Highlights
-
•
The replication of S-intact PEDV was enhanced during coinfection with an S1 NTD-del PEDV in neonatal pigs.
-
•
The S1 NTD-del PEDV was unable to outcompete the S-intact PEDV during coinfection in piglets.
-
•
Certain concentrations of mucin, bile and bile acids increased the replication of S-intact but not S1 NTD-del PEDV.
Keywords: Porcine epidemic diarrhea virus, Spike protein, N-terminal domain, S1 NTD-del, Coinfection, Pathogenicity
Abstract
Porcine epidemic diarrhea virus (PEDV) variants having a large deletion in the N-terminal domain of the S1 subunit of spike (S) protein were designated as S1 NTD-del PEDVs. They replicate well in experimentally infected pigs. However, on farms they often co-infect pigs with the PEDV containing an intact S protein (S-intact PEDV). We aimed to characterize viral replication and pathogenesis in neonatal gnotobiotic pigs infected simultaneously with the two types of PEDV using two recombinant PEDVs: icPC22A and its S1 NTD-del form icPC22A-S1Δ197. Additionally, viral replication was compared in Vero and IPEC-DQ cells at the presence of bovine mucin (BM), porcine gastric mucin (PGM), swine bile and bile acids during inoculation. In the pigs coinfected with icPC22A and icPC22A-S1Δ197, icPC22A replicated to a higher peak titer than its infection of pigs without the presence of icPC22A-S1Δ197. The severity of diarrhea and intestinal atrophy were similar between icPC22A and the coinfection groups, but were significantly higher than icPC22A-S1Δ197 group. In Vero and IPEC-DQ cells, certain concentrations of BM, PGM, bile and bile acids increased significantly the infectivity of icPC22A but had no or negative effects on icPC22A-S1Δ197. These results indicated that the replication of the S-intact PEDV was enhanced during coinfection in piglets. This observation may be explained partially by the fact that mucin, bile and bile acids in gastrointestinal tract had facilitating effects on the infection of S-intact PEDV, but no/inhibition effects on S1 NTD-del PEDV.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) belongs to the genus Alphacoronavirus in the family Coronaviridae. It causes severe gastroenteritis in neonatal pigs worldwide. The genome size of PEDV is 28 kb. It contains ORF1a and ORF1b encoding 14–16 non-structural proteins, an ORF3 encoding an accessory protein, and four ORFs encoding structural proteins: spike (S), membrane (M), envelop (E), and nucleocapsid (N) proteins.
PEDV S protein mediates the essential functions of receptor binding (via S1 subunit) and subsequent fusion of the viral and cellular membranes (via S2 subunit) (Li, 2016). S1 subunit contains two domains, the N-terminal domain (S1 NTD, residues 21–324 based on PEDV CV777) and the C-terminal domain (S1 CTD, residues 253–638). However, the cellular receptor of PEDV is still unknown (Li et al., 2017b; Shirato et al., 2016). Although the sialic acid binding activity of S1-NTD was observed in many coronaviuses and facilitated virus replication (Schwegmann-Wessels et al., 2011; Li et al., 2016; Desmarets et al., 2014), S1-NTD is dispensable for some mutants of transmissible gastroenteritis coronavirus (TGEV) and PEDV (Schwegmann-Wessels et al., 2011; Oka et al., 2014; Suzuki et al., 2016; Hou et al., 2017). PEDV variants with a big deletion in the S1-NTD have been designated as S1 NTD-del PEDVs (Hou et al., 2017). They have been detected in field pig samples, indicating sufficient replication of these S1 NTD-del PEDVs in vivo (Suzuki et al., 2015; Diep et al., 2017; Zhang et al., 2018a; Su et al., 2018). Interestingly, most field S1 NTD-del PEDVs exist as a member of coinfection of pigs with the S-intact PEDV (Diep et al., 2017; Su et al., 2018). However, the mechanisms and consequences of the coinfection have not been investigated.
Studies showed that under unfavorable conditions as encountered in the intestinal tract, sialic acid-binding of TGEV played an important role in viral replication. When absorption time was reduced, TGEV infectivity was also reduced in the cells whose sialic acids were removed (Schwegmann-Wessels et al., 2011). Consequently, sialic acid binding activity may facilitate the infection of TGEV by helping the virus resist detergent-like substances encountered during passing through the gastrointestinal tract (Krempl et al., 2000). Therefore, we hypothesized that mucin, rich in sialic acid, may enhance the infection of PEDV. Interestingly, bile acids can induce or promote the replication of some enteric viruses like a porcine sapovirus (Chang et al., 2004) and human noroviruses (Ettayebi et al., 2016). Since intestinal epithelial cells are exposed to bile acids, we hypothesized that bile acids promote PEDV infection.
Previously, we generated two recombinant PEDVs, PEDV icPC22A and icPC22A-S1Δ197 (Hou et al., 2017). PEDV icPC22A is an infectious cDNA clone-derived virus of the emerging highly virulent PEDV PC22A strain. On the other hand, PEDV icPC22A-S1Δ197 is a mutant version of icPC22A bearing a 197-aa deletion (residues 34–230) in the S1 NTD region, corresponding to the S1 domain 0 (S1°) of the spike protein (Li et al., 2017a; Hou et al., 2017). In this study, we investigated the replication and pathogenesis of the two viruses during coinfection in neonatal gnotobiotic (Gn) piglets. Some intestinal substances (mucin, bile and bile acids) were tested for their effects on the infection of the individual viruses in vitro. We selected two cell lines, Vero and IPEC-DQ cells. Vero cells are the most commonly used cell line for PEDV isolation and propagation (Teeravechyan et al., 2016). IPEC-DQ cell is a subclone of porcine small intestinal cell line IPEC-J2 (Zhang et al., 2018b). Although PEDV grows less efficiently in IPEC-DQ cells than in Vero cells, PEDV infection of IPEC-DQ cells is more relevant physiologically to PEDV natural infection in the intestines of pigs.
2. Materials and methods
2.1. Cell lines, recombinant viruses, reagents and antibodies
Vero cells (ATCC number CCL81) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Carlsbad, CA, USA) supplemented with 5% fetal bovine serum (FBS, HyClone, Logan, UT, USA) and 1% penicillin/streptomycin (Gibco). For virus propagation in Vero cells, the maintenance medium was DMEM supplemented with 0.3% tryptose phosphate broth solution (TPB, Sigma, St. Louis, MO, USA), 1% Penicillin/Streptomycin (Gibco) and 10 μg/mL trypsin (2.5% Trypsin, Gibco). IPEC-DQ cell line was a gift from Dr. Dongwan Yoo, University of Illinois at Urbana-Champaign, Urbana IL, USA. It was a subclone of IPEC-J2 cells, a porcine small intestinal cell line, and maintained in Roswell Park Memorial Institute 1640 Medium (RPMI-1640, Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin (Zhang et al., 2018b). For virus propagation in IPEC-DQ cells, maintenance medium was RPMI-1640 containing 0.3% TPB, 1% Penicillin/Streptomycin and 1 μg/mL trypsin. Two recombinant PEDV, icPC22A and icPC22A-S1Δ197, were generated previously by our lab using the infectious clone of PEDV PC22A strain (Hou et al., 2017).
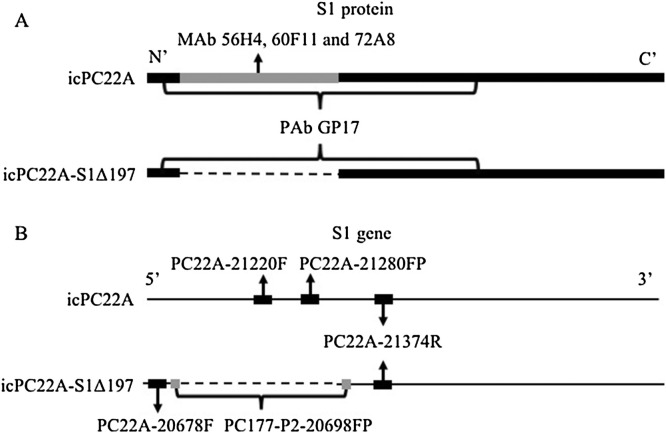
Neuraminidase (NA, from Clostridium welchii), porcine gastric mucin (PGM) and several bile acids [(glycochenodeoxycholic acid (GCDCA), chenodeoxycholic acid (CDCA), deoxycholic acid (DCA), and ursodeoxycholic acid (UDCA)] were all purchased from Sigma. Bovine mucin (BM) was purchased from Worthington (Worthington Biochemical Corp., Lakewood, NJ, USA). Swine bile was collected from a pregnant sow after C-section surgery. The mouse monoclonal antibodies (MAb) 56H4, 60F11 and 72 A8 targeting the S1° domain of PEDV S protein (Fig. S1A) was provided by Dr. Berend Jan Bosch, Utrecht University, Utrecht, Netherlands (Li et al., 2017a). The MAb SD6-29 targeting PEDV nucleocapsid (N) protein was provided by Drs. Eric Nelson and Steven Lawson, South Dakota State University. The guinea pig polyclonal antibody (PAb) GP17 targeting the S1 subunit of the S protein of PEDV PC177 strain (S1Δ197) (Fig. S1A), rabbit PAb RB9 targeting PEDV N protein and swine PEDV-positive serum (virus neutralization titer 1:1024) were generated by our laboratory previously (Hou et al., 2019).
