analysis was collected in 8 ml Vacuette Serum Clot Activator Tubes (Greiner Bio-One) and incubated for at least 4 h at room temperature to allow for clotting. Serum samples were subsequently collected by centrifugation at 700 × g for 15 min at 4 °C and stored at −20 °C until further use. To determine EAV neutralizing antibody titres in the serum, a virus neutralization assay was performed. Briefly, duplicate two-fold serial dilutions (1:2 to 1:4096) of serum samples were made in 96-well plates and mixed with a 50% tissue culture infective dose (TCID50) of 800 of EAV030 (van Dinten et al., 1997). After a 1-h incubation at 37 °C, 1.2 × 104 Vero cells were added to each well. Plates were subsequently incubated for 4 days at 37 °C after which each well was scored (positive or negative) for cytopathic effect (CPE) by visual inspection. The EAV neutralizing antibody titre was finally determined as the reciprocal value of the highest serum dilution at which no CPE was observed.
2.4. Quantitative reverse transcriptase PCR assay
Total RNA was isolated from whole blood collected in Tempus Blood RNA Tubes (Greiner Bio-One) using the MagMAX for Stabilized Blood Tubes RNA isolation kit (Life Technologies) according to the manufacturer's instructions. Isolated RNA was converted to cDNA using RevertAid H Minus reverse transcriptase (Fermentas) and oligo(dT)20 primer. Samples were subsequently analyzed by quantitative reverse-transcriptase (qRT) PCR on a CFX384 Touch Real-Time PCR detection system (BioRad) using iTaq Sybr Green Supermix (BioRad). Primers targeting mRNAs encoding equine β-actin (NM_001081838, 5′CCACGCCATCCTGCGTCTGG-3′, 5′ACCGCTCGTTGCCGATGGTG-3′), ISG15 (XM_001496658, 5′GAATTCCTGGTGCCCCTGAA-3′, 5′CAGTTCTGCACGACAAGCAC-3′), and MX1 (NM_001082492, 5′CGGCCAGCAGCTGCAGAAGT-3′, 5′GGCCTCCGCTCCCTGGAGAT-3′), or EAV RNA (NC_002532 and AF107279, 5′GGTTCGCGGCAACGGGTACA-3′, 5′GGTGGCGCGCTCCTGTTGAT-3′) were designed using Primer3 (Rozen and Skaletsky, 1998). The EAV-specific primer set amplifies cDNA derived from both genomic and subgenomic viral mRNAs and the forward primer includes one mismatch with KY84. The real-time PCR program was performed in triplicate, starting with 3 min at 95 °C, 30 s at 65 °C, followed by 40 cycles at 95 °C for 10 s, 65 °C for 10 s, and 72 °C for 30 s. All runs included a standard dilution series and were followed by a melting-curve analysis to confirm the identity of the reaction products.
3. Results
3.1. Vaccine viruses do not differ in the induction of fever or neutralizing antibodies
To assess whether a vaccine virus lacking PLP2 DUB activity would provide better protection against heterologous challenge than its DUB-positive parental counterpart, we performed a vaccination-challenge trial in horses. Twenty female Shetland horses were randomly assigned to one of three groups. Animals from two groups were subsequently vaccinated intramuscularly with cell culture-adapted DUB-competent (Group 1, n = 9) or DUB-negative (Group 2, n = 9) EAV. Horses from Group 3 (n = 2) were not vaccinated and served as challenge controls. All horses were intranasally challenged with virulent EAV KY84 at 34 dpv. A schematic representation of the experimental set-up including timing of vaccination, challenge, and sampling is depicted in Fig. 1. The identity of the virus used for vaccination was confirmed by sequencing of viral RNA isolated from whole blood at 4 dpv for a subset of four horses per group from Groups 1 and 2 (data not shown).
