Highlights
-
•
The circulation of G8 RVA strains in calves with diarrhea detected first time in Turkey.
-
•
First report on whole genome of G8P[5] RVA strains from calves with diarrhea in Turkey.
-
•
VP7 gene of the both Turkish bovine RVA strains showed the closest with human RVA strains detected in Europe and Africa.
-
•
The genotype constellation of the strains is G8-P[5]-I2-R2-C2-M2-A3-N2-T6-E2-H3.
-
•
The findings raise provocative questions related to strain-specific vaccine effectiveness in herds where commercial RVA vaccines are routinely utilized.
Keywords: Calves, Rotavirus, G8P[5], Whole genome sequencing, Vaccine
Abstract
Group A rotaviruses (RVA) are regarded as major enteric pathogens of large ruminants, including cattle. Rotavirus vaccines administered to pregnant cows are commonly used to provide passive immunity that protects newborn calves from the clinical disease. In this study we report the detection of RVA from calves with severe diarrhea in a herd regularly vaccinated to prevent enteric infections including RVA. Diarrheic disease was observed in newborn calves aged 4–15 days, with high morbidity and mortality rates, but no diarrhea was seen in adult animals. Rotavirus antigen was detected by enzyme-immunoassay in the intestinal content or the fecal samples of all examined animals. Besides RVA, bovine coronavirus and bovine enteric calicivirus were detected in some samples. Selected RVA strains were characterized by whole genome sequencing. Two strains, RVA/Cow-wt/TUR/Amasya-1/2015/G8P[5] and RVA/Cow-wt/TUR/Amasya-2/2015/G8P[5] were genotyped as G8-P[5]-I2-R2-C2-M2-A3-N2-T6-E2-H3 and showed >99% nucleotide sequence identity among themselves. This genomic constellation is fairly common among bovine RVA strains; however, phylogenetic analysis of the G8 VP7 gene showed close genetic relationship to some European human RVA strains (up to 98.4% nt identity). Our findings is the first indication regarding the circulation of G8 RVA strains in Turkey. Given that the administered RVA vaccines contained type G6 and G10 VP7 antigens some concerns raised with regard to the level of heterotypic protection elicited by the vaccine strains against circulating bovine G8 RVA strains. Enhancement of surveillance of circulating RVA strains in calves across Turkey is needed to support ongoing vaccination programs.
1. Introduction
Neonatal diarrhea in calves is an important disease, which causes economic loss in the cattle industry due to high mortality and morbidity (Cho et al., 2013). There are various infectious agents which cause diarrhea and various factors including herd management, animal nutrition and environmental conditions affect the severity of the disease (Cho et al., 2013, Kaplon et al., 2013). Rotaviruses (RV) are one of the major causative agents of neonatal diarrhea in calves worldwide (Okada and Matsumoto, 2002, Alkan et al., 2010, Martella et al., 2010, da Silva Medeiros et al., 2015). Vaccines containing RV antigens are commercially available and commonly used to prevent RV diarrhea in young calves.
With a 11-segmented dsRNA genome that is enclosed in a triple-layered protein capsid RVs are members of the Reoviridae family (Lee et al., 2000, Reidy et al., 2006). Based on the genetic and antigenic characteristics of the inner capsid protein, VP6, RVs are divided into eight groups (or species; designated from A to H) (Matthijnssens et al., 2009) and new data suggest that a candidate ninth group can also be distinguished (Mihalov-Kovács et al., 2015). Group A rotaviruses (RVA) are the most commonly detected RV species in the gastrointestinal infection in cattle, although Group B and C rotaviruses are also regularly identified. RVAs are classified into G (glycoprotein) and P (protease sensitive) types according to the sequence diversity of the outer capsid proteins VP7 and VP4, respectively, which independently generate serotype-specific neutralizing antibodies in vivo (Monini et al., 2008, Matthijnssens et al., 2011, Papp et al., 2013). Increased diversity in RVA strains is generated principally by accumulation of point mutations and by reassortment of cognate genes. In addition, interspecies transmission is another important way to increase viral diversity in a host species (Papp et al., 2013). To date, at least 27 G and 37 P genotypes have been defined in mammals and avians (da Silva Medeiros et al., 2015). Among these, the most common G and P genotypes in cattle are G6, G8, G10 (for VP7) and P[1], P[5], P[11] P[15], and P[21] (for VP4) (Martella et al., 2010, Papp et al., 2013). More recently, the classification system of RVAs was extended to include the remaining capsid and non-structural protein genes; this new system denotes the VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5/6 genes of a rotavirus strain by the descriptor Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx, where x indicates the genotype number (Matthijnssens et al., 2011).