2.2. Duplex RT-qPCR method for the detection and differentiation of PEDV icPC22A and icPC22A-S1Δ197
Based on the nucleotide sequence differences in the S1 NTD coding region between the icPC22 A and icPC22 A-S1Δ197, a duplex TaqMan real-time reverse-transcription quantitative PCR (RT-qPCR) was developed. We designed the primers and probes (Table 1 , Fig. S1B) and had them synthesized by Integrated DNA Technologies (Skokie, IL, USA). The RNA of the two viruses was extracted using RNeasy Mini Kit (QIAGEN, Valencia, CA, USA). To generate a standard DNA, a kit of SuperScript™ III First-Strand Synthesis SuperMix (Thermo Fisher Scientific, Carlsbad, CA, USA) was used for cDNA synthesis following manufacturer’s instructions. Primer set PEDV-20320F and PEDV-21816R (Oka et al., 2014) was used to amplify a 1.5 kb or a 0.9 kb fragment covering PEDV partial nsp16 gene and S1 gene of icPC22 A and icPC22 A-S1Δ197, respectively. The PCR products were purified using Gel Extraction Kit (QIAGEN) and used as standard DNA. The reaction consisted with 4 μL 5 × PCR Buffer, 0.8 μL 10 mM dNTPs (QIAGEN), 1.2 μL each primer (10 μM), 0.4 μL each probe (5 μM), 0.2 μL RNAsin (PROMEGA, WI, USA), 0.8 μL Enzyme Mix (QIAGEN), 1.6 μL each DNA/RNA and 6.6 μL sterilized water. The RT-qPCR procedure was: 50 °C for 30 min, 95 °C for 15 min and 45 cycles of 95 °C for 15 s and 60 °C for 60 s. We generated two standard curves using LightCycler® 96 System (Roche Life Science, Indianapolis, IN, USA). The two standard curves for the serially diluted DNA fragments of icPC22A and icPC22A-S1Δ197 showed regressions with coefficients of -0.2873 and -3.009, respectively, and the corresponding correlation coefficient (R2) of 0.9987 and 0.9963, respectively. The primers and probes were further tested using individual and the mixture of two viruses. We tested the correlation between S gene titers and infectious titers of PEDV icPC22 A and icPC22 A-S1Δ197 using serially diluted, cell cultured viruses (Table S1 and S2). For the samples of mixed viruses, the duplex RT-qPCR was performed to differentiate the S gene of each virus. The detection limit was 5 log10 S gene copies/mL for both icPC22 A and icPC22 A-S1Δ197.
Table 1.
Primers and probes of the duplex RT-qPCR.
| Name | Nucleotide sequence (5’-3’) |
|---|---|
| PC22A-21220F (for icPC22A) | GTTACAACAGTGGAGGTTGT |
| PC22A-21280FP (for icPC22A) | CY5-ATGTTACTAGTGCTGGTGAGGATGGT-BHQ2 |
| PC22A-20678F (for icPC22A-S1Δ197) | AACACTTAGCCTACCACAA |
| PC177-P2-20698FP (for icPC22A-S1Δ197) | FAM-TGCTCAGCTAACACTAATTTTAGGACAGCT-BHQ3 |
| PC22A-21374R (for both viruses) | ATTGGGCTCAGTAGCAAAT |
2.3. Study design of experimental infection of Gn piglets with PEDV
The Gn piglets were derived and raised as described previously (Saif et al., 1996). Thirty-one Gn piglets were assigned to five experimental groups: (1) icPC22 A (n = 14; 100 PFU/pig); (2) icPC22A-S1Δ197 (n = 7; 100 PFU/pig); (3) Coinfection of icPC22A and icPC22 A-S1Δ197 (n = 7; 100 PFU/pig of each virus); (4) Mock (n = 2; PBS); (5) Second passage of coinfection [n = 1; small intestinal contents (SIC) (sample #PE1158) of one piglet in group 3 collected at 2 day post inoculation (dpi), at a dose of 10 log10 copies/mL based on the N gene, 9 log10 copies/mL and 7 log10 copies/mL based on the icPC22A-specific S gene and icPC22A-S1Δ197-specific S gene, respectively, corresponding to 4 log10 PFU/mL of infectious PEDV] (Jung et al., 2014). At 5 days of age, piglets in groups 1–4 were orally inoculated with individual or the mixture of the viruses. The group 5 pig was inoculated at 12 days of age and was euthanized at 1 dpi. To examine histopathological changes, one or two piglets of groups 1–4 were euthanized at 0.5 dpi and 2–3 dpi, respectively.
After inoculation, all piglets were evaluated daily for clinical signs, such as diarrhea, vomiting, anorexia, and depression. Fecal consistency was examined by collecting rectal swabs and scored as follows: 0, solid; 1, pasty; 2, semiliquid (mild diarrhea); and 3, liquid (severe diarrhea). Total fecal RNA was extracted using MagMAX-96 RNA isolation kit (Thermo Fisher Scientific), and the viral RNA was titrated by the RT-qPCR targeting the N gene (Jung et al., 2014) and the duplex RT-qPCR targeting the S gene of each virus. Fecal infectious PEDV shedding titers were tested using Vero cells in 96-well plates and determined as 50% tissue culture infective doses (TCID50) (Reed and Muench, 1938).
2.4. Histopathological examination, immunohistochemical (IHC) and immunofluorescent (IF) staining of PEDV-specific antigens
At necropsy, three sections of the jejunum (proximal, middle, and distal) and one section of the ileum were collected and fixed in 3.7% formalin (Fisher scientific co LLC, Florence, KY, USA). All tissues were trimmed, processed, embedded in paraffin, and sectioned using routine procedures (Jung et al., 2014). To compare the PEDV-specific histopathological changes among different groups of pigs, IHC staining was performed as described previously for the detection of PEDV N proteins using MAb SD6-29 as the primary antibody (Hou et al., 2017). The IHC staining was carried out using a nonbiotin polymerized horseradish peroxidase system (BioGenex Laboratories, San Ramon, CA, USA). Finally, tissues were counterstained with hematoxylin. Images of tissues were observed and captured by Olympus IX-70 fluorescent microscope (Center Valley, PA, USA). Ratios of villous height to crypt depth (VH/CD) of the jejunum and ileum of individual piglets were measured using a computerized imaging system (MetaMorph, Olympus, Japan) as described previously (Jung et al., 2014). For each intestinal section, 10 villi and crypts were measured.
To detect and differentiate icPC22 A- and icPC22 A-S1Δ197-infected cells in the coinfected pigs, IF staining was performed. The slide preparation steps were as same as those of the above IHC procedure. The mixture of mouse MAbs 56H4, 60F11 and 72A8 (1: 100 in PBS) (Li et al., 2017a) targeting the S1° domain of PEDV S protein was used as the primary antibody for the staining of icPC22 A-infected cells only. Tissues were incubated at 4℃ overnight, after three washings with PBST (PBS with 0.1% tween20), Alexa Fluor 594-conjugated goat anti-mouse IgG (H + L) (Thermo Fisher Scientific) (1: 500 diluted in PBS) was used as the second antibody and tissues were incubated at room temperature for 2 h. After washing with PBST, guinea pig PAb GP17 (1: 200 diluted in PBS), targeting the S1Δ197 and was for the staining of both viruses, was added. Then, the slides were incubated at room temperature for 2 h. After washing three times with PBST, Alexa Fluor 488-conjugated goat anti-guinea pig IgG (H + L) (Thermo Fisher Scientific) (1: 500 diluted in PBS) was used as the 2nd antibody and the slides were incubated at room temperature for 2 h. After washing with PBST, 1 mg/mL DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride, Roche Life Science, Indianapolis, IN, USA) was added for nucleic acid staining. Finally, an autofluorescence quenching kit (Vector® TrueVIEW™, Burlingame, CA, USA) was used to quench the autofluorescence of the tissues. Images of stained tissues were captured by Olympus IX-70 fluorescent microscope. Red, green and blue fluorescence staining was merged by ImageJ software (https://imagej.nih.gov).
2.5. Isolation of the pig-passaged icPC22 A and icPC22 A-S1Δ197 from the SIC of a pig coinfected with the viruses
The SIC (sample #PE1158) of a coinfected piglet in group 3 was positive for both viruses by the duplex RT-qPCR. Virus isolation was performed using plague assays (Oka et al., 2014). The SIC was diluted in DMEM and the 10% suspension was vortexed briefly followed by centrifugation at 15,000 ×g for 3 min at 4℃. A series of 10-fold dilutions (10−1–10−5) of the supernatants were prepared in DMEM (containing 7.5 μg/mL trypsin) and were used immediately for inoculation of Vero cell monolayers in 6-well plates (200 μL per well). After incubating at 37℃ for 1 h, the inoculum was removed and the monolayers were washed once with PBS. Then 2 mL/well of agarose overlay was added. The plate was kept for 3 days before picking up clones. For propagating each clone, Vero cell monolayers in 96-well plates were prepared and individual plagues were picked and added into individual wells. At 3 dpi, RNA was extracted and the duplex RT-qPCR method specific for both viruses was performed.