Upon vaccination, animals from Groups 1 and 2 (DUB-competent and DUB-negative virus, respectively) developed comparable levels of mild fever which reached an average maximum of 38.8 °C at 3 dpv and subsided approximately 8 dpv in both groups (Fig. 2 A). Upon challenge, only the non-vaccinated animals in Group 3 developed a fever, which reached higher levels (average maximum 40.3 °C) and lasted approximately two days longer than the mild temperature increase observed after vaccination. Neutralizing antibodies could be detected in both Groups 1 and 2 from 6 dpv onwards and no difference in titres was observed between the two groups (Fig. 2B). Titres remained stable during the course of the experiment and did not show an increase upon challenge. In the unvaccinated controls, neutralizing antibody titres could be detected at 8 days post challenge (dpc) and reached similar titres as observed after vaccination.
Fig. 2.
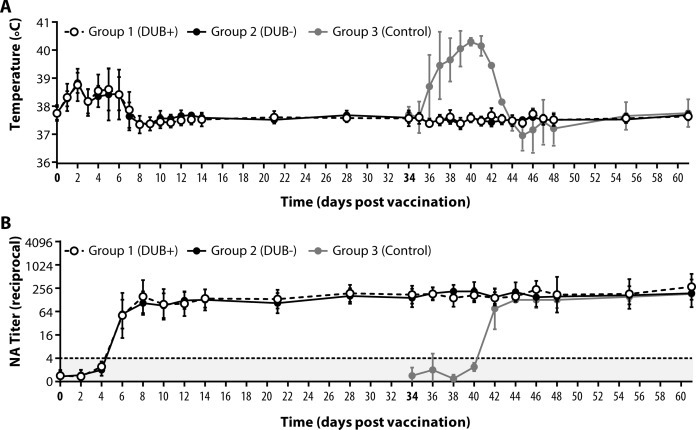
Rectal temperatures and neutralizing antibody titres. (A) Rectal temperatures were taken daily for the first 14 days post vaccination/challenge and at 21, 28, 55, and 61 dpv. (B) Neutralizing antibody (NA) titres in serum were determined using a virus neutralization assay for samples obtained every other day during the first 14 days post vaccination/challenge and at 21, 28, 55, and 61 dpv. White, black, and grey circles represent data from animals in Group 1, 2, and 3, respectively. The days of vaccination (0) and challenge (34) are depicted in bold face. The dotted line in panel B represents the limit of detection. Measurements/samples taken on the day of vaccination/challenge were obtained before infection and horses in Group 3 were included in the experiment starting at the day of challenge. Abbreviations: DUB+, deubiquitinase-competent vaccine virus; DUB−, deubiquitinase-negative vaccine virus; NA, neutralizing antibody.
From 0 to 14 dpc, clinical signs (including for example fever, nasal secretions, and loss of appetite) were recorded daily for each animal. An overall clinical signs score was subsequently determined for each animal by scoring these clinical signs according to Table 1. For example, a horse that has reduced appetite on day 5, mucopurulent eye discharge and a temperature of 39 °C on day 8, and no abnormalities on any of the other days has an overall clinical signs score of 4 (1 + 2 + 1). This resulted in an average score per animal per day of 0.28 (±0.22, Group 1) and 0.36 (±0.22, Group 2), which did not differ significantly between the two groups (Student's t-test; p > 0.05; Table 2 ). The horses in Group 3 reached an average score of 3.4 (±0.33) per animal per day, which is consistent with the fact that these animals had not been vaccinated. Thus, vaccination with either virus provided a similar high degree of protection against clinical disease.
Table 2.
Clinical signs score per animal at 0–14 dpc.
