Highlights
-
•
VSV M protein interacts with the i subunit of eIF3.
-
•
The region of M that interacts with eIF3i is located within the 122- to -181 amino acids.
-
•
M–eIF3i interaction affects VSV growth.
Keywords: VSV, Matrix protein, EIF3i, Akt1, Interaction
Abstract
The matrix protein of vesicular stomatitis virus (VSV) performs multiple functions during viral genome replication and virion production and is involved in modulating multiple host signaling pathways that favor virus replication. To perform numerous functions within infected cells, the M protein needs to recruit cellular partners. To better understand the role of M during VSV replication, we looked for interacting partners by using the two-hybrid system. The eukaryotic translation initiation factor 3, subunit i (eIF3i) was identified to be an M-binding partner, and this interaction was validated by GST pull-down and laser confocal assays. Through a mutagenesis analysis, we found that some mutants of M between amino acids 122 and 181 impaired but did not completely abolish the M–eIF3i interaction. Furthermore, the knockdown of eIF3i by RNA interference decreased viral replication and transcription in the early stages but led to increase in later stages. VSV transcription was increased at 4 h post-infection but was not changed at 8 and 12 h post-infection after the over-expression of eIF3i. Finally, we also demonstrated that VSV could inhibit the activity of Akt1 and that the knockdown of eIF3i inhibited the expression of the ISGs regulated by phospho-Akt1. These results indicated that eIF3i may affect VSV growth by regulating the host antiviral response in HeLa cells.
1. Introduction
Vesicular stomatitis (VS) is a highly contagious disease in swine, horses, cattle and other mammals. It is caused by the vesicular stomatitis virus (VSV) and characterized by widely erosive vesicles on the surface of the lips, tongue, gums and teats (Letchworth et al., 1999). Vesicular stomatitis virus (VSV) is a member of the Vesiculovirus genus, which belongs to the Rhabdoviridae family. The VSV genome is composed of a single negative polarity RNA strand that encodes five proteins: nucleocapsid (N), phosphoprotein (P), matrix (M) protein, glycoprotein (G) and large (L) viral polymerase (Barr et al., 2002). The nucleocapsid (N) protein tightly binds to the viral genomic and nascent RNA. The polymerase L is an RNA-dependent RNA polymerase (RdRP) that associates with phosphoprotein (P), nucleocapsid (N) and genomic RNA to form the transcriptionally active nucleocapsid (Barr et al., 2002). This complex is condensed by matrix protein (M) to generate a coiled helical structure and then enclosed within a lipid bilayer directed by the integral transmembrane glycoprotein (G) (Ge et al., 2010, Beilstein et al., 2015).
The matrix (M) protein of VSV is a multifunctional protein that plays an important role in the induction of cell rounding (Blondel et al., 1990), the modulation of apoptosis (Kopecky et al., 2001), the inhibition of host transcription (Black and Lyles, 1992), virion assembly/budding (Gaudin et al., 1995, Harty et al., 1999) and the blocking of nuclear-cytoplasmic transport of host RNAs (Her et al., 1997). To perform numerous functions, viral proteins may need to interact with specific host proteins. A number of M-interacting cellular proteins have been identified that contribute to these various functions during viral infection. It has been demonstrated that host E3 ubiquitin ligase Nedd4 interacts with M protein to promote efficient virus egress (Harty et al., 2001). In addition to facilitating virus egress, VSV M protein can deregulate nucleo-cytoplasmic transport to inhibit cellular gene expression by targeting Nup98 (von Kobbe et al., 2000). Furthermore, M protein can also interact with dynamin and inhibit clathrin-mediated endocytosis. Disrupting the M-dynamin interaction blocks VSV assembly and budding and affects the cellular localization of the N and G proteins (Raux et al., 2010). In addition, M protein has been shown to be associated with host factor-Rae1, and this complex serves as a “platform” to recruit cellular proteins involved in host transcription (Rajani et al., 2012). Thus, further exploration of the interaction of M with host cellular protein(s) is essential for understanding the roles of M in the replication and pathogenesis of VSV. In this study, we identified the host protein eIF3i as a novel M-interacting partner that regulates the growth of VSV.
EIF3i is a subunit of the eukaryotic translation initiation factor 3 complex, which was initially isolated as an interactor of TGF β-receptor type II (Chen et al., 1995). The EIF3 complex has been shown to affect the growth of different groups of viruses. For example, the rabies virus (RV) M protein can inhibit eukaryotic translation via a protein interaction with eIF3 h (Komarova et al., 2007). Furthermore, eIF3f can also interact with the S protein of SARS-CoV and infectious bronchitis virus (IBV) to inhibit expression of host genes (Xiao et al., 2008). In addition to inhibiting host translation, eIF3f can inhibit HIV replication by specifically impeding the 3′ end processing of HIV-1 mRNAs (Valente et al., 2009). However, no studies have investigated the role of eIF3 complex in the VSV life cycle. Here, we demonstrated that VSV M protein interacts with eIF3i and that this interaction affects the growth of VSV.