2.6. Multi-step growth kinetics and IF staining of coinfection of PEDV icPC22A and icPC22A-S1Δ197 in Vero and IPEC-DQ cells
The growth kinetics of icPC22A-S1Δ197 and icPC22A during the co-infection of the two viruses were investigated in Vero and IPEC-DQ cells. Cell monolayers on 96-well plates were inoculated with one of the three inocula: (1) 0.01 multiplicity of infection (MOI) of icPC22A; (2) 0.01 MOI of icPC22A-S1Δ197; and (3) 0.01 MOI of icPC22A and 0.01 MOI of icPC22 A-S1Δ197. Viruses were diluted in DMEM containing 10 μg/mL trypsin and were added to cell monolayers. After incubating at 37℃ for 2 h, the inoculum was removed and the cell monolayers were washed for three times. Then the plates were added with the maintenance medium and incubated at 37℃. The samples were collected at different time points (2 h, 8 h, 12 h, 36 h, 48 h) and frozen (-80℃) and thawed once before testing. RNA was extracted using the MagMAX-96 RNA isolation kit (Thermo Fisher Scientific) and virus titers were tested by the duplex RT-qPCR specific for both viruses. For IF staining of S proteins at 8 h post infection (hpi), the procedure in 2.4 section was performed.
2.7. Infection of PEDV icPC22A and icPC22A-S1Δ197 in NA-treated Vero and IPEC-DQ cells
Vero/IPEC-DQ cell monolayers in 96-well plates were treated with 250 mU NA in DMEM/RPMI-1640 or mock DMEM/RPMI-1640 at 37℃ for 2 h. After three washings with DMEM/RPMI-1640, cells were inoculated with icPC22 A or icPC22 A-S1Δ197 at a MOI of 0.02 with 10 μg/mL trypsin for 2 h. Next, the inoculum was removed followed by three washings. DMEM/RPMI-1640 containing 50 μg/mL soybean trypsin inhibitor (SBTI) and swine PEDV positive serum (virus neutralization titer 1:1024, 1:500 dilution) was added to prevent virus spreading. Cells were fixed with acetone-methanol (1:4) at 6 hpi. The numbers of PEDV-infected cell foci were determined by IF assays as described for the staining of PEDV N proteins (Hou et al., 2017). Briefly, rabbit PAb RB9 (1:1000) was used as the primary antibody. After incubating at 4℃ overnight, cells were washed three times. Then Alexa Fluor 488-conjugated goat anti-rabbit IgG (H + L) (Thermo Fisher Scientific) (1:1000) was used as the 2nd antibody and the cells were incubated at room temperature for 1 h. After washing and adding mounting medium, cells were observed and picture were captured by using Olympus IX-70 fluorescent microscope. The numbers of fluorescent focus units (FFU) were counted by ImageJ software. The percentages of FFU were calculated by the value of mock group as 100%.
2.8. Effect of mucin, bile and bile acids on the infection of PEDV icPC22A and icPC22A-S1Δ197 in Vero and IPEC-DQ cells
Viruses (icPC22A or icPC22A-S1Δ197) were mixed with different concentrations of BM (0, 0.1, 0.3, 0.5 mg/mL) or PGM (0, 0.5, 1.0, 2.5, 5.0 mg/mL). Then 10 μg/mL trypsin were added to the mixture. Monolayer of Vero/IPEC-DQ cells in 96-well plates were inoculated with each virus mixture at a MOI of 0.02. The plates were incubated at 37℃ for 2 h, then the inoculum was removed. After three washings, DMEM/RPMI-1640 containing 50 μg/mL SBTI and swine PEDV positive serum (1:500 dilution) was added. Cells were fixed at 6 hpi. The numbers of FFU were determined by IF assay for the staining of PEDV N proteins (see section 2.7). The percentages of FFU were calculated by the value of mock group as 100%. Similarly, different concentration of swine bile (0, 0.1, 0.3, 0.5, 1.0%) and bile acids (GCDCA, CDCA, DCA, and UDCA) at 0, 25, 50, 100, 200 μM were tested.
2.9. Statistical analysis
Statistical analysis was performed using GraphPad Prism 7 software (GraphPad Software, La Jolla, CA, USA). The experimental data were analyzed by one-way ANOVA and Student`s t-test. Data are shown as M ± SD, and a P < 0.05 was considered as significant.
3. Results
3.1. The replication of icPC22A was eventually enhanced during coinfection with icPC22A-S1Δ197 in neonatal Gn piglets
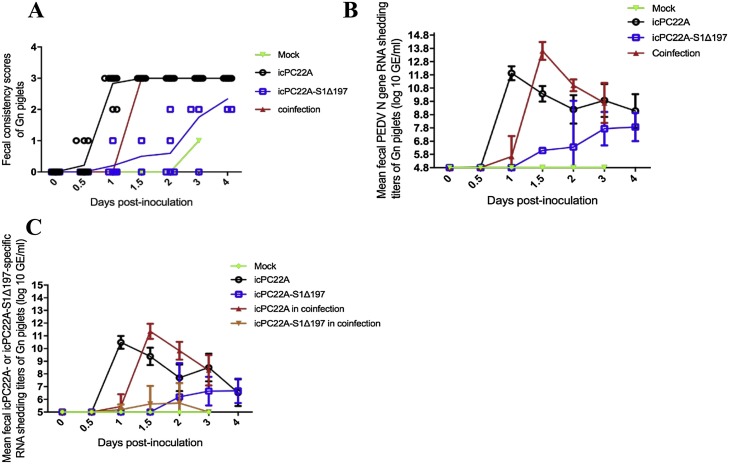
As shown in Fig. 1 , all PEDV-inoculated pigs but no pigs in the mock group had diarrhea and shed PEDV. The fecal consistency scores and peak fecal PEDV N and S RNA shedding titers of piglets in the coinfection group reached the peak at 1.5 dpi, which was delayed half a day compared with the icPC22A group (1 dpi) but earlier than the icPC22A-S1Δ197 group (> 4 dpi). By 4 dpi, all pigs in the icPC22A and coinfection groups but no pigs in the icPC22A-S1Δ197 group died or showed moribund. Compared with the peak fecal PEDV N gene shedding titer (11.6 ± 0.2 log10 copies/mL) of piglets in the icPC22A group (1 dpi), pigs in the coinfection group had a significantly higher peak titer (13.6 ± 0.7 log10 copies/mL) (Fig. 1B and Table 2 ) at a delayed time point (1.5 dpi). In addition, the peak fecal infectious PEDV shedding titer of the coinfection group (5.8 ± 0.4 log10 TCID50/mL) were significantly higher than that of the single infection groups (4.9 ± 0.1 and 1.8 ± 1.8 log10 TCID50/mL of icPC22 A and icPC22 A-S1Δ197 groups, respectively) (Table 2). Interestingly, the peak S gene shedding titers of icPC22 A (11.4 ± 0.5 log10 copies/mL) in the coinfection group at 1.5 dpi was significantly higher than that of icPC22 A group (10.5 ± 0.5 log10 copies/mL) at 1 dpi (Fig. 1C, Table 2). However, the peak S gene shedding titers (5.8 ± 1.2 log10 copies/mL) of icPC22 A-S1Δ197 in the coinfection group was significantly lower than that of the icPC22 A-S1Δ197 group (7.6 ± 1.7 log10 copies/mL) (Fig. 1C, Table 2). In the second passage of coinfection, only icPC22 A but not icPC22 A-S1Δ197 S gene was detected in the large intestinal contents (LIC) of the piglet, which was orally inoculated with the SIC (sample #PE1158) of one pig in the coinfection group and was positive for both viruses. The LIC had a virus titer of 8.8 log10 TCID50/mL for infectious PEDV, 12.3 log10 S gene copies/mL and 14.6 log10 N gene copies/mL for icPC22A.
Fig. 1.
Evaluation of the replication of icPC22A and icPC22A-S1Δ197 in Gn pigs infected with individual or both viruses. (A) Fecal consistency scores of individual Gn piglets and the mean of each group were shown. The scores were named as follows: 0, solid (normal); 1, pasty (normal); 2, semiliquid (mild diarrhea); and 3, liquid (severe diarrhea). Two piglets from icPC22A, icPC22A-S1Δ197 and coinfection groups, and one pig from mock group were euthanized at 12 hpi and 2–3 dpi, respectively, for histopathological examination. (B) Fecal PEDV N gene RNA shedding titers (for both icPC22A and icPC22A-S1Δ197) of each group. Data are shown as mean (M) ± standard deviations (SD) of pigs in each group. (C) Fecal PEDV S gene RNA shedding titles of each virus. Data are shown as M ± SD.
Table 2.