| Group 1 (DUB+) |
Group 2 (DUB−) |
Group 3 (Control) |
||||||
|---|---|---|---|---|---|---|---|---|
| Horse ID | Total score | Score/day | Horse ID | Total score | Score/day | Horse ID | Total score | Score/day |
| 1 | 5 | 0.33 | 11 | 2 | 0.13 | 10 | 54 | 3.6 |
| 2 | 2 | 0.13 | 12 | 10 | 0.67 | 20 | 47 | 3.1 |
| 3 | 0 | 0.00 | 13 | 4 | 0.27 | |||
| 4 | 4 | 0.27 | 14 | 4 | 0.27 | |||
| 5 | 0 | 0.00 | 15 | 7 | 0.47 | |||
| 6 | 3 | 0.20 | 16 | 3 | 0.20 | |||
| 7 | 8 | 0.53 | 17 | 1 | 0.07 | |||
| 8 | 9 | 0.60 | 18 | 7 | 0.47 | |||
| 9 | 7 | 0.47 | 19 | 10 | 0.67 | |||
| Average | 4.2 (±3.3) | 0.28 (±0.22) | Average | 5.3 (±3.3) | 0.36 (±0.22) | Average | 50.5 (±4.9) | 3.4 (±0.33) |
3.2. PLP2 DUB-negative vaccine virus appears to replicate slightly less efficiently than parental virus in vivo
We then assessed viral replication from 0 to 10 dpv by performing real-time qRT-PCR analysis on total RNA isolated from whole blood. As can be seen in Fig. 3 A, all vaccinated horses showed viral replication which, on average, peaked at 4 dpv in both groups. Whereas viral RNA could be detected on multiple days in all Group 1 horses (DUB-competent vaccine), in four Group 2 horses (DUB-negative vaccine), viral RNA could only be detected on a single day, i.e. 4 dpv. In addition, when comparing the average amounts of viral RNA between groups at each day post vaccination, these were consistently lower in Group 2 compared to Group 1. However, this difference was only statistically significant at 10 dpv (Student's t-test; p < 0.05).
Fig. 3.
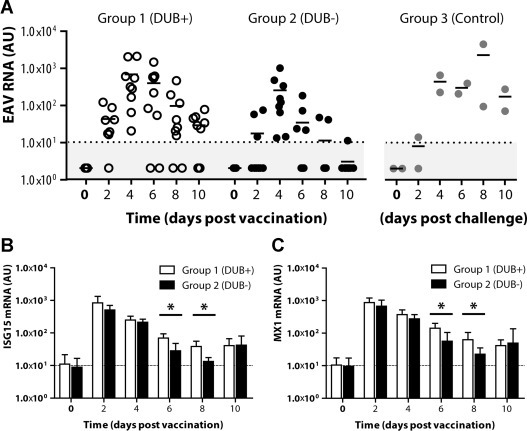
Quantitative RT-PCR analysis of viral RNA and cellular mRNA encoding ISG15 and MX1. (A) Viral replication from 0 to 10 dpv (Groups 1 and 2) or 0 to 10 dpc (Group 3) was assessed by real-time qRT-PCR analysis on total RNA isolated from whole blood using EAV-specific primers. White, black, and grey circles represent animals from Group 1, 2, and 3, respectively. Horizontal bars represent the mean for all animals on that day and the dotted line represents the limit of detection. Activation of the innate immune response was assessed by real-time qRT-PCR on total RNA isolated from whole blood using primers specific for B) ISG15- or (C) MX1-encoding mRNA. Values obtained for ISG15 and MX1 mRNA levels were normalized to the amount of β-actin mRNA. The dotted lines represent average baseline levels, error bars represent standard deviations, and asterisks indicate statistical significance (Student's t-test; p < 0.05). Abbreviations: AU, arbitrary units; DUB+, deubiquitinase-competent vaccine virus; DUB−, deubiquitinase-negative vaccine virus.
Upon challenge with the virulent EAV KY84 strain, both non-vaccinated control horses (Group 3) showed virus replication, with amounts of viral RNA reaching levels comparable to those produced by the vaccine viruses (Fig. 3A). In contrast, amounts of challenge virus RNA remained below the detection limit of the assay in all vaccinated horses from Groups 1 and 2. This result was consistent with the observed appearance of neutralizing antibodies after vaccination in all horses and the low clinical sign scores in both vaccinated groups after challenge.