2. Materials and methods
2.1. Cells and virus
HeLa and BSR-T7/5 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (GIBCO, Invitrogen), supplemented with 10% fetal bovine serum (FBS; HyClone), 2 mM l-glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin and maintained in a 37 °C, 5% CO2 incubator. Vesicular stomatitis virus serotype Indiana (VSV-IND) was propagated in HeLa cells.
2.2. Construction of expression vectors
The eIF3i gene (GenBank accession no. NM_003757. 3) was amplified by using cDNA extracted from HeLa cells and inserted into the pcmv-flag vector (catalog no. 635688; Clontech) to generate the pcmv-flag-eIF3i plasmid. A Flag-M eukaryotic expression vector was constructed using the pMD-18T backbone vector that contains the T7 polymerase promoter, the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) and Flag-M, followed by a T7 terminator, which was designated pT-Flag-M. Other EGFP-M, EGFP, Dsred-eIF3i and Dsred eukaryotic expression plasmids were constructed as described above and designated pT-EGFP-M, pT-EGFP, pT-Dsred-eIF3i and pT-Dsred, respectively. The eIF3i and M ORFs were cloned into the C-terminus of GST in the vector pGEX-4T-1 to construct the prokaryotic expression plasmids. The primers sequences are listed in Table 1 .
Table 1.
Primers used in this study.
| Primer | Sequence(5′–3′)a | Purpose |
|---|---|---|
| BD-M-F | CCGGAATTCCGGATGAGTTCCTTAAAGAAGATTCTC | Amplification and cloning of M in PGBKT7 vector |
| BD-M-R | CGCGGATCCGCGTCATTTGAAGTGGCTGACAG | Amplification and cloning of M in PGBKT7 vector |
| AD-eIF3i-F | CCGGAATTCCGGATGAAGCCGATCCTACTGC | Amplification and cloning of eIF3i in pGADT7 vector |
| AD-eIF3i-R | CCGCTCGAGCGGTTAAGCCTCAAACTCAAATTC | Amplification and cloning of eIF3i in pGADT7 vector |
| GST-M-F | CGCGGATCCGCGATGAGTTCCTTAAAGAAGATTCTC | Amplification and cloning of M in PGEX-4T-1 vector |
| GST-M-R | CCGGAATTCCGGTCATTTGAAGTGGCTGACAG | Amplification and cloning of M in PGEX-4T-1 vector |
| GST-eIF3i-F | CCGGAATTCCGGATGAAGCCGATCCTACTGC | Amplification and cloning of eIF3i in pGEX-4T-1 vector |
| GST-eIF3i-R | CCGCTCGAGCGGTTAAGCCTCAAACTCAAATTC | Amplification and cloning of eIF3i in PGEX-4T-1 vector |
| Flag-eIF3i-F | CGGAATTCCGATGAAGCCGATCCTACTGC | Amplification and cloning of eIF3i in pcmv-flag vector |
| Flag-eIF3i-R | GGGGTACCCCTTAAGCCTCAAACTCAAATTC | Amplification and cloning of eIF3i in pcmv-flag vector |
| VSV P-F | GTGACGGACGAATGTCTCATAA | Amplification of VSV P mRNA |
| VSV P-R | TTTGACTCTCGCCTGATTGTAC | Amplification of VSV P mRNA |
| VSV 2795-F | GTGACGGACGAATGTCTCATAA | Amplification of VSV antigenomic RNA |
| VSV2955-R | TGATGAATGGATTGGGATAACA | Amplification of VSV antigenomic RNA |
| Actin-F | TGACGTGGACATCCGCAAAG | Amplification of actin mRNA |
| Actin-R | CTGGAAGGTGGACAGCGAGG | Amplification of actin mRNA |
Underlined sequences indicate restriction sites.