Summary of pig group information, inoculum, pig diarrhea outcomes and virus peak shedding titers after inoculation.
| Pig groupa (Pig number) |
Severe diarrhea rate (%) b, c, g | Mortality rate by 4 dpi (%)c, g | Onset of diarrhea (dpi)g | Peak fecal shedding of the S gene of icPC22 A (log10 copies/mL) d, g | Peak fecal shedding of the S gene of icPC22 A-S1Δ197 (log10 copies/mL)d, g | Peak fecal shedding of N gene (log10 copies/mL) e, g | Peak fecal infectious PEDV shedding titers (log10 TCID50/mL) g |
|---|---|---|---|---|---|---|---|
| icPC22 A (n = 14) |
100 (12/12)f A | 100 (12/12)f A | 1.00 ± 0.00 C | 10.5 ± 0.5 B | — | 11.6 ± 0.2 B | 4.9 ± 0.1 B |
| icPC22 A S1Δ197 (n = 7) |
20 (1/5)f B | 0 (0/5)f C | 3.25 ± 0.50 A | — | 7.6 ± 1.7 A | 8.9 ± 1.9 C | 1.8 ± 1.8 C |
| Co-infection (n = 7) |
100 (5/5)f A | 100 (5/5)f A | 1.50 ± 0.00 B | 11.4 ± 0.5 A | 5.8 ± 1.2 B | 13.6 ± 0.7 A | 5.8 ± 0.4 A |
| Mock (n = 2) |
0 (0/1)f C | 0 (0/1)f C | — | — | — | — | — |
Piglets in group 1 were inoculated with 100 PFU/pig of icPC22 A; Piglets in group 2 were inoculated with 100 PFU/pig of icPC22A-S1Δ197; Piglets in group 3 were inoculated with 100 PFU/pig of each virus; Piglets in group 4 were inoculated with PBS; Piglets in group 5 were inoculated with the SIC of piglet in group 3 with a dose of 4 log10 PFU/mL targeting N gene of PEDV.
Fecal consistency (FC) was scored as follows: 0, solid, 1, pasty, 2, semiliquid; and 3, liquid. An FC score of 2 and 3 were considered diarrhea and severe diarrhea, respectively.
Values in parentheses are the number of positive results/number of animals tested.
The detection limit of PEDV RT-qPCR targeting the S gene of icPC22A or icPC22 A-S1Δ197 is 5 log10 copies/mL.
The detection limit of PEDV RT-qPCR targeting the N gene of both icPC22A and icPC22 A-S1Δ197 is 4.8 log10 copies/mL (Jung et al., 2014).
The pig number for diarrhea observation is 1 or 2 less than the total pig number because 1 or 2 pigs of each group were euthanized at 12 hpi.
The experimental data were analyzed by Student`s t-test. Letters ‘A, B and C’ indicate a mean significant difference between groups (P < 0.05). ‘—’, not detected.
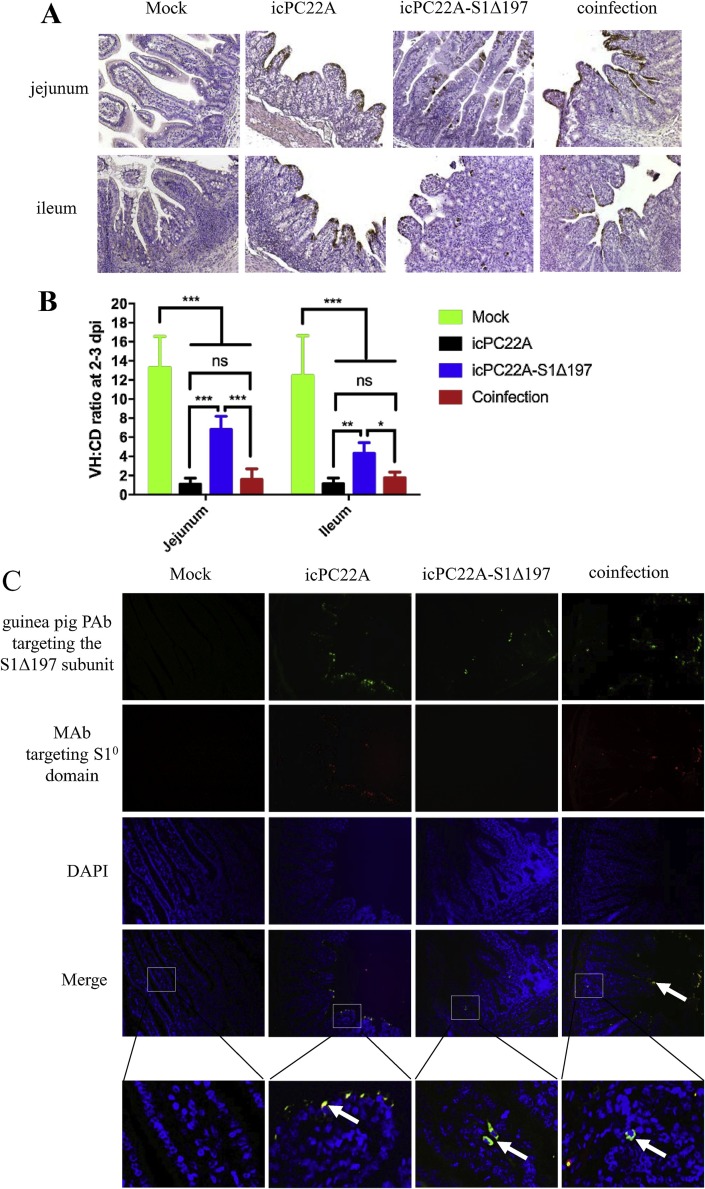
Histopathological examination was performed and villous atrophy was observed in the jejunum and ileum of piglets in all PEDV-inoculated groups (Fig. 2 A and B). The small intestinal villi in the icPC22A-S1Δ197 group had a milder villous atrophy than those in the icPC22A and coinfection groups. The VH/CD ratios of jejunum and ileum of all the PEDV-inoculated groups were significantly lower than those of the mock group (Fig. 2B). The ratios of the icPC22 A and coinfection groups were similar and were significantly lower than that of the icPC22 A-S1Δ197 group. IHC staining showed that no PEDV-positive intestinal epithelial cells were observed in all PEDV-inoculated pigs at 0.5 dpi (data not shown). At 2–3 dpi, a few PEDV N antigens were observed in the small intestine of the icPC22A-S1Δ197-infected piglets, while extensive antigens were stained in those of the icPC22A and coinfection piglets (Fig. 2A). By IF staining for the PEDV S1 proteins, a few PEDV antigens were observed in the jejunum of the icPC22A-S1Δ197-inoculated piglets (green alone, positive for S1Δ197 only), while many antigens were observed in those of the icPC22 A-inoculated piglets [yellow, representing positive for both S1Δ197 (green) and S1° domain (red)] and the coinfection piglets (yellow) (Fig. 2C).
Fig. 2.
Histopathological examination of the Gn piglets infected with individual or both icPC22A and icPC22A-S1Δ197. (A) IHC staining of PEDV N proteins in the jejunal and ileal sections of piglets that died or were euthanized at 2–3 dpi (magnification, 200×). The brown signals represented the PEDV N antigens in enterocytes. (B) Villous height to crypt depth (VH/CD) ratios of jejunum and ileum of the Gn piglets euthanized at 2–3 dpi. For each intestinal section, 10 villi and crypts were measured. Data are shown as the M ± SD. Number of asterisks indicate significant difference between groups (*, p < 0.05; **, P < 0.01; ***, P < 0.001). ‘ns’, not significant. (C) IF staining of PEDV S proteins in the jejunal sections of piglets that died or were euthanized at 2–3 dpi (magnification, 200 x). Tissue sections were stained for the detection of icPC22A and icPC22A-S1Δ197 antigens targeting the S1Δ197 protein (green), and for icPC22A antigens targeting the S1° domain (red), and counterstained for cell nuclei (blue). In the merged images, yellow dots represented icPC22A-infected (or co-infected in coinfection group) cells (merged from red and green) and the green dots represented icPC22A-S1Δ197 infection alone.
3.2. icPC22A alone, but not icPC22A-S1Δ197 alone was isolated from the SIC of one of the pigs infected with the two viruses simultaneously
The SIC of the one piglet in the coinfection group, which was positive for both viruses by the duplex RT-qPCR were used for virus isolation in Vero cells using plague assays. We picked up 72 plagues and found that 55 plagues were positive for icPC22A and 17 plagues were positive for both viruses by the duplex RT-qPCR. No plagues were positive for S1 NTD-del PEDV alone.