Determination of the genotype(s) of bovine RVA strains that causes the infection is amongst the most important factors for the implementation of effective vaccination to protect newborn calves from RVA infection and severe dehydrating diarrhea (Saif and Fernandez, 1996, Dulgheroff et al., 2012). Calves could be protected from diarrhea by appropriate management techniques and vaccination strategies. However, the potential of gene reassortment among various strains, the antigenic and genetic diversity between vaccine and field strains, the insufficient protection raised by vaccine strains against heterotypic strains and inappropriate vaccination strategy might be reasons for vaccine failure and thus remain a concern (Papp et al., 2013, da Silva Medeiros et al., 2015).
In Turkey, RVAs have been associated repeatedly with diarrhea in calves (Alkan et al., 2004, Alkan et al., 2010, Can-Sahna and Alkan, 2003, Ozkul et al., 2002) and RVA-specific antibodies have been demonstrated in the sera of adult bovines (Alkan et al., 2004, Can-Sahna and Alkan, 2003). In a study on VP4 and VP7 diversity of the RVAs circulating in Turkey, RVAs isolated from diarrheic calves from several Turkish geographical areas between 1997 and 2008 (Alkan et al., 2010). In this study, the majority (40/53) of the RVAs were characterized as G6, followed by G10 (15/53), while G8 viruses were not detected. The most common VP4 type was P[11] followed by P[5]. Additionally, a large outbreak of enteric disease with high morbidity and mortality rates in young kids was reported as associated with the RVA strain (RVA/goat-tc/TUR/Kirklareli/2007/G8P[1]) characterized as G8P[1], with E2 NSP4 and VP6 I2 genotype (Alkan et al., 2012).
In this study, we detected and characterized RVA strains from a herd regularly vaccinated against RVA and found differences in the virion components between the vaccine strains and the field strains. This finding may have implications for specific prevention of RVA disease in cattle herds.
2. Material and methods
2.1. History of the herd and the outbreak
During January and February 2015, an outbreak of diarrhea occurred in newborn calves at age 4–15 days in a large Turkish cattle herd (no. of cows, 917). The outbreak was associated with high mortality among newborn calves born in January and February 2015 (14 of 73 calves died). In this herd a systematic RV vaccination program was carried on in the past 3 years. Due to economic reasons the management decided to change the vaccine type in 2014. ‘Vaccine A’ was used in 2012–2013 and contained two RVA genotypes, G6 and G10, (in addition to coronavirus, Escherichia coli having the K99 pili adherence factor and Clostridium perfringens type C), whereas ‘vaccine B’ contained a single RVA genotype, G6P[5], (in addition to coronavirus, Escherichia coli with K99 pilus type),
2.2. Diagnosis of the infection
In February, specimens from the diarrhea outbreak were obtained for diagnostic investigation. The intestinal content from a dead calf and four fecal samples from calves with diarrhea were tested for rotavirus, coronavirus, E. coli K99 antigens by using a commercial ELISA kit (IDEXX Rota-Corona-K99, Ag Test) according to manufacturer’s instructions. For this reason, microplates coated with a mix of antibodies against 3 mentioned antigens were used. The faeces diluted (1/10) in dilution buffer were plated triplicate into the wells and incubated on the microplate for 30 min at room temperature (approximately 25 °C). After this first incubation step, the plate was washed by manually using the washing solution, and then each pathogen specific conjugates, peroxidase-labelled anti-pathogen monoclonal antibodies, were added to the related wells. The plate was then incubated for 30 min at room temperature again. Following the final wash, the chromogen (tetramethylbenzidine) was added to each well and the plate was held at room temperature away from light for 10 min. A stop solution was added and the optical densities were measured at 450 nm using an ELISA Reader. The ELISA reader optical density data was calculated according to the manufacturer’s instruction. The samples were also tested for bovine enteric caliciviruses (BEC) by RT-PCR (Park et al., 2007, Park et al., 2008).
2.3. Genotyping and genome sequencing
All ELISA positive samples were tested for possible RVA G genotypes by RT-PCRs using oligonucleotides specific for RVA G6 and G10 genotypes. (Iturizza-Gomara et al., 1999, Gouvea et al., 1990, Gouvea et al., 1994, Isegawa et al., 1993). Briefly, synthesis of cDNA was achieved following denaturation of RNA at 70 °C for 5 min. The cDNA was synthesized using Moloney Murine Leukemia Virus (MMLV) reverse transcriptase (RT) (Fermentas, Lithuania) and random hexamers (Fermentas, Lithuania), by incubating at 25 °C for 10 min, and thereafter at 37 °C for 1 h. MMLV-RT was therefore inactivated at 70 °C for 10 min (Iturizza-Gomara et al., 1999). Detection and characterization of BRV was performed as described elsewhere (Gouvea et al., 1990, Gouvea et al., 1994, Isegawa et al., 1993) with minor modifications using oligonucleotide primers designed by same researchers. The resulting amplicons were analyzed on 1.5% agarose gel after electrophoresis at 80 V for 30 min and visualized under ultraviolet light.