2.3. Yeast two-hybrid assay
VSV M was inserted into a pGBKT7 plasmid, fused to the DNA-binding domain and transformed into the yeast strain AH109 using LiAc. The auto activation and toxicity of the bait were verified on SDO/X (SD/-Trp/X-a-Gal) and SDO (SD/-Trp) plates. The normalized mouse brain cDNA library fused to the sequence encoding the GAL4 activation domain, pre-transformed in the Y187 yeast strain, was purchased from Clontech (catalog no. 630488; Clontech). Two-hybrid screens were performed using bait yeast mating with the library yeast strain and selected on DDO/X/A (SD/-Leu/-Trp/X-a-Gal/AbA) plates. Positive colonies were confirmed on QDO/X/A (SD/-Ade/-His/-Leu/-Trp/X-a-Gal/AbA) plates. Inserts of all putative positive clones were isolated, sequenced and analyzed through NCBI BLAST searches. To eliminate false-positive clones, the bait and prey plasmids were co-transformed into the AH109 yeast strain. AH109 co-transformed with pGBKT7-p53 (BD-p53) and pGADT7-T (AD-T) was used as a positive control, and AH109 co-transformed with pGBKT7-Lambda (BD-Lam) and AD-T was used as a negative control.
2.4. Plasmid transfection
Briefly, the cells were transfected with 1 ml of serum-free DMEM containing 4 μg of plasmids and 12 μl of Lipofectamine 2000 transfection reagent. At 4 h post-transfection, the transfection mixture was replaced with DMEM supplemented with 2% FBS and incubated for an additional 24 h before being assayed.
2.5. GST pull-down assay
For expression of the GST-tagged or GST-M fusion protein, E. coli BL21 (DE3) bacteria were transformed with the pGEX-4T-1 or pGEX-4T-M plasmids, and expression was induced by addition of 1 mM IPTG for 18 h at 16 °C. The collective bacteria pellet was resuspended in cold phosphate-buffered saline (PBS) containing 1 mg/ml protease inhibitor PMSF, followed by gentle sonication. The soluble proteins were obtained by centrifugation at 12,000 × g for 10 min at 4 °C. Subsequently, the soluble GST or GST-M protein was incubated with glutathione Sepharose 4B beads (catalog no. 17075601; GE Healthcare) for 6 h at 4 °C. The beads were washed three times with cold PBS. Cells transfected with the pcmv-flag-eIF3i or pT-M plasmid were washed with cold PBS and lysed with NP-40 (catalog no. P0013F; Beyotime, Shanghai, China). The cell lysate was centrifuged at 12,000 × g for 10 min at 4 °C, and the supernatant was incubated with glutathione Sepharose 4 B beads for 3 h at 4 °C. The beads were washed three times again, and the bead-bound protein complexes were separated by SDS-PAGE followed by western blotting using an anti-flag MAb (1:2000) (catalog no. 66008-2-Ig; Proteintech) and an anti-GST MAb (1:2000) (catalog no. HRP-66001; Proteintech). GST-eIF3i expressed in E. coli BL21(DE3) and Flag-M expressed in BSR-T7/5 cells were also included in the GST pull-down assay to further validate the interaction as described above.
2.6. Confocal microscopy
BSR-T7/5 cells seeded on glass cover slips were co-transfected with the indicated plasmids using Lipofectamine 2000 (Life Technology; USA). At 24 h post-transfection, the cells were washed with PBS and fixed with 4% paraformaldehyde for 20 min. After being stained with Hoechst (0.2 mg/ml, 10 min) and washed with PBS, the cells were visualized by laser scanning confocal microscopy.
2.7. Western blotting
Proteins were separated by SDS-PAGE in 12% gels, followed by electrotransfer to PVDF membranes. The blots were then blocked with 5% skim milk and incubated with the respective primary antibodies diluted in PBST (0.05% Tween-20 in PBS). After washing with PBST, the blots were incubated with secondary goat anti-rabbit (mouse) IgG-HRP. Finally, they were washed three times with PBST and visualized by enhanced chemiluminescence (ECL) (Thermo Fisher Scientific).
2.8. Gene knockdown by siRNAs
The siRNAs for silencing the eIF3i gene were purchased from Guangzhou RiboBio, Corp. China. The siRNA target sequences were as follows: sieIF3i 1 (sense), 5′-GACCCACAGUACUUCGAAUTT-3′; sieIF3i 2 (sense), 5′-CCACAACUCUUGAACAUCATT-3′; sieIF3i 3 (sense), 5′- GGACAGAACGUCCUGUCAATT-3′, The control siRNA was provided by Guangzhou RiboBio, Corp. (China). Monolayers of HeLa cells grown in 6-well plates (Corning, USA) were transfected with 50 nM of mixed eIF3i-specific siRNAs (sieIF3i) using the Lipofectamine RNAiMax Transfection Reagent (Invitrogen). The knockdown efficiency was examined by western blotting. To investigate the effect of eIF3i knockdown on the replication of VSV, the siRNA-treated HeLa cells were then infected with VSV at an MOI of 1 at 48 h post-transfection. At 4, 8 and 12 h post-infection, the viral RNA copies in the cells were detected.