3.3. During coinfection with icPC22A, icPC22A-S1Δ197 S gene titers were significantly lower than those in its single infection both in Vero cells and in IPEC-DQ cells
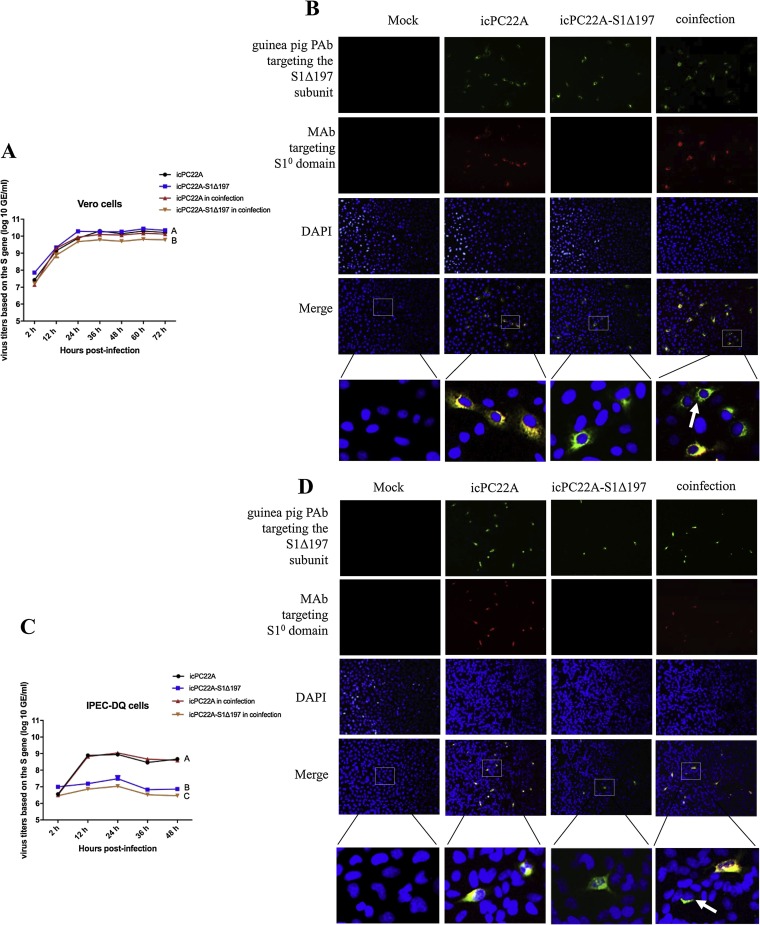
During coinfection in Vero cells, icPC22A replicated to a titer of 10.10 ± 0.05 log10 copies/mL at 72 hpi, which was similar to that of the single infection (10.20 ± 0.09 log10 copies/mL) (Fig. 3 A). However, icPC22A-S1Δ197 S gene peak titer was 9.80 ± 0.03 log10 copies/mL, which was significantly lower than that of the single infection (10.30 ± 0.04 log10 copies/mL). In Vero cells, icPC22A alone and icPC22A-S1Δ197 alone replicated to similar titers. Similarly, during coinfection in IPEC-DQ cells, icPC22A replicated to a similar titer to its single infection (8.60 ± 0.18 vs. 8.70 ± 0.11 log10 copies/mL at 48 hpi (Fig. 3C). icPC22A-S1Δ197 S gene peak titer was 7.03 ± 0.12 log10 copies/mL, which was significantly lower than that of the single infection (7.49 ± 0.25 log10 copies/mL). In contrast to those results in Vero cells, icPC22A alone replicated to significantly higher titers than icPC22A-S1Δ197 alone in IPEC-DQ cells. By IF staining, more icPC22A-S1Δ197-infected cells (green) were observed in Vero cells than in IEPC-DQ cells in both icPC22A-S1Δ197 alone and co-infection conditions (Fig. 3B and D). At the second passage of the coinfection, icPC22 A RNA and antigens were detected in both cells; however, icPC22 A-S1Δ197 RNA and antigens were detected exclusively in Vero cells but not in IPEC-DQ cells (data not shown).
Fig. 3.
Growth kinetics and IF staining of icPC22A and icPC22A-S1Δ197 in individual virus infection and coinfection conditions in Vero and IPEC-DQ cells. (A, C) 0.01 MOI of the two viruses was used to individually infect or coinfect Vero or IPEC-DQ cell monolayers in 96-well plates. Samples (supernatant and cells) were collected at different time points and the virus titers were detected using the duplex RT-qPCR. (B, D) IF staining of PEDV S proteins of the two viruses in Vero or IPEC-DQ cells (magnification, 200 x). Cells were stained for the detection of icPC22A and icPC22 A-S1Δ197 antigens targeting the S1Δ197 protein (green), and for icPC22A antigens targeting the S1° domain (red), and counterstained for cell nuclei (blue). In the merged images, yellow dots represented icPC22A-infected (or co-infected in coinfection condition) cells and the green dots represented icPC22A-S1Δ197 infection alone. The experimental data were analyzed by Student`s t-test. Letters ‘A, B and C’ indicate a mean significant difference between groups (P < 0.05).
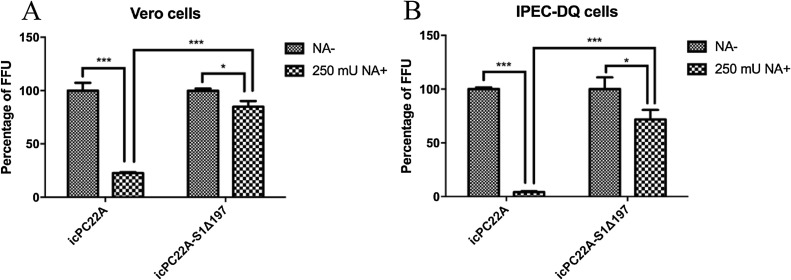
3.4. The percentage of infectivity of icPC22A was significantly lower than that of icPC22A-S1Δ197 in NA-treated Vero and IPEC-DQ cells
After removing sialic acids from the cell surface using 250 mU NA in Vero and IPEC-DQ cells, the percentages of infectivity of icPC22A and icPC22A-S1Δ197 were reduced significantly (Fig. 4 ). However, in both cells treated with NA, the percentages of infectivity of icPC22A were significantly lower than those of icPC22A-S1Δ197.
Fig. 4.
Infection of PEDV icPC22A and icPC22A-S1Δ197 in neuraminidase (NA)-treated Vero and IPEC-DQ cells. Vero or IPEC-DQ cell monolayers in 96-well plates were treated with 250 mU NA or mock at 37℃ for 2 h. After three washings with DMEM/RPMI-1640, cells were inoculated with equal amounts of virus at a MOI of 0.02 diluted in the medium containing 10 μg/mL trypsin. After incubation at 37℃ for 2 h, the inoculum was removed followed by three washings. DMEM/RPMI-1640 containing 50 μg/mL soybean trypsin inhibitor (SBTI) and swine PEDV positive serum (virus neutralization titer 1:1024, 1:500 dilution) was added. At 8 hpi, cells were fixed with acetone-methanol (1:4) and infectivity was tested by IF assay. Each Experiment was performed three times. Data are shown as the M ± SD, and number of asterisks indicate significant difference between groups (*, p < 0.05; **, P < 0.01; ***, P < 0.001).
3.5. Mucin enhanced the infectivity of icPC22A but had no or inhibition effects on icPC22A-S1Δ197 in both Vero and IPEC-DQ cells
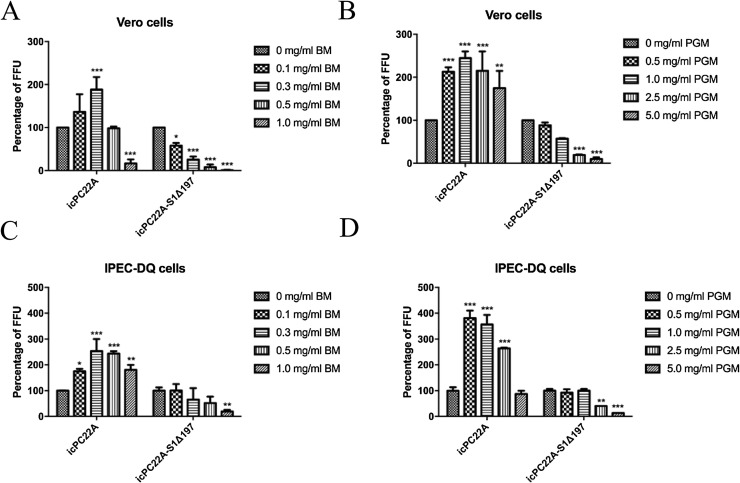
In Vero cells, 0.1–0.3, 0.5 and 1.0 mg/mL BM increased, had no effects and decreased the infectivity of icPC22A, respectively (Fig. 5 A). 0.5–5.0 mg/mL PGM increased the infectivity of icPC22A 1.7–2.5 fold (Fig. 5B). However, the infectivity of icPC22A-S1Δ197 was unaffected or inhibited significantly by BM and PGM at the tested concentration (Fig. 5A and 5B).
Fig. 5.
Effect of bovine mucin (BM) and porcine gastric mucin (PGM) on the infection of PEDV icPC22A and icPC22A-S1Δ197 in Vero (A and B) and IPEC-DQ cells (C and D). Monolayers of Vero or IPEC-DQ cells in 96-well plates were inoculated with a MOI of 0.02 icPC22A or icPC22A-S1Δ197 in medium containing 10 μg/mL trypsin and different concentrations of BM or PGM. After incubation at 37℃ for 2 h, the inoculum was removed followed by three washings. DMEM/RPMI-1640 containing 50 μg/mL soybean trypsin inhibitor (SBTI) and swine PEDV positive serum (virus neutralization titer 1:1024, 1:500 dilution) was added. At 8 hpi, cells were fixed with acetone-methanol (1:4) and infectivity was tested by IF assay. Each Experiment was performed three times. Data are shown as M ± SD, and number of asterisks indicate significant difference between groups (*, p < 0.05; **, P < 0.01; ***, P < 0.001).