3.3. No major differences between vaccine viruses in the induction of ISG15 and MX1 mRNA expression could be detected in RNA extracted from whole blood
Finally, we assessed the induction of an innate immune response by parental and PLP2 DUB-negative virus from 0 to 10 dpv by means of real-time qRT-PCR on RNA extracted from whole blood. Since we were unable to detect mRNA encoding type I interferons (a hallmark of innate immune activation), we instead focussed on two highly expressed interferon-stimulated genes encoding ISG15 and MX1, respectively. Both of these genes showed an approximately 2-log increase of expression at 2 dpv, which did not significantly differ between animals from Groups 1 and 2 (Fig. 3B and C). At 6 and 8 dpv, the expression of ISG15 and MX1 mRNA was significantly lower in the animals vaccinated with the PLP2 DUB-negative virus (Group 2) than in those vaccinated with the parental virus (Group 1) (Student's t-test; p < 0.05), which might be explained by the slight decrease in replication efficiency of the former virus.
4. Discussion
We have previously shown that mutant EAV lacking PLP2 DUB activity induces a more potent innate immune response than its DUB-competent parental virus upon infection in cell culture (van Kasteren et al., 2013), and hypothesized that this feature might be included in an improved arterivirus vaccine. In order to assess whether a PLP2 DUB-negative vaccine virus provides better protection than its DUB-competent counterpart against challenge with a virulent EAV strain, we performed a vaccination-challenge trial in horses. The data obtained in this study showed that in contrast to what was previously seen in cell culture experiments (van Kasteren et al., 2013), the PLP2 mutant virus appears to replicate slightly less efficiently than its parental virus in vivo (Fig. 3A). Even though the DUB-negative virus is clearly replication competent, this finding does suggest a role for PLP2 DUB activity in supporting replication in vivo, e.g. by inhibiting the innate immune response. In addition, despite this slight decrease in replication efficiency, the levels of protection against clinical disease (Table 2), the antibody response (Fig. 2B), and the innate immune response (assessed based on the expression of two interferon-stimulated genes; Fig. 3B and C) induced by the PLP2 DUB-negative virus was comparable to that induced by the parental virus. Surprisingly, we did not observe an increase in EAV-neutralizing antibody titres upon challenge infection of vaccinated animals (Fig. 2B). A possible explanation for this finding is that the virus dose used for challenge was too low, resulting in its efficient clearance before (re-)activation of the antibody response. Taken together, the present study could not be used to substantiate (or refute) our hypothesis that a DUB-negative virus provides better protection against challenge than its DUB-positive counterpart. The main reason for this seems to be the fact that the parental virus already provides a very high degree of protection, which as it turned out could not be detectably improved using this experimental set-up. In hindsight, one solution might have been to perform a more severe challenge, for example by using a higher virus dose for inoculation or a more heterologous and/or virulent strain than KY84. As PRRSV vaccines are in general less efficacious than EAV vaccines, another option would be to perform a similar trial with a PRRSV vaccine in pigs, but this will first require the design of viable PRRSV PLP2 DUB-negative mutants.
In contrast to what was previously found in cell culture-based assays (van Kasteren et al., 2013), we did not detect an increased activation of innate immunity by the PLP2 DUB-negative virus compared to its parental virus in vivo. However, it needs to be noted that we have thus far assessed the expression of only two interferon-stimulated genes, whereas many more exist. It therefore remains a possibility that differences do exist in the expression of other interferon-stimulated genes. Similarly, the induction of interferon-stimulated genes is only one of several consequences of the activation of innate immune signalling. It therefore remains possible that the DUB-negative vaccine virus differs from the parental virus in some other respect pertaining to immunity, for example the activation of cell-mediated adaptive immunity. Also, PLP2 DUB activity likely constitutes only one of several innate immune evasion strategies employed by arteriviruses. For example, nonstructural protein 1 (nsp1) of EAV was recently shown to inhibit the induction of interferon beta in a luciferase reporter assay (Go et al., 2014) and the PRRSV nsp4 main protease was recently shown to cleave the innate immune signalling factor NEMO (Huang et al., 2014). Also nsp1α (Chen et al., 2010, Han et al., 2013, Kim et al., 2010, Song et al., 2010), nsp1β (Beura et al., 2010, Chen et al., 2010, Patel et al., 2010, Wang et al., 2013a), and the nucleocapsid protein (Sagong and Lee, 2011) have been proposed to counteract innate immune activation. Taken together, the PLP2 DUB activity appears to constitute only a part of the total repertoire of arterivirus innate immune evasion strategies. For this reason, its contribution to the inhibition of innate immunity perhaps cannot be readily detected in the in vivo experimental set-up used here. For example, detecting more prominent consequences of its inactivation may depend on the details of the animal study, including specific properties of viruses and horses used. Nevertheless, the apparently limited negative effect on viral replication of PLP2 DUB-mutations does open up possibilities for combining mutations in multiple domains involved in immune evasion, thereby potentially synergistically increasing the overall immunogenicity of the virus.