2.9. Real-Time RT-PCR
Total RNA was extracted from the VSV-infected HeLa cells using a TriPure RNA Isolation reagent (catalog no. 11667165001; Roche). First-strand cDNA was synthesized with 1 μg of total RNA by use of the M-MLV reverse transcriptase (catalog no. 2641A; TaKaRa, Dalian, China) according to the manufacturer’s protocol. Oligo (dT) primers were used for synthesis of all cDNAs except the VSV antigenome, for which the VSV2955R primer was used. VSV P mRNA and the antigenome RNA of VSV were quantified by a real-time RT-PCR assay as described by Das et al. (2014).
2.10. In vitro transcription and luciferase assay
Capped and polyadenylated luciferase and M RNA were synthesized by using the mMESSAGE mMACHINE T7 Ultra kit (Ambion, Austin, TX) according to the manufacturer’s protocol. HeLa cells cultured in 6-well plates were co-transfected with either 2 μg of luciferase mRNA and 2 μg of pT-M or 2 μg of M mRNA and decreasing amounts of luciferase mRNA (1, 0.5, 0.25, 0.125 and 0.06 μg). Luciferase activity was determined by using a luciferase assay system (Promega, USA).
2.11. Statistical analysis
The data were analyzed for statistical significance by using Student’s t-test.
3. Results
3.1. VSV M protein interacts with the i subunit of eIF3
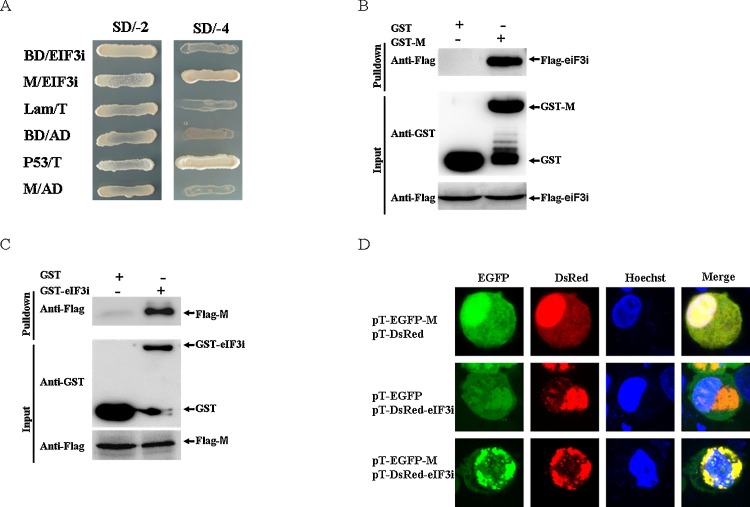
To search for cellular proteins that interact with M, we performed a two-hybrid screen in Saccharomyces cerevisiae (S. cerevisiae). Full-length VSV M was used as bait against a commercial yeast pre-transformed with a normalized mouse brain cDNA library. From 6 million yeast transformants screened, positive clones were isolated, sequenced and subjected to a BLAST search against the NCBI database. Three of the positive clones encoding the translation initiation factor 3 subunit i (eIF3i) were chosen for further investigation. To preclude possible self-activation, a yeast co-transformation assay was conducted, and only BD-M/AD-eIF3i and the positive control co-transformed AH109 strain were alive on the SD/–4 medium (Fig. 1 A). Taken together, these data indicated that eIF3i might be a potential cellular partner for VSV M.
Fig. 1.
VSV M protein interacts with the i subunit of eIF3.
(A) Binding of the VSV-M protein to eIF3i in a yeast two-hybrid system. Yeast strain AH109 was co-transformed with a bait plasmid (BD-M) and a prey plasmid (AD-eIF3i), which encodes the full-length eIF3i fused to the GAL4 activation domain. The empty vector BD was co-transformed with AD-eIF3i to exclude self-activation. Co-transformation of BD-Lambda/AD-T, BD-p53/AD-T and BD/AD were used as negative, positive and blank controls, respectively. (B) GST-M pull-down assay. The GST and GST-M proteins expressed in E. coli BL21 (DE3) were purified with glutathione Sepharose 4B beads (catalog no. 17075601; GE Healthcare). The beads conjugated with GST or GST-M were incubated with the recombinant Flag-eIF3i protein. After washing with cold PBS, the bound proteins were separated by SDS-PAGE (12%) and detected by western blotting. (C) GST- eIF3i pull-down assay. The GST or GST-eIF3i proteins expressed in E. coli BL21 (DE3) were purified with glutathione Sepharose 4B beads (catalog no.17075601; GE Healthcare). The beads conjugated with GST or GST-eIF3i proteins were incubated with the recombinant Flag-M protein. (D) Co-localization of M with eIF3i in the cells. The cells were co-transfected with pT-EGFP-M (or pT-EGFP, as control) and pT-DsRed-eIF3i (or pT-DsRed, as control) and fixed with 4% paraformaldehyde at 24 h post-transfection. The results were obtained with a laser confocal scanning microscope. All the experiments were repeated three times. SD/-2, SD/-Leu/-Trp; SD/-4, SD/-Ade/-His/-Leu/-Trp.