In IPEC-DQ cells, the infectivity of icPC22A was increased 1.7–2.6 fold and 2.6–3.8 fold by BM (0.1–1.0 mg/mL) and PGM (0.5–2.5 mg/mL), respectively. However, there was no effects at the highest PGM concentration tested (5.0 mg/mL). On the other hand, low to medium concentrations of BM (0.1-0.5 mg/mL) and PGM (0.5–1.0 mg/mL) had no effects on the infectivity of icPC22 A-S1Δ197. Only high concentrations of BM (1.0 mg/mL) and PGM (2.5–5.0 mg/mL) reduced the infectivity of icPC22 A-S1Δ197 significantly (Fig. 5C and 5D).
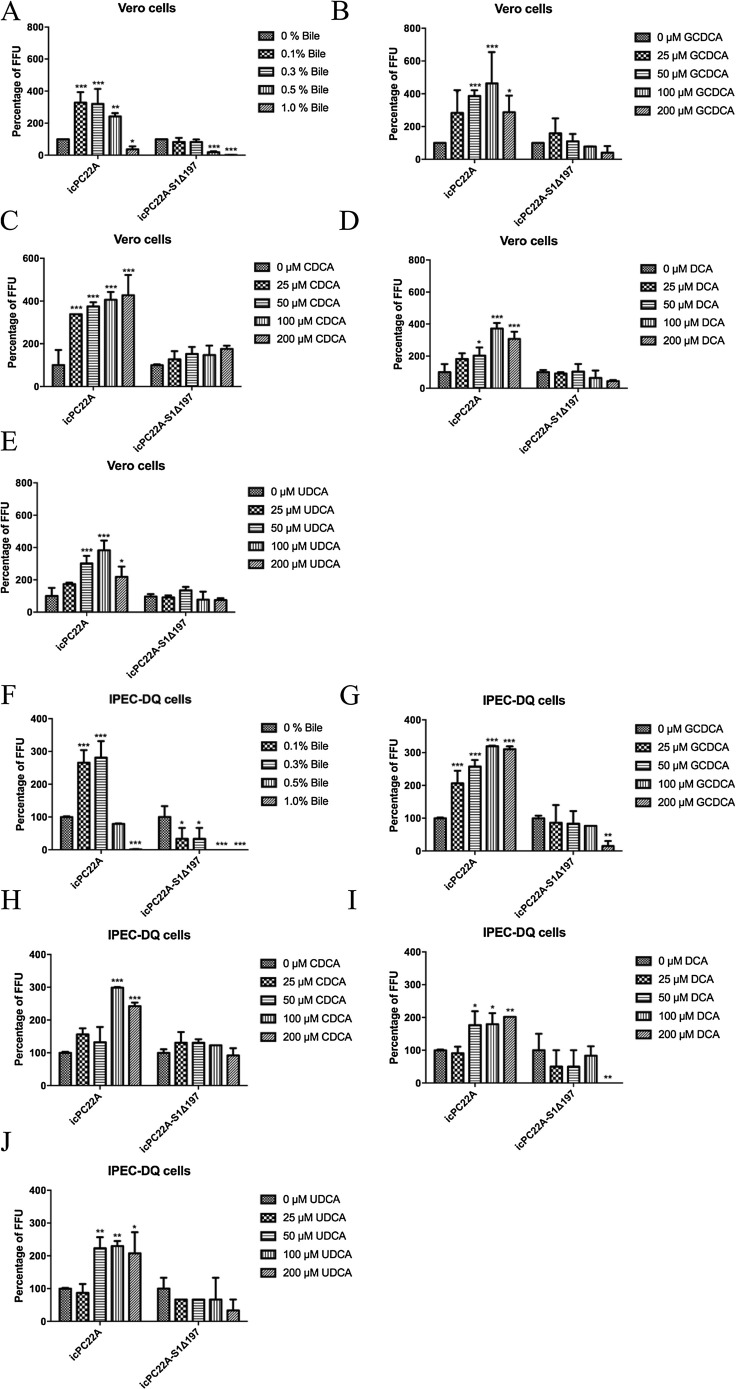
3.6. Bile and bile acids enhanced the infectivity of icPC22A but had no or inhibition effects on icPC22A-S1Δ197
In Vero cells, low to medium concentrations of swine bile (0.1-0.5%) increased 2.4–3.3 fold of the infectivity of icPC22A (Fig. 6 A). High concentration of bile (1.0%) inhibited icPC22 A infection. Bile acids GCDCA (25–200 μM), CDCA (25–200 μM), DCA (50–200 μM) and UDCA (50–200 μM) facilitated the infectivity of icPC22A (Fig. 6B–E). However, the infectivity of icPC22A-S1Δ197 was unaffected (0.1-0.3%) or inhibited significantly by bile (0.5–1.0%) by bile (Fig. 6A). Bile acids at the tested concentrations had no effects on the infection of icPC22 A-S1Δ197 (Fig. 6B–E).
Fig. 6.
Effect of bile and bile acids on the infection of PEDV icPC22A and icPC22A-S1Δ197 in Vero (A–E) and IPEC-DQ cells (F–J). Monolayers of Vero or IPEC-DQ cells in 96-well plates were inoculated with a MOI of 0.02 icPC22 A or icPC22 A-S1Δ197 in medium containing 10 μg/mL trypsin and different concentrations of bile or bile acids. After incubation at 37℃ for 2 h, the inoculum was removed followed by three washings. DMEM/RPMI-1640 containing 50 μg/mL SBTI and swine PEDV positive serum (virus neutralization titer 1:1024, 1:500 dilution) was added. At 8 hpi, cells were fixed with acetone-methanol (1:4) and infectivity was tested by IF assay. Each Experiment was performed three times. Data are shown as M ± SD, and number of asterisks indicate significant difference between groups (*, p < 0.05; **, P < 0.01; ***, P < 0.001).
Similarly, in IPEC-DQ cells, the infectivity of icPC22A was increased significantly by 0.1-0.3% bile but was decreased significantly by the concentration of 1.0% (Fig. 6F). However, 0.1–1.0% bile decreased the infectivity of icPC22A-S1Δ197 significantly. Bile acids GCDCA (25–200 μM), CDCA (100–200 μM), DCA (50–200 μM) and UDCA (50–200 μM) facilitated the infectivity of icPC22A (Fig. 6G–J). No significant effects of bile acids on icPC22A-S1Δ197 were observed except for that the highest concentration (200 μM) of GCDCA and DCA decreased its infectivity significantly (Fig. 6G–J).
4. Discussion
Previously, our laboratory isolated the first S1 NTD-del PEDV strain, PC177, from Vero cell culture (Oka et al., 2014). Recent studies revealed that S1 NTD-del PEDVs naturally evolved in the field and often coinfected pigs with the S-intact PEDV (Suzuki et al., 2015; Diep et al., 2017; Zhang et al., 2018a; Su et al., 2018). The S1 NTD-del PEDV had no tissue tropism change compared with the S-intact PEDV (Suzuki et al., 2016; Hou et al., 2017). This differs from porcine respiratory coronavirus (PRCV) that is a S1 NTD-del version of TGEV and changes the major tissue tropism from intestines to respiratory tract (Zhang et al., 2007). In this study, we investigated the replication of S1 NTD-del and S-intact PEDVs during coinfection in pigs.
We developed a duplex RT-qPCR targeting the S gene of PEDV to differentiate the two viruses. We found that the duplex RT-qPCR assay had different sensitivities for the detection of infectious icPC22 A and icPC22 A-S1Δ197 (Table S1) although it had a similar detection limit (5 log10 copies/mL) for both viruses. For example, 9 log10 S gene copies/mL corresponded to 6.2 and 4.2 log10 FFU/mL for icPC22A and icPC22A-S1Δ197, respectively. It may reflect the fact that icPC22A-S1Δ197 replicates to lower peak infectious titers (∼5.0 log10 FFU/mL) than that (∼7.0 log10 FFU/mL) of icPC22A in Vero cells although both viruses replicated to similar peak titers based on the S gene (∼10.0 log10 copies/mL) (Fig. 3A). In the mixture of two viruses, the detection sensitivity of each virus differed significantly (Tables S2). For the detection of icPC22 A at high titers (6.2 log10 FFU/mL) and as the predominant virus in the mixture of two viruses (icPC22A: icPC22 A-S1Δ197 = 102-105:1), the S gene titers (∼8.8–9.1 log10 S gene copies/mL) (Table S2) were similar to that (∼9.0 log10 S gene copies/mL) in the single virus alone (Table S1). Because icPC22 A replicated to high titers and was the predominant virus during the coinfection in pigs, we can predict that the peak infectious titer of icPC22A in coinfection was ∼1.0 log10-higher than that in its single infection based on the duplex RT-qPCR results (∼11.4 vs ∼10.5 log10 S gene copies/mL) (Table 2). So, we conclude that the replication of icPC22A was interfered at the beginning based on delayed timing (1.5 vs 1.0 dpi) to reach peak titers but enhanced eventually in coinfection compared with its single infection in pigs. On the other hand, the sensitivity for the detection of icPC22A-S1Δ197 decreased significantly and icPC22A-S1Δ197 became undetectable when icPC22A was high (6.2 log10 FFU/mL) and icPC22 A-S1Δ197 infectious titers were lower than 3.2 log10 FFU/mL in the mixture of two viruses (Table S2). Therefore, the decreased peak S gene titer (∼5.8 log10 copies/mL) of icPC22A-S1Δ197 in coinfection of pigs compared with that (∼7.6 log10 copies/mL) in its single infection may be due to inhibition of replication or the decreased detection sensitivity of the duplex RT-qPCR for the detection of icPC22A-S1Δ197 during coinfection (Table 2).