Following these laboratory diagnosis, two RVA strains were selected for whole-genome sequencing using the protocol from published studies (Dóró et al., 2014, Mihalov-Kovács et al., 2015). In brief, after nuclease treatment, viral RNA was extracted by using Direct-zol RNA Mini Prep Kit (Zymo Research) according to manufacturer’s instructions. Random RT-PCR was carried out and then gel extraction of products was processed to start barcoded library preparation for Ion Torrent New Generation sequencing. Sequencing was carried out on a 316 chip using the 200 bp sequencing protocol. Raw sequencing data were evaluated by the CLC Genomics Workbench version 7 (CLC Bio-Qiagen, Aarhus, Denmark). By using a combination of de novo assembly and reference sequence mapping a single consensus sequence was obtained for all 11 viral gene segments of both bovine RVA strains, RVA/Cow-wt/TUR/Amasya-1/2015/G8[5] and RVA/Cow-wt/TUR/Amasya-2/2015/G8[5].
For each genome segment, multiple sequence alignments were prepared by the Muscle algorithm as implemented in Aliview Software (Edgar, 2004, Larsson, 2014). Cognate sequences of reference RVAs representing different genotypes for all genes were retrieved from GenBank through the Blast engine. Phylogenetic analyses of the full-length nucleotide sequences of the VP1-4, VP6, VP7 and NSP1-5 genes of the two field RVA strains, RVA/Cow-wt/TUR/Amasya-1/2015/G8[5] and RVA/Cow-wt/TUR/Amasya-2/2015/G8[5], were performed using MEGA 6.06 software (Tamura et al., 2013) The phylogenetic trees were constructed using the neighbor-joining method and p-distance nucleotide substitution model and the statistical significance was estimated by bootstrap analysis (500 replicates). The nucleotide identity table was computed in CLC Main Workbench (CLC Bio-Qiagen, Aarhus, Denmark)
Genome sequences of RVA/Cow-wt/TUR/Amasya-1/2015/G8[5] and RVA/Cow-wt/TUR/Amasya-2/2015/G8[5] were deposited in GenBank under the following accession numbers: KX212865-KX212886.
3. Results
3.1. Laboratory diagnosis
Diagnostic tests indicated that all samples were positive for bovine RVA. In addition, one sample was found positive for bovine coronavirus and two samples tested positive for BEC (data not shown). All samples were negative for enterotoxigenic E.coli K99 antigens. In addition, Clostridium perfringens and Cryptosporidium spp. were not detected in an independent laboratory where samples from the outbreak were also submitted for laboratory diagnosis.
3.2. Genotyping
All samples positive for RVA were tested in genotyping assay for G6 and G10 genotypes that are the only genotypes reported in Turkey to identify the VP7 genotype. However, the samples gave no specific signals with the G6- and G10-specific primers. Previous studies indicated that a random-primed amplification method combined with next generation sequencing is useful in the determination of whole genotype constellation of RVA strains. Thus, a published method was implemented to identify and characterize the genotypes of the epizootic bovine RVA strains (Dóró et al., 2014, Mihalov-Kovács et al., 2015).
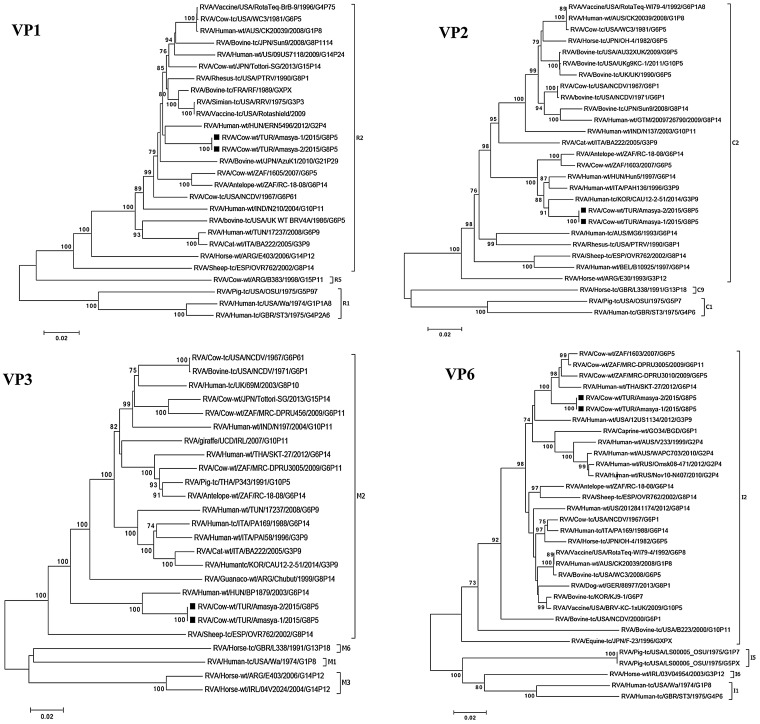
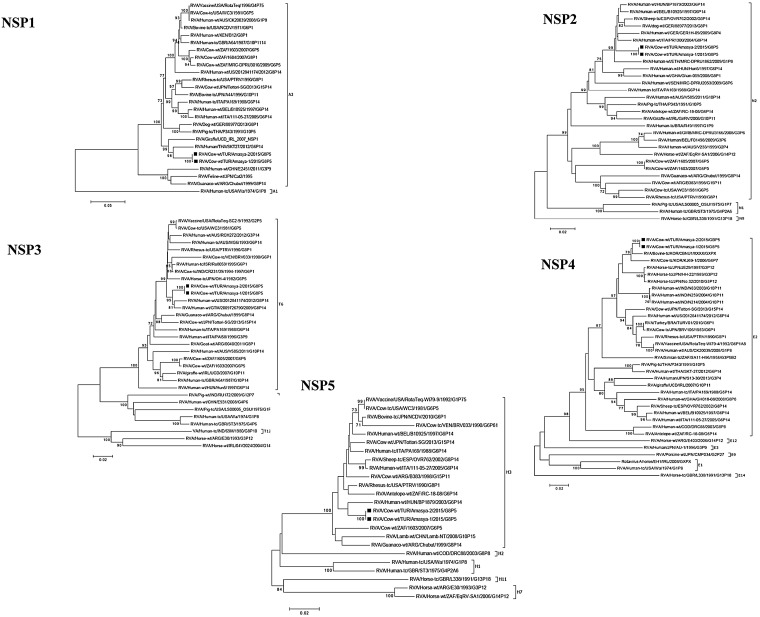