To further validate the eIF3i-M interaction, a GST pull-down assay was conducted with the GST-tagged M protein expressed in E. coli and the Flag-tagged eIF3i protein expressed in BSR-T7/5 cells. The results showed that GST-M, but not GST, interacted with Flag-eIF3i (Fig. 1B). GST-eIF3i expressed in E. coli and the Flag-M expressed in BSR-T7/5 cells were also used in the pull-down assay. The results indicated that GST-eIF3i, but not GST, interacted with Flag-M (Fig. 1C).
To test the co-localization of M protein with eIF3i, the subcellular localization of EGFP-M and Dsred-eIF3i was examined by confocal microscopy in BSR-T7/5 cells. When co-expressed with Dsred, the EGFP-M exhibited homogeneous distribution in the cytoplasm and nucleus. In addition, Dsred-eIF3i distributed in a punctuate manner in the cytoplasm. However, EGFP-M showed a punctuate co-localization with Dsred-eIF3i in the cytoplasm when co-expressed with Dsred-eIF3i (Fig. 1D). Taken together, these results demonstrated that eIF3i was an interacting partner of the VSV M protein.
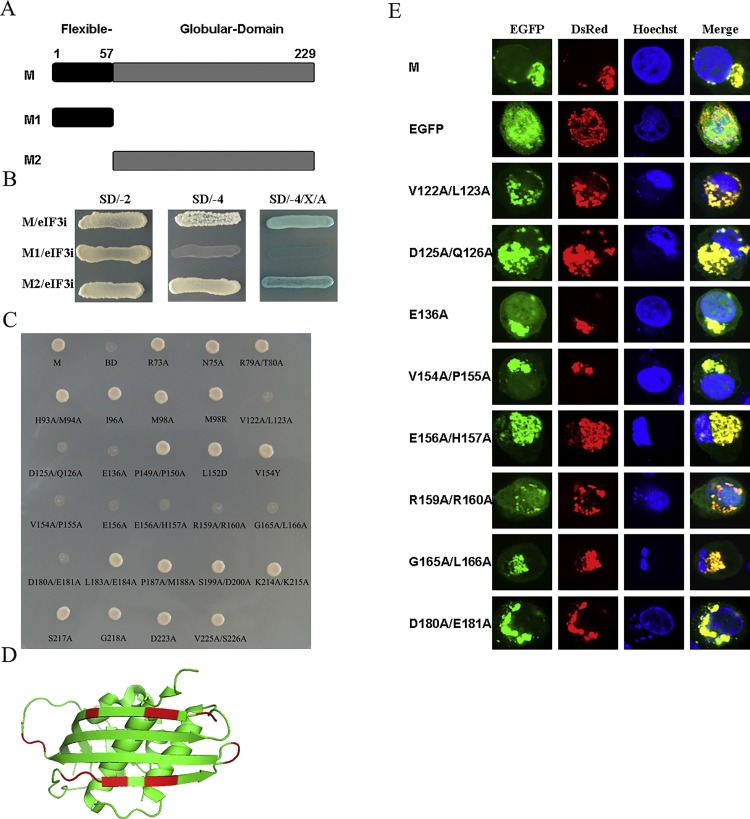
The region of M that interacts with eIF3i is located within the 122- to -181 amino acids
The VSV M protein is composed of two domains, a globular carboxy-terminal domain and a flexible amino-terminal domain (comprising the 57 first residues) (Gaudier et al., 2002, Beilstein et al., 2015) (Fig. 2 A). To assess which domain was necessary for the interaction with eIF3i, each domain of M was fused to the GAL4 DNA-binding domain and examined for their binding ability using the Y2H system. After co-transformation, the globular domain of M grew well on all three selective media and formed blue colonies when grown on SD/-4/X/A (SD/-Ade/-His/-Leu/-Trp/X −α-Gal/Aba) medium; However, the flexible domain of M did not form colonies on SD/-4 (SD/-Ade/-His/-Leu/-Trp) and SD/-4/X/A medium (Fig. 2B).