In the first passage of coinfection, we observed much lower numbers of S1 NTD-del PEDV-infected cells than the S-intact PEDV-infected cells in the small intestines of pigs infected with the same dose of both viruses simultaneously. In addition, plaque assays were performed to isolate and purify individual viruses from the SIC of one of the coinfection pigs. However, only S-intact PEDV was isolated alone. In contrast, S1 NTD-del PEDV was exclusively detected together with the S-intact PEDV from the plagues. In the second passage of the coinfection in pigs, S-intact PEDV but not S1 NTD-del PEDV RNA was detected in the LIC of the pig inoculated with the SIC of the pig inoculated with both viruses. This finding suggests that S1 NTD-del PEDV had no replication advantage and was either outcompeted or coexisted with S-intact PEDV in pigs. This conclusion is in agreement with what were observed in the field (Diep et al., 2017; Su et al., 2018). S1 NTD-del PEDV replicated to a lower peak titer in coinfection than that in single virus infection in both Vero cells and IPEC-DQ cells. These in vitro results were similar to those in the pig studies. In the second passage of coinfection, S1 NTD-del PEDV RNA was detected in Vero cells but not in IPEC-DQ cells, probably due to the lower replication efficiency of the S1 NTD-del PEDV in IPEC-DQ cells than in Vero cells (Fig. 3A and C).
Similar to other sialic acid-binding coronaviruses, the S1 NTD of PEDV has a sialic acid binding activity, and the sugar-binding activities of a field isolate PEDV variant CHGD-01 was stronger than that of the prototype strain PEDV CV777 using BM (Deng et al., 2016). The sialic acid binding activity occurred within the N-terminal 249 residues and the capacity of sialic acid binding differs among PEDV strains using an hemagglutination assay (Li et al., 2016). The recombinant virus used in this study, PEDV icPC22 A-S1Δ197, is a S1 NTD-del version of icPC22A. It lost most sialic acid binding activity tested in Vero cells (Hou et al., 2017). In this study, we comparatively tested icPC22 A-S1Δ197 and icPC22 A in Vero and IPEC-DQ cells treated with NA. Similar to the previous reports, our results confirmed that the binding of S1 NTD of PEDV to sialic acids on cell surface enhanced virus infectivity. BM is a mixture of highly glycosylated proteins containing sugar moieties, such as 5-N-acetyl-9-O-acetylneuraminic acid (Neu5,9Ac2), 5-N-glycolylneuraminic acid (Neu5Gc), and 5-N-acetylneuraminic acid (Neu5Ac). Among them, Neu5Gc and Neu5Ac can serve as receptors or co-receptors for some alphacoronaviruses (e.g., TGEV) and gammacoronaviruses [e.g., infectious bronchitis virus (IBV)] (Cavanagh and Davis, 1986; Krempl et al., 1997). Similar to TGEV, which uses Neu5Gc and Neu5 Ac as co-receptors (Krempl et al., 1997; Schultze et al., 1996; Schwegmann-Wessels and Herrler, 2006), the S1 NTD of PEDV interacted with sugar (Deng et al., 2016). PGM contains 0.5–1.5% N-Acetylneuraminic acid (NeuAc). Our data indicated that low-medium concentrations of BM or PGM enhanced the infectivity of S-intact PEDV in both Vero and IPEC-DQ cells. In contrast, the S1 NTD-del PEDV was not affected or inhibited, probably due to the covered mucin that blocked virus binding to the receptors. The intestinal epithelial cells are covered by a layer of mucus that is rich in sialic acids. These results suggest that low-medium amount of mucin binding to S-intact PEDV may help the virus particles approach the receptors via sticking to the sialic acids on the cell surface. However, the receptor binding domain (RBD) on the S protein of S1 NTD-del PEDV, which lacks most sialic acid binding activity, is probably covered with mucin and blocked its binding to the receptors. When high concentration of mucin saturates the S1 NTD of S-intact PEDV, it can also cover viral RBD and block its binding to the receptors, resulting in decreased infectivity as observed in 1.0 mg/mL BM effects on S-intact PEDV in Vero cells (Fig. 5A).
The major components of bile include bile acids, cholesterol, lipids, bilirubin, proteins and carbohydrates. The bile acids are stored in the gall bladder (at ∼300 mM) and released into the small intestine (McLeod and Wiggins, 1968). In the small intestine, the total bile acids have a concentration of 2–30 mM (Dowling, 1973). We selected two primary bile acids (GCDCA and CDCA) and two secondary bile acids (DCA and UDCA) to test the effects of bile acids on PEDV infection. The secondary bile acids are dehydroxylated ones from the primary bile acids by intestinal bacteria (Björkhem, 1985). Our data indicated that a broad range of concentrations of bile and bile acids enhanced the infectivity of S-intact PEDV in both Vero and IPEC-DQ cells. However, bile and bile acids cannot increase the infectivity of S1 NTD-del PEDV. These results suggest that the bile- and bile acid-mediated promotion of PEDV infection is related to the S1 NTD. It was reported that GCDCA may facilitate the adaptation of a S-intact PEDV to trypsin-free growth in Vero cells at the early passages (< P10) (Kim et al., 2017). However, the mechanisms of the bile acid function on the PEDV infection is unknown and consequently need to be further investigated.
The inability of the S1 NTD-del PEDV to outcompete the S-intact PEDV may account for the fact that S1 NTD-del PEDVs were exclusively detected from coinfection with the S-intact PEDV (Diep et al., 2017; Su et al., 2018). Consequently, the disease caused by the coinfection was as severe as that by the highly virulent S-intact PEDV alone. This situation is quite different from what occurred for PRCV and TGEV: the wild spread of clinically mild PRCV, which often infects pigs asymptomatically and repeatedly, inducing protective herd immunity against TGEV outbreaks (VanCott et al., 1994; Brim et al., 1995). PRCV functions as a natural effective vaccine against TGEV. However, for PEDV, the clinically mild S1 NTD-del PEDV variants do not infect pigs alone. Therefore, safe and effective vaccines are still desired to control the deadly PEDV infection in neonatal piglets. Due to the 100% mortality rates of piglets in the icPC22 A-infected pigs (Hou et al., 2017) and coinfection groups, we currently cannot determine whether the pathogenicity of PEDV icPC22 A is enhanced during the coinfection. That should be investigated in older pigs (weaned pigs, sows and boars) which are more resistant to PEDV infection and diseases (Niederwerder and Hesse, 2018). In addition, the emergence of S1 NTD-del PEDV variants may complicate PEDV disease pattern on farms, which needs to be further investigated.
5. Conclusion
In this study, we showed that the replication of the S-intact PEDV was enhanced during coinfection with an S1 NTD-del PEDV in pigs. We found that mucin, bile and bile acids can all increase the infection of S-intact PEDV but not the S1 NTD-del PEDV. This feature may help explain why S-intact PEDV outcompetes S1 NTD-del PEDV in vivo. Further studies are needed to understand the mechanisms of mucin-, bile- and bile acid-mediated enhancement or inhibition effects on the infection of different PEDV variants.
Ethics in animal experimentation
All animal experiments were performed according to the protocols approved by Institutional Animal Care and Use Committee (IACUC) of the Ohio State University (OSU).
Conflict of interest statement
The authors declare no conflict of interest.
Declarations of interest
None.