Two samples positive for bovine RVA strains selected for next generation sequencing represented a common bovine RVA genotype constellation, G8-P[5]-I2-R2-C2-M2-A3-N2-T6-E2-H3 and these two strains shared very high genetic similarities in all 11 genes (Fig. 1, Fig. 2, Fig. 3 ). The Turkish RVA strains were named as RVA/Cow-wt/TUR/Amasya-1/2015/G8[5] and RVA/Cow-wt/TUR/Amasya-2/2015/G8[5].
Fig. 1.
Phylogenetic trees of the full-length nucleotide sequences of VP4 and VP7 genes of RVA strains identified in this study with those of other RVAs selected in GenBank. The study strains are indicated by a black square. The statistical significance was estimated by bootstrap method (500 pseudo-replicates) and values of <70% are omitted.
Fig. 2.
Phylogenetic trees of the full-length nucleotide sequences of VP1, VP2, VP3 and VP6 genes RVA strains identified in this study with those of other RVAs selected in GenBank. The study strains are indicated by a black square. The statistical significance was estimated by bootstrap method (500 pseudo-replicates) and values of <70% are omitted.
Fig. 3.
Phylogenetic trees of the full-length nucleotide sequences of NSP1, NSP2, NSP3, NSP4 and NSP5 genes of RVA strains identified in this study with those of other RVAs selected in GenBank. The study strains are indicated by a black square. The statistical significance was estimated by bootstrap method (500 pseudo-replicates) and values of <70% are omitted.
3.3. Phylogenetic analysis
3.3.1. Structural genes
In the analysis of the neutralization antigen coding genes, VP7 and VP4, we observed an inverse relationship with homologous and heterologous RVA strains. For VP7 gene, both Turkish bovine RVA strains showed the closest relationship and shared up to 98.4% nt identity with human RVA strains detected in Europe and Africa (e.g., GER1H-09, SI-885, and 6810/ARN). In contrast, in the analysis of the VP4 gene, we found that both outbreak strains shared the closest genetic relationship with other Turkish bovine RVA strains (nt, 98.3%) and also clustered with the common bovine strain, UK, and its descendants.
Concerning the VP1 gene both Turkish RVA strains showed fairly close genetic relationship with other typical bovine RVA strains, such as RF (95.8% nt identity), WC3 (95.3% nt identity) and NCDV (94.7% nt identity), and reassortant heterologous RVA strains, including the simian origin RRV (the parental strain of the Rota shield human vaccine) (95.5% nt identity) and a human G2P[4] strain (ERN5496) (95.2% nt identity) detected in Hungary. In the VP2 gene phylogeny the Turkish strains grouped with African bovine (e.g., RC-18-08, 95.1% nt identity) and European/Asian bovine-like human RVA strains (e.g., Hun5 96.9% nt identity) detected in humans within the C2 genotype. In the VP3 gene phylogeny we observed that our strains formed a common cluster with the Hungarian zoonotic human G6P[14] RVA strain, BP1879 (94.9% nt identity). Analysis of the VP6 gene revealed close genetic relationship among the Turkish outbreak strains and African bovine RVA strains (up to 97.1% nt identity). In addition, this branch included zoonotic human G6P[14] RVA strain from Thailand.