Fig. 2.
The region of M that interacts with eIF3i is located within the 122- to -181 amino acids.
(A) Schematic diagram showing the M domains tested in this study. (B) Y2H screens were performed to confirm the interaction between the eIF3i protein and the flexible domain (amino acids 1–57) and the globular domain (amino acids 58 to 229) of M protein. (C) Analysis of the interaction between the mutants of M with eIF3i in a yeast two-hybrid system. AH109 yeast cells co-transfected with the indicated bait and prey plasmids were streaked onto QDO (SD/–Ade/–His/–Leu/–Trp) plates, and the clones were determined after 72 h. (D) The residues involved in the M-eIF3i interaction were shown in red. (E) Analysis of the interaction between mutants of M with eIF3i by confocal microscopy assay. The cells were co-transfected with the indicated bait and pT-DsRed-eif3i plasmids and then fixed with 4% paraformaldehyde at 24 h post-transfection. The results were obtained with a laser confocal scanning microscope. Each experiment was repeated at least three times. BD, pGBKT7 empty vector; SD/-2, SD/-Leu/-Trp; SD/-4, SD/-Ade/-His/-Leu/-Trp; SD/-4/X/A, SD/-Ade/-His/-Leu/-Trp/X-a-Gal/Aureobasidin A.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To further determine the key resides of M protein participating in this interaction, amino acids exposed at the surface of the globular domain of M protein were chosen for mutagenesis analysis (Gaudier et al., 2002, Beilstein et al., 2015). As shown in the results, all other mutants grew as well as wild type M on SD/-4 medium except for V122A/L123A, D125A/Q126A, E136A, V154A/P155A, E156A, E156A/H157A, R159A/R160A, G165A/L166A and D181A/E182A (Fig. 2C). According to the crystal structure of VSV M, these resides were rather clustered on the side of the big 4-stranded beta-sheet (Fig. 2D). However, these mutants could interact with eIF3i when detected by confocal assay (Fig. 2E). The results indicated that these mutants of M impaired but did not completely abolish the M–eIF3i interaction.
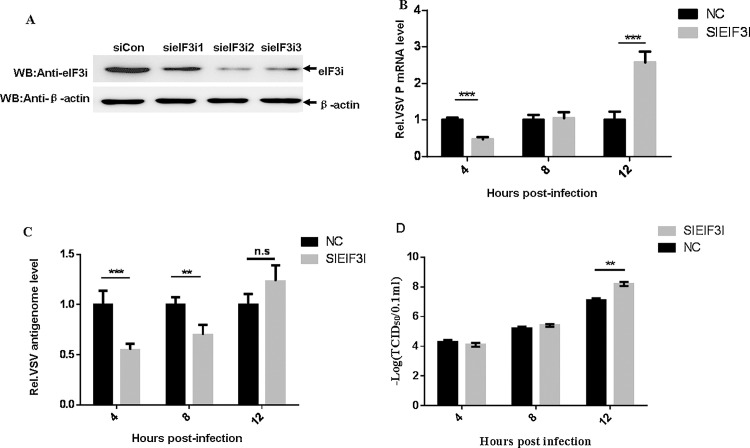
3.2. Knockdown of eIF3i by siRNAs affects VSV growth
To investigate whether eIF3i was involved in VSV propagation, HeLa cells were transfected with the indicated siRNAs resulting in the efficient knockdown of eIF3i (Fig. 3 A), and then infected with VSV. At 4, 8 and 12 h post-infection, VSV P mRNA (measure of transcription) and the antigenome (measure of replication) RNA levels were quantified by quantitative real-time PCR. The results showed that the silencing of eIF3i decreased VSV mRNA transcription and replication at 4 h post-infection but increased VSV mRNA transcription at 12 h post-infection (Fig. 3B and C). To further confirm the above results, virus titers were examined by TCID50 assay. The results showed that silencing of eIF3i increased VSV progeny production at 12 h post-infection (Fig. 3D).
Fig. 3.
Knockdown of eIF3i by siRNAs effect VSV growth.