5. Conclusion
Notably, arteriviruses are not the only group of viruses that were shown to harbour DUB activity. For example, coronaviruses (including SARS- and MERS-CoV) encode papain-like proteases that display DUB activity (for a review, see Mielech et al., 2014). The OTU proteases of nairoviruses have also been found to possess DUB activity (Frias-Staheli et al., 2007). Like arterivirus PLP2, these viral DUBs have been implicated in innate immune evasion (Frias-Staheli et al., 2007, Mielech et al., 2014) and are therefore potential targets for the design of novel vaccines. Interestingly, we have recently been able to show that the DUB and polyprotein processing functions of the papain-like protease encoded by MERS-CoV can also be separated by targeted mutagenesis (Bailey-Elkin et al., 2014). Taken together, the data obtained in this study definitely warrant further in vivo examination of the consequences of knocking out arterivirus PLP2 DUB activity as well as other immune-evasive activities to improve vaccine efficacy. The data obtained in such studies could subsequently also be used as preliminary proof of principle for the design of novel vaccines for other viruses, including corona- and nairoviruses.
Conflict of interest statement
MK, EJS, and PBK have filed a provisional patent application that relates to some aspects of this work.
Acknowledgements
We would like to thank the animal caretakers at the MSD-AH animal facility for their efforts. This research was supported in part by the Division of Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW) through ECHO grant 700.59.008 to MK and EJS.
References
- Bailey-Elkin B.A., Knaap R.C., Johnson G.G., Dalebout T.J., Ninaber D.K., van Kasteren P.B., Bredenbeek P.J., Snijder E.J., Kikkert M., Mark B.L. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasuriya U.B., Go Y.Y., Maclachlan N.J. Equine arteritis virus. Vet. Microbiol. 2013;167:93–122. doi: 10.1016/j.vetmic.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Sarkar S.N., Kwon B., Subramaniam S., Jones C., Pattnaik A.K., Osorio F.A. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J. Virol. 2010;84:1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Lawson S., Sun Z., Zhou X., Guan X., Christopher-Hennings J., Nelson E.A., Fang Y. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology. 2010;398:87–97. doi: 10.1016/j.virol.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Staheli N., Giannakopoulos N.V., Kikkert M., Taylor S.L., Bridgen A., Paragas J., Richt J.A., Rowland R.R., Schmaljohn C.S., Lenschow D.J., Snijder E.J., Garcia-Sastre A., Virgin H.W.T. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y.Y., Li Y., Chen Z., Han M., Yoo D., Fang Y., Balasuriya U.B. Equine arteritis virus does not induce interferon production in equine endothelial cells: identification of nonstructural protein 1 as a main interferon antagonist. Biomed. Res. Int. 2014;2014:420658. doi: 10.1155/2014/420658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Du Y., Song C., Yoo D. Degradation of CREB-binding protein and modulation of type I interferon induction by the zinc finger motif of the porcine reproductive and respiratory syndrome virus nsp1alpha subunit. Virus Res. 2013;172:54–65. doi: 10.1016/j.virusres.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Huang C., Zhang Q., Guo X.K., Yu Z.B., Xu A.T., Tang J., Feng W.H. Porcine reproductive and respiratory syndrome virus nonstructural protein 4 antagonizes IFNbeta expression by targeting NEMO. J. Virol. 2014 doi: 10.1128/JVI.01396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O., Sun Y., Lai F.W., Song C., Yoo D. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology. 2010;402:315–326. doi: 10.1016/j.virol.2010.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimman T.G., Cornelissen L.A., Moormann R.J., Rebel J.M., Stockhofe-Zurwieden N. Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine. 2009;27:3704–3718. doi: 10.1016/j.vaccine.2009.04.022. [DOI] [PubMed] [Google Scholar]