3.3.2. Non-structural genes
The NSP1 gene of strains RVA/Cow-wt/TUR/Amasya-1/2015/G8P[5] and RVA/Cow-wt/TUR/Amasya-2/2015/G8P[5] exhibited the maximum nucleotide sequence identity (up to 94.2% respectively) with the cognate gene of the human G6P[14] strain, SKT-27 from Thailand. Of interest, in the phylogenetic analysis of NSP2, NSP3, and NSP5 genes, similarly close relationship was identified with other zoonotic human P[14] strains as seen in other gene analysis (e.g., NSP2. BP1879, 97.4% nt identity; NSP3, 2012841174, 97.4%; NSP5, BP1879, 97.1%) (Bányai et al., 2009, Mijatovic-Rustempasic et al., 2015). However, in the NSP4 gene phylogeny, the Turkish strains tended to cluster with other bovine RVA strains from Korea sharing up to 98.1% nt identity with those strains.
4. Discussion
In this study, the causative agent of an outbreak of calf diarrhea, which occurred in the Black Sea region of Turkey during early 2015 was investigated. In addition to routine bacteriological procedures, molecular detection methods were utilized to detect some of the major enteric viruses (ie. RVA, BEC and BCoV). As a result, while RVA was detected in all samples examined, BEC or BCoV was detected only in a few samples. Altogether, results of the diagnostic investigations and epidemiological remarks as age of affected calves and severe clinical symptoms, etc. suggested that this high mortality outbreak of bovine neonatal diarrhea was caused by RVA.
This finding was somewhat unexpected given that the herd was vaccinated against RVA. Thus we sought to investigate the genetic features of the causative RVA strains. Random primed RT-PCR coupled with high-throughput sequencing detected two G8P[5] RVA strains from fecal specimen or intestinal content. The two strains, RVA/Cow-wt/TUR/Amasya-1/2015/G8P[5] and RVA/Cow-wt/TUR/Amasya-2/2015/G8P[5], were essentially identical, suggesting the outbreak was caused by the introduction of a single strain. Phylogenetic analysis showed that these two strains presented a G8-P[5]-I2-R2-C2-M2-A3-N2-T6-E2-H3 genomic constellation, which is a typical bovine genotype constellation of RVAs. However, only the VP4, VP6, and NSP4 gene phylogenies were consistent with the bovine origin of the identified strains. In other genes (such as VP1, VP2, VP3 and NSP2), we observed genetic relationship between the Turkish strains and other ruminant RVAs and zoonotic human RVA strains. This apparent genetic relationship between bovine and ovine RVA strains and zoonotic human RVA strains shows the complex interaction and pattern of gene flow among heterologous RVA strains, which may be multidirectional among ruminants and typically leads to dead-end infection from a ruminant host to human beings. Moreover, the VP7 gene clustered with human G8 RVA strains reported from Croatia, Germany and Slovenia (Steyer et al., 2007, Pietsch et al., 2009, Delogu et al., 2013) and they represented a separate branch on the phylogenetic tree clearly distinct from other bovine-like G8 strains and caprine RVA strains detected previously in Turkey (represented by RVA/goat-tc/TUR/Kirklareli/2007/G8P[1]) (Alkan et al., 2012). Although it has been hypothesized for a while that in many instances the G8 VP7 genotype may have originated from the bovine host through interspecies transmission and reassortment (Adah et al., 2003, Chitamber et al., 2011, Ghosh et al., 2011) the putative heterologous RVA strain is seldom identified in these studies. The new sequence information generated in our study helps identify the host species origin of the contemporary European lineage of human G8 RVA strains by providing some evidence that the VP7 gene of these human-animal reassortant strains originate from a bovine host. At present the spatial connection between the Turkish bovine and the European human cases is not clear; however Turkey imports large number of live animals from Europe each year (http://ec.europa.eu/agriculture/beef-veal/presentations/market-situation_en.pdf) and it is possible that the identified Turkish G8 strain was imported from a European country more recently. The hypothesis of a relatively recent introduction is supported by findings of past RVA strain surveillance data conducted over a 12-year period from 1997 to 2008 that indicated a lack of circulation of G8 strains in Turkish cattle herds. Thus, this is the first description of the G8 genotype in Turkish bovine RVAs, even though this same genotype was reported as a cause of high mortality diarrhea outbreak in two goat flocks (Alkan et al., 2012). It remains to be studied whether there are some factors responsible for restriction of host species jump of G8 RVAs from goats to cattle that could have prevented the transmission of caprine G8 RVA strains to emerge in calf herds.