(A) HeLa cells were transfected with 50 nM control siRNA or three different siRNAs targeting eIF3i. At 48 h post-transfection, the levels of the eIF3i protein were determined by immunoblotting with an anti-eIF3i polyclonal antibody (catalog no. 11287-1-AP; Proteintech). Actin served as the loading control. (B and C) HeLa cells were transfected with 50 nM control siRNA or sieIF3i 2. Subsequently, the cells were infected with VSV (MOI = 1). At 4, 8 and 12 h post-infection, VSV P mRNA (B) and VSV antigenomic RNA (C) levels were determined by qRT-PCR. (D) HeLa cells were transfected with 50 nM control siRNA or sieIF3i 2. Subsequently, the cells were infected with VSV (MOI = 1). At 4, 8 and 12 h post-infection, virus titers were examined by TCID50 assay. The data were averaged from three replicates of three independent experiments. The data are shown as the means ± SD (***p < 0.01; **p < 0.05; n.s., no significance).
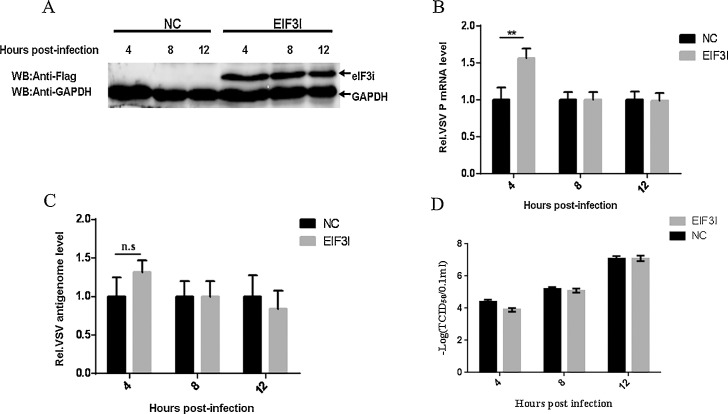
Over-expression of eIF3i has no significant effect on VSV replication in the late stages
To further verify the functional relevance of the M-eIF3i interaction to the VSV life cycle, HeLa cells were transfected with the pcmv-flag-eIF3i or pcmv-flag plasmid to up-regulate expression of eIF3i (Fig. 4 A) and then infected with VSV. Compared with the control group, the levels of VSV P mRNA were increased at 4 h post-infection but were not significantly changed at 8 and 12 h post-infection in eIF3i-overexpressed group (Fig. 4B). However, overexpression of eIF3i displayed no significant effect on replication and progeny production of VSV (Fig. 4C and D).
Fig. 4.
Over-expression of eIF3i has no significant effect on VSV replication in the late stages.
(A) HeLa cells grown in a 6-well plate were transfected with pcmv-flag-eIF3i or pcmv-flag plasmid (4 μg). At 24 h post-transfection, the levels of the eIF3i protein were determined by immunoblotting with an anti-Flag MAb (1:2000) (catalog no. 66008-2-Ig; Proteintech). GAPDH served as the loading control. (B) HeLa cells were transfected with pcmv-flag-eIF3i or pcmv-flag plasmid (4 μg). At 24 h post-transfection, the cells were infected with VSV (MOI = 1). At 4, 8 and 12 h post-transfection, VSV P mRNA (B) and VSV antigenomic (C) RNA were determined by qRT-PCR. (D) HeLa cells were transfected with pcmv-flag-eIF3i or pcmv-flag plasmid (4 μg). At 24 h post-transfection, the cells were infected with VSV (MOI = 1). At 4, 8 and 12 h post-transfection, virus titers were examined by TCID50 assay. The data were averaged from three replicates of three independent experiments. The data are shown as the means ± SD (**p < 0.05; n.s., no significance).
3.3. M protein does not inhibit host translation
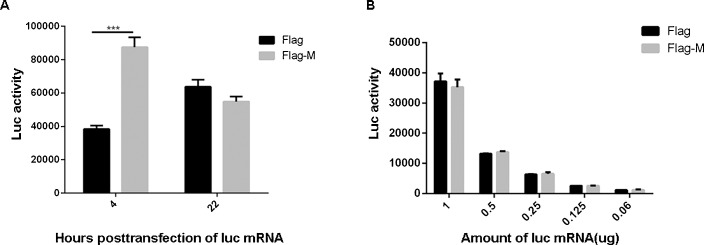
To investigate whether M could inhibit host translation by interaction with eIF3i, HeLa cells were transfected with in vitro transcribed mRNA derived from the luciferase gene. At 4 and 22 h post-transfection with luciferase mRNA, the cells were transfected with M mRNA and lysed at 28 h post-transfection with luciferase mRNA to measure the luciferase activities. Compared with the control group, M caused an increase in luciferase translation at 4 h post-transfection with luciferase mRNA but did not cause significant changes at 22 h post-transfection (Fig. 5 A). To obtain further information on these observations, HeLa cells were transfected with decreasing amounts of luciferase mRNA. At 22 h post-transfection with luciferase mRNA, the cells were transfected with M mRNA and lysed at 28 h post-transfection with luciferase mRNA to measure the luciferase activities. The results showed that M did not inhibit luciferase mRNA translation at the different concentrations (Fig. 5B).