Acknowledgements
We thank Juliette Hanson, Megan Strother, Dennis Hartzler, Ronna Wood, Sara Tallmadge, and Jeff Ogg for animal care assistance and Xiaohong Wang for technical assistance. IPEC-DQ cells were provided by Dr. Dongwan Yoo, University of Illinois at Urbana-Champaign, Urbana IL, USA. The MAb SD6-29 was provided by Drs. Eric Nelson and Steven Lawson, South Dakota State University. The MAbs 56H4, 60F11 and 72A8 were provided by Dr. Berend Jan Bosch, Utrecht University, Utrecht, The Netherlands. This work was partially funded by National Institute of Food and Agriculture, U.S. Department of Agriculture, under award number 2015-67015-23067 (QW, PI). Salaries and research support were provided by state and federal funds appropriated to OARDC, OSU. Yunfang Su’s stipend is supported by China Scholarship Council.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.vetmic.2018.11.025.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Björkhem I. Mechanism of bile acid biosynthesis in mammalian liver. New Compr. Biochem. 1985;12:231–278. doi: 10.1016/S0167-7306(08)60685-7. [DOI] [Google Scholar]
- Brim T.A., VanCott J.L., Lunney J.K., Saif L.J. Cellular immune responses of pigs after primary inoculation with porcine respiratory coronavirus or transmissible gastroenteritis virus and challenge with transmissible gastroenteritis virus. Vet. Immunol. Immunopathol. 1995;48:35–54. doi: 10.1016/0165-2427(94)05416-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986;67(Pt. 7):1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- Chang K.-O., Sosnovtsev S.V., Belliot G., Kim Y., Saif L.J., Green K.Y. Bile acids are essential for porcine enteric calicivirus replication in association with down-regulation of signal transducer and activator of transcription 1. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8733–8738. doi: 10.1073/pnas.0401126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F., Ye G., Liu Q., Navid M.T., Zhong X., Li Y., Wan C., Xiao S., He Q., Fu Z.F., Peng G. Identification and comparison of receptor binding characteristics of the spike protein of two porcine epidemic diarrhea virus strains. Viruses. 2016;8:55. doi: 10.3390/v8030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarets L.M.B., Theuns S., Roukaerts I.D.M., Acar D.D., Nauwynck H.J. Role of sialic acids in feline enteric coronavirus infections. J. Gen. Virol. 2014;95:1911–1918. doi: 10.1099/vir.0.064717-0. [DOI] [PubMed] [Google Scholar]
- Diep N.V., Norimine J., Sueyoshi M., Lan N.T., Yamaguchi R. Novel porcine epidemic diarrhea virus (PEDV) variants with large deletions in the spike (S) gene coexist with PEDV strains possessing an intact S gene in domestic pigs in Japan: a new disease situation. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling R.H. The enterohepatic circulation of bile acids as they relate to lipid disorders. J. Clin. Pathol. Suppl. (Assoc. Clin. Pathol.) 1973;5:59–67. doi: 10.1136/jcp.s1-5.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettayebi K., Crawford S.E., Murakami K., Broughman J.R., Karandikar U., Tenge V.R., Neill F.H., Blutt S.E., Zeng X.-L., Qu L., Kou B., Opekun A.R., Burrin D., Graham D.Y., Ramani S., Atmar R.L., Estes M.K. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Lin C.-M., Yokoyama M., Yount B.L., Marthaler D., Douglas A.L., Ghimire S., Qin Y., Baric R.S., Saif L.J., Wang Q. Deletion of a 197-Amino-Acid region in the N-Terminal domain of spike protein attenuates porcine epidemic diarrhea virus in piglets. J. Virol. 2017;91 doi: 10.1128/JVI.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Meulia T., Gao X., Saif L.J., Wang Q. The deletion of both tyrosine-based endocytosis signal and endoplasmic reticulum-retrieval signal in the cytoplasmic tail of spike protein attenuates PEDV in pigs. J. Virol. 2019;93:e01758–18. doi: 10.1128/JVI.01758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Wang Q., Scheuer K.A., Lu Z., Zhang Y., Saif L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014;20:662–665. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Oh C., Shivanna V., Hesse R.A., Chang K.-O. Trypsin-independent porcine epidemic diarrhea virus US strain with altered virus entry mechanism. BMC Vet. Res. 2017;13:356. doi: 10.1186/s12917-017-1283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C., Schultze B., Laude H., Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 1997;71:3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C., Ballesteros M.L., Zimmer G., Enjuanes L., Klenk H.D., Herrler G. Characterization of the sialic acid binding activity of transmissible gastroenteritis coronavirus by analysis of haemagglutination-deficient mutants. J. Gen. Virol. 2000;81:489–496. doi: 10.1099/0022-1317-81-2-489. [DOI] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., van Kuppeveld F.J.M., He Q., Rottier P.J.M., Bosch B.-J. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016;226:117–127. doi: 10.1016/j.virusres.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li W., Lucio de Esesarte E., Guo H., van den Elzen P., Aarts E., van den Born E., Rottier P.J.M., Bosch B.-J. Cell attachment domains of the porcine epidemic diarrhea virus spike protein are key targets of neutralizing antibodies. J. Virol. 2017;91 doi: 10.1128/JVI.00273-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Luo R., He Q., van Kuppeveld F.J.M., Rottier P.J.M., Bosch B.-J. Aminopeptidase N is not required for porcine epidemic diarrhea virus cell entry. Virus Res. 2017;235:6–13. doi: 10.1016/j.virusres.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod G.M., Wiggins H.S. Bile-salts in small intestinal contents after ileal resection and in other malabsorption syndromes. Lancet Lond. Engl. 1968;1:873–876. doi: 10.1016/s0140-6736(68)90235-3. [DOI] [PubMed] [Google Scholar]
- Niederwerder M.C., Hesse R.A. Swine enteric coronavirus disease: A review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound. Emerg. Dis. 2018;65:660–675. doi: 10.1111/tbed.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Saif L.J., Marthaler D., Esseili M.A., Meulia T., Lin C.-M., Vlasova A.N., Jung K., Zhang Y., Wang Q. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet. Microbiol. 2014;173:258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- Saif L.J., Ward L.A., Yuan L., Rosen B.I., To T.L. The gnotobiotic piglet as a model for studies of disease pathogenesis and immunity to human rotaviruses. Arch. Virol. Suppl. 1996;12:153–161. doi: 10.1007/978-3-7091-6553-9_17. [DOI] [PubMed] [Google Scholar]
- Schultze B., Krempl C., Ballesteros M.L., Shaw L., Schauer R., Enjuanes L., Herrler G. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J. Virol. 1996;70:5634–5637. doi: 10.1128/jvi.70.8.5634-5637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconj. J. 2006;23:51–58. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Bauer S., Winter C., Enjuanes L., Laude H., Herrler G. The sialic acid binding activity of the S protein facilitates infection by porcine transmissible gastroenteritis coronavirus. Virol. J. 2011;8:435. doi: 10.1186/1743-422X-8-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Maejima M., Islam M.T., Miyazaki A., Kawase M., Matsuyama S., Taguchi F. Porcine aminopeptidase N is not a cellular receptor of porcine epidemic diarrhea virus, but promotes its infectivity via aminopeptidase activity. J. Gen. Virol. 2016;97:2528–2539. doi: 10.1099/jgv.0.000563. [DOI] [PubMed] [Google Scholar]
- Su Y., Hou Y., Prarat M., Zhang Y., Wang Q. New variants of porcine epidemic diarrhea virus with large deletions in the spike protein, identified in the United States. Arch. Virol. 2018:2016–2017. doi: 10.1007/s00705-018-3874-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Murakami S., Takahashi O., Kodera A., Masuda T., Itoh S., Miyazaki A., Ohashi S., Tsutsui T. Molecular characterization of pig epidemic diarrhoea viruses isolated in Japan from 2013 to 2014. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015;36:363–368. doi: 10.1016/j.meegid.2015.10.017. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Shibahara T., Yamaguchi R., Nakade K., Yamamoto T., Miyazaki A., Ohashi S. Pig epidemic diarrhoea virus S gene variant with a large deletion non-lethal to colostrum-deprived newborn piglets. J. Gen. Virol. 2016;97:1823–1828. doi: 10.1099/jgv.0.000513. [DOI] [PubMed] [Google Scholar]
- Teeravechyan S., Frantz P.N., Wongthida P., Chailangkarn T., Jaru-Ampornpan P., Koonpaew S., Jongkaewwattana A. Deciphering the biology of porcine epidemic diarrhea virus in the era of reverse genetics. Virus Res. 2016;226:152–171. doi: 10.1016/j.virusres.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanCott J.L., Brim T.A., Lunney J.K., Saif L.J. Contribution of antibody-secreting cells induced in mucosal lymphoid tissues of pigs inoculated with respiratory or enteric strains of coronavirus to immunity against enteric coronavirus challenge. J. Immunol. Baltim. Md. 1994;1950(152):3980–3990. [PubMed] [Google Scholar]
- Zhang X., Hasoksuz M., Spiro D., Halpin R., Wang S., Stollar S., Janies D., Hadya N., Tang Y., Ghedin E., Saif L. Complete genomic sequences, a key residue in the spike protein and deletions in nonstructural protein 3b of US strains of the virulent and attenuated coronaviruses, transmissible gastroenteritis virus and porcine respiratory coronavirus. Virology. 2007;358:424–435. doi: 10.1016/j.virol.2006.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yim-Im W., Chen Q., Zheng Y., Schumacher L., Huang H., Gauger P., Harmon K., Li G. Identification of porcine epidemic diarrhea virus variant with a large spike gene deletion from a clinical swine sample in the United States. Virus Genes. 2018;54:323–327. doi: 10.1007/s11262-018-1542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ke H., Blikslager A., Fujita T., Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 2018:92. doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.