The finding of G8 RVAs in a vaccinated cattle herd raises concerns related to strain-specific vaccine effectiveness. It is well known that VP7 and VP4 are involved in viral neutralization in vitro and likely in vivo, and the binary combination classifying these antigens has been used in many molecular epidemiological studies to describe the distribution of field RVA strains and to develop vaccination strategy in animals and also humans (Delogu et al., 2013). Detection of a G8P[5] RVA strain in a herd routinely vaccinated against RVA infection by using commercial, heterotypic strains is an interesting finding and raise some questions. Firstly, which were the circulating RVA antigen type specificities during and before the implementation of vaccination? Secondly, at what extent did commercial vaccines provide heterologous protection against RVA strains in Turkey? Thirdly, did vaccine strain specificity influence the emergence of a heterotypic strain? There are many notifications from different parts of the world that indicate the occurrence of neonatal diarrhea after vaccination caused by genotypes either identical to or different from that of represented by RVA vaccines (Lorenzetti et al., 2011). Lu et al. (1994) reported that due to the various level of cross protection between different P genotypes, protection of calves from mothers which were vaccinated with particular P type(s) that is different from that of seen in the wild-type strains, was failed. It is reported that persistence of rotaviral diarrhea could be caused by different P genotypes identified from the specimens although using G6P[1] vaccine strains for years in America (Lu et al., 1994). In the present study we identified a genotype G8P[5] field strain that caused severe diarrhea and deaths in a herd immunized with inactivated vaccine strain containing genotype G6P[5]. Thus, the P type specificity was shared between the vaccine and the field strain, while the G type specificity was different, pointing out the possible importance of the other neutralization antigen, the VP7.
In summary, current rotavirus vaccination strategies in cattle are mainly based on inactivated vaccines that are composed of the virion proteins only and are administered to pregnant cows to protect newborn calves at the early age when clinical manifestations of RVA diarrhea are the most severe. Reasons behind insufficient heterotypic immunity induced by a mono- or bivalent commercial RVA vaccines in calves are to be clarified. It remains important to perform rotavirus strain surveillance program to reveal relevant genetic changes in the rotavirus genome, to detect possible new genotypes and to evaluate the effectiveness of vaccination in herds where commercial RVA vaccines are routinely utilized.
Conflicts of interest
None.
Acknowlegments
IK was supported by the International Doctoral Research Fellowship Program (2214A) of Scientific and Technological Research Council of Turkey (TUBITAK) at Institute for Veterinary Medical Research, Centre for Agricultural Research, Hungarian Academy of Sciences, Budapest, Hungary.
References
- Adah M.I., Nagashima S., Wakuda M., Taniguchi K. Close relationship between G8-serotype bovine and human rotaviruses isolated in Nigeria. J. Clin. Microbiol. 2003;41:3945–3950. doi: 10.1128/JCM.41.8.3945-3950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan F., Burgu I., Can-Sahna K., Cokcalıskan C. Yeni dogan buzagı ishallerine karşı ticari ası ile asılanan sıgırlardan dogan yavrularda pasif bagışıklık duzeyi. Ankara Univ Vet Fak Derg. 2004;51:47–53. [Google Scholar]
- Alkan F., Ozkul A., Oguzoglu T.C., Timurkan M.O., Caliskan E., Martella V., Burgu I. Distribution of G (VP7) and P (VP4) genotypes of group A bovine rotaviruses from Turkish calves with diarrhea, 1997–2008. Vet. Microbiol. 2010;141:231–237. doi: 10.1016/j.vetmic.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Alkan F., Gulyaz V., Timurkan M.O., Iyisan S., Ozdemir S., Turan N., Buonavoglia C., Martella V. A large outbreak of enteritis in goat flocks in Marmara, Turkey, by G8P[1] group A rotaviruses. Arch. Virol. 2012;157:1183–1187. doi: 10.1007/s00705-012-1263-5. [DOI] [PubMed] [Google Scholar]