- McCollum W.H., Timoney P.J., Roberts A.W., Willard J.E., Carswell G.D. Response of vaccinated and non-vaccinated mares to artificial insemination with semen from stallions persistently infected with equine arteritis virus. Fifth International Conference on Equine Infectious Diseases, 1988; Lexington, KY; 1987. pp. 13–18. [Google Scholar]
- Mielech A.M., Chen Y., Mesecar A.D., Baker S.C. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014 doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D., Nan Y., Shen M., Ritthipichai K., Zhu X., Zhang Y.J. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J. Virol. 2010;84:11045–11055. doi: 10.1128/JVI.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H.J. Primer3 on the WWW for general users and for biologist programmers. In: Misener S.K.S., editor. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press; Totowa, NJ: 1998. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sagong M., Lee C. Porcine reproductive and respiratory syndrome virus nucleocapsid protein modulates interferon-beta production by inhibiting IRF3 activation in immortalized porcine alveolar macrophages. Arch. Virol. 2011;156:2187–2195. doi: 10.1007/s00705-011-1116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Krell P., Yoo D. Nonstructural protein 1alpha subunit-based inhibition of NF-kappaB activation and suppression of interferon-beta production by porcine reproductive and respiratory syndrome virus. Virology. 2010;407:268–280. doi: 10.1016/j.virol.2010.08.025. [DOI] [PubMed] [Google Scholar]
- Sun Z., Chen Z., Lawson S.R., Fang Y. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J. Virol. 2010;84:7832–7846. doi: 10.1128/JVI.00217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dinten L.C., den Boon J.A., Wassenaar A.L., Spaan W.J., Snijder E.J. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. U. S. A. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren P.B., Bailey-Elkin B.A., James T.W., Ninaber D.K., Beugeling C., Khajehpour M., Snijder E.J., Mark B.L., Kikkert M. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E838–E847. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren P.B., Beugeling C., Ninaber D.K., Frias-Staheli N., van Boheemen S., Garcia-Sastre A., Snijder E.J., Kikkert M. Arterivirus and nairovirus ovarian tumor domain-containing deubiquitinases target activated RIG-I to control innate immune signaling. J. Virol. 2012;86:773–785. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Nan Y., Yu Y., Zhang Y.J. Porcine reproductive and respiratory syndrome virus Nsp1beta inhibits interferon-activated JAK/STAT signal transduction by inducing karyopherin-alpha1 degradation. J. Virol. 2013;87:5219–5228. doi: 10.1128/JVI.02643-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xiao Y., Opriessnig T., Ding Y., Yu Y., Nan Y., Ma Z., Halbur P.G., Zhang Y.J. Enhancing neutralizing antibody production by an interferon-inducing porcine reproductive and respiratory syndrome virus strain. Vaccine. 2013;31:5537–5543. doi: 10.1016/j.vaccine.2013.09.023. [DOI] [PubMed] [Google Scholar]
- Zhang J., Go Y.Y., Huang C.M., Meade B.J., Lu Z., Snijder E.J., Timoney P.J., Balasuriya U.B. Development and characterization of an infectious cDNA clone of the modified live virus vaccine strain of equine arteritis virus. Clin. Vaccine Immunol. 2012;19:1312–1321. doi: 10.1128/CVI.00302-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Yang H. Porcine reproductive and respiratory syndrome in China. Virus Res. 2010;154:31–37. doi: 10.1016/j.virusres.2010.07.016. [DOI] [PubMed] [Google Scholar]