- Bányai K., Martella V., Molnár P., Mihály I., Van Ranst M., Matthijnssens J. Genetic heterogeneity in human G6P [14] rotavirus strains detected in Hungary suggests independent zoonotic origin. J. Infect. 2009;59:213–215. doi: 10.1016/j.jinf.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Can-Sahna K., Alkan F. Sıgırlarda rotavirus enfeksiyonunun epidemiyolojisinde gebeligin rolu. Fırat Üniv Sağlık Bil Derg. 2003;17:203–209. [Google Scholar]
- Chitamber S.D., Arora R., Kolpe A.B., Yadav M.M., Raut C.G. Molecular characterization of unusual bovine group A rotavirus G8P[14] strains identified in western India: emergence of Pgenotype., 2016 genotype. Vet. Microbiol. 2011;148:384–388. doi: 10.1016/j.vetmic.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Cho Y.I., Han J.I., Wang C., Cooper V., Schwartz K., Engelken T., Yoon K.J. Case–control study of microbiological etiology associated with calf diarrhea. Vet. Microbiol. 2013;166:375–385. doi: 10.1016/j.vetmic.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Medeiros T.N., Lorenzetti E., Alfieri A.F., Alfieri A.A. Phylogenetic analysis of a G6P[5] bovine rotavirus strain isolated in a neonatal diarrhea outbreak in a beef cattle herd vaccinated with G6P[1] and G10P[11] genotypes. Arch. Virol. 2015;160:447–451. doi: 10.1007/s00705-014-2271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dóró R., Mihalov-Kovács E., Marton S., László B., Deák J., Jakab F., Juhász Á., Kisfali P., Martella V., Melegh B., Molnár P., Sántha I., Schneider F., Bányai K. Large-scale whole genome sequencing identifies country-wide spread of an emerging G9P[8] rotavirus strain in Hungary, 2012. Infect. Genet. Evol. 2014;28:495–512. doi: 10.1016/j.meegid.2014.09.016. [DOI] [PubMed] [Google Scholar]
- Delogu R., Lo Presti A., Ruggeri F.M., Cella E., Giovanetti M., Ciccozzi M., Ljubin-Sternak S., Bukovski-Simonoski S., Lukic-Grlic A., Ianiro G., Fiore L. Full-genome characterization of a G8P[8] rotavirus that emerged among children with diarrhea in Croatia in 2006. J. Clin. Microbiol. 2013;51:1583–1588. doi: 10.1128/JCM.00396-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulgheroff A.C.B., Figueiredo E.F., Moreira L.P., Moreira K.C., Moura L.M.S., Gouvêa V.S., Domingues A.L.S. Distribution of rotavirus genotypes after vaccine introduction in the Triângulo Mineiro region of Brazil: 4-year follow-up study. J. Clin. Virol. 2012;55:67–71. doi: 10.1016/j.jcv.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Gather Z., Nyangao J., Adachi N., Urushibara N., Kobayashi N. Full genomic analysis of a G8P[1] rotavirus strain isolated from an asymptomatic infant in Kenya provides evidence for an artiodactyl-to-human interspecies transmission event. J. Med. Virol. 2011;83:367–376. doi: 10.1002/jmv.21974. [DOI] [PubMed] [Google Scholar]
- Gouvea V., Glass R.I., Woods P., Taniguchi K., Clark H.F., Forrester B., Fang Z.Y. Polymerase chain reaction amplification and typing of rotavirus nucleic acids from stool specimens. J. Clin. Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea V., Santos N., Timenetsky C. Mdo. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 1994;32:1338–1340. doi: 10.1128/jcm.32.5.1338-1340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isegawa Y., Nakagomi O., Nakagomi T., Ishida S., Uesugi S., Ueda S. Determination of bovine rotavirus G and P serotypes by polymerase chain reaction. Mol. Cell. Probes. 1993;7:277–284. doi: 10.1006/mcpr.1993.1041. [DOI] [PubMed] [Google Scholar]
- Iturizza-Gomara M., Green J., Brown D.W., Desselberger U., Gray J.J. Comparison of specific and random priming in the reverse transcription polymerase chain reaction for genotyping group A rotaviruses. J. Virol. Methods. 1999;78:93–103. doi: 10.1016/s0166-0934(98)00168-2. [DOI] [PubMed] [Google Scholar]
- Kaplon J., Fremy C., Bernard S., Rehby L., Aho S., Pothier P., Ambert-Balay K. Impact of rotavirus vaccine on rotavirus genotypes and caliciviruses circulating in French cattle. Vaccine. 2013;31:2433–2440. doi: 10.1016/j.vaccine.2013.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.N., Wang Y.L., Kao C.L., Zao C.L., Lee C.Y., Chen H.N. NSP4 gene analysis of rotaviruses recovered from infected children with and without diarrhea. J. Clin. Microbiol. 2000;38:4471–4477. doi: 10.1128/jcm.38.12.4471-4477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzetti E., da Silva Medeiros T.N., Alfieri A.F., Alfieri A.A. Genetic heterogeneity of wild-type G4P[6] porcine rotavirus strains detected in a diarrhea outbreak in a regularly vaccinated pig herd. Vet. Microbiol. 2011;154:191–196. doi: 10.1016/j.vetmic.2011.06.026. [DOI] [PubMed] [Google Scholar]
- Lu W., Duhamel G.E., Benfield D.A., Grotelueschen D.M. Serological and genotypic characterization of group A rotavirus reassortants from diarrheic calves born to dams vaccinated against rotavirus. Vet. Microbiol. 1994;42:159–170. doi: 10.1016/0378-1135(94)90015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Bányai K., Matthijnssens J., Buonavoglia C., Ciarlet M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010;140:246–255. doi: 10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J., Bilcke J., Ciarlet M., Martella V., Bányai K., Rahman M., Zeller M., Beutels P., Van Damme P., Van Ranst M. Rotavirus disease and vaccination: impact on genotype diversity. Future Microbiol. 2009;4:1303–1316. doi: 10.2217/fmb.09.96. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J., Ciarlet M., McDonald S.M., Attoui H., Bányai K., Brister J.R., Buesa J., Esona M.D., Estes M.K., Gentsch J.R., Iturriza-Gómara M., Johne R., Kirkwood C.D., Martella V., Mertens P.P., Nakagomi O., Parreño V., Rahman M., Ruggeri F.M., Saif L.J., Santos N., Steyer A., Taniguchi K., Patton J.T., Desselberger U., Van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch. Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalov-Kovács E., Gellért Á., Marton S., Farkas S.L., Fehér E., Oldal M., Jacab F., Martella V., Bányai K. Candidate new rotavirus species in sheltered dogs, Hungary. Emerg. Infect. Dis. 2015;21:660–663. doi: 10.3201/eid2104.141370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijatovic-Rustempasic S., Roy S., Sturgeon M., Rungsrisuriyachai K., Reisdorf E., Cortese M.M., Bowen M.D. Full-genome sequence of the first G8P [14] rotavirus strain detected in the United States. Genome Announc. 2015;3:e00677–15. doi: 10.1128/genomeA.00677-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monini M., Cappuccini F., Battista P., Falcone E., Lavazza A., Ruggeri F.M. Molecular characterization of bovine rotavirus strains circulating in Northern Italy, 2003–2005. Vet. Microbiol. 2008;129:384–389. doi: 10.1016/j.vetmic.2007.11.036. [DOI] [PubMed] [Google Scholar]
- Okada N., Matsumoto Y. Bovine rotavirus G and P types and sequence analysis of the VP7 gene of two G8 bovine rotaviruses from Japan. Vet. Microbiol. 2002;84:297–305. doi: 10.1016/s0378-1135(01)00445-x. [DOI] [PubMed] [Google Scholar]
- Ozkul A., Yesilbag K., Karaoglu T., Burgu I. Elecrophoretypes of bovine rotavirus detected in Turkey. Turk. J. Vet. Anim. Sci. 2002;26:359–362. [Google Scholar]
- Papp H., László B., Jakab F., Ganesh B., De Grazia S., Matthijnssens J., Ciarlet M., Martella V., Bányai K. Review of group A rotavirus strains reported in swine and cattle. Vet. Microbiol. 2013;165:190–199. doi: 10.1016/j.vetmic.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.I., Jeong C., Kim H.H., Park S.H., Park S.J., Hyun B.H., Yang D.K., Kim S.K., Kang M.I., Cho K.O. Molecular epidemiology of bovine noroviruses in South Korea. Vet. Microbiol. 2007;124:125–133. doi: 10.1016/j.vetmic.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.I., Jeong C., Park S.J., Kim H.H., Jeong Y.J., Hyun B.H., Chun Y.H., Kang M.I., Cho K.O. Molecular detection and characterization of unclassified bovine enteric caliciviruses in South Korea. Vet. Microbiol. 2008;130:371–379. doi: 10.1016/j.vetmic.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietsch C., Petersen L., Patzer L., Liebert U.G. Molecular characteristics of German G8P[4] rotavirus strain GER1H-09 suggest that a genotyping and subclassification update is required for G8. J. Clin. Microbiol. 2009;47:3569–3576. doi: 10.1128/JCM.01471-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy N., Lennon G., Fanning S., Power E., O'Shea H. Molecular characterization and analysis of bovine rotavirus strains circulating in Ireland 2002–2004. Vet. Microbiol. 2006;117:242–247. doi: 10.1016/j.vetmic.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Saif L.J., Fernandez F.M. Group A rotavirus veterinary vaccines. J. Infect. Dis. 1996;174:98–106. doi: 10.1093/infdis/174.Supplement_1.S98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyer A., Poljšak-Prijatelj M., Bufon T.L., Marčun-Varda N., Marin J. Rotavirus genotypes in Slovenia: unexpected detection of G8P[8] and G12P[8] genotypes. J. Med. Virol. 2007;79:626–632. doi: 10.1002/jmv.20811. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]