Fig. 5.
M protein did not inhibit host translation.
(A) HeLa cells grown in a 6-well plate were transfected with 2 μg of luciferase mRNA. At 4 and 22 h post-transfection with luciferase mRNA, cells were transfected with 2 μg of M mRNA and lysed at 28 h post-transfection with luciferase mRNA to measure the luciferase activities. (B) HeLa cells grown in a 6-well plate were transfected with decreasing amounts of luciferase mRNA (1, 0.5, 0.25, 0.125 and 0.06 μg). At 22 h post-transfection with luciferase mRNA, the cells were transfected with 2 μg M mRNA and lysed at 28 h post-transfection with luciferase mRNA to measure the luciferase activities. The data were averaged from three independent experiments. The data are shown as the means ± SD (***p < 0.01; **p < 0.05).
3.4. EIF3I may affect VSV growth by modulating host antiviral response in HeLa cells
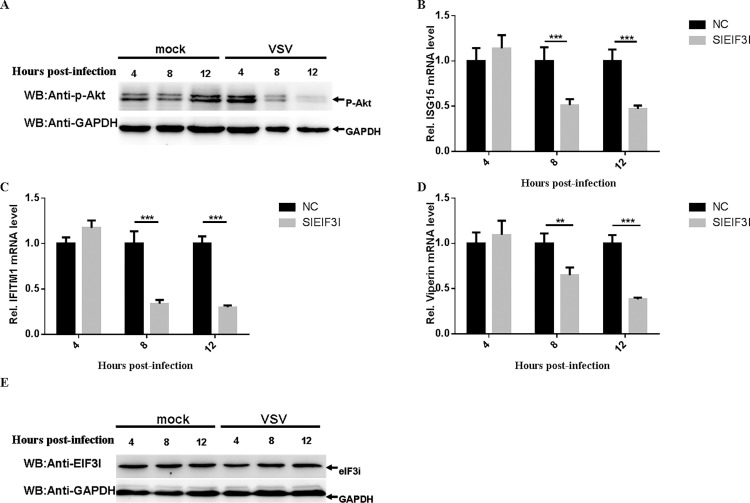
A previous study has demonstrated that eIF3i could interact with Akt1 and prevent PP2A dephosphorylation of Akt1 to activate Akt1 oncogenic signaling (Wang et al., 2013). Furthermore, Akt1 could modulate antiviral immunity through phosphorylation of the transcriptional repressor EMSY (Ezell et al., 2012). To determine whether eIF3i exploited these mechanisms to affect VSV replication in HeLa cells, the cells were infected with VSV and the level of phospho-Akt1 was detected at 4, 8 and 12 h post-infection. As shown in the results, VSV inhibited Akt phosphorylation at 8 and 12 h post-infection (Fig. 6 A). To further confirm this hypothesis, we investigated the effect of eIF3i on expression of Akt/EMSY-regulated ISGs after infection with VSV, and the results showed that the knockdown of eIF3i inhibited expression of these ISGs (Fig. 6B–D). We also demonstrated that VSV did not affect the level of the cellular eIF3i protein (Fig. 6E). These results indicated that eIF3i might affect VSV growth by modulating host antiviral response in HeLa cells.
Fig. 6.
EIF3I may affect VSV growth by modulating host antiviral response in HeLa cells.
(A) VSV inhibited Akt phosphorylation at threonine 308. HeLa cells were infected or mock infected with VSV (MOI = 10). The level of Akt phosphorylation was determined by immunoblotting with an anti-phospho-Akt (Thr308) mAb (catalog no. 4056; Cell Signaling Technology). (B–D) Knockdown of eIF3i inhibited the expression of Akt/EMSY-regulated ISGs after infected with VSV. HeLa cells were transfected with 50 nM control siRNA or sieIF3i 2. At 50 h post-transfection, cells were infected with VSV (MOI = 1). At 4, 8 and 12 h post-infection, ISG15 (B), IFITM1 (C) and Viperin (D) mRNA were determined by qRT-PCR. (E) VSV did not affect the expression of eIF3i protein. HeLa cells were infected or mock infected with VSV at an MOI of 10. The level of eIF3i protein was determined by immunoblotting with an anti-eIF3i polyclonal antibody (catalog no. 11287-1-AP; Proteintech). Each experiment was repeated at least three times. The data are shown as the means ± SD (***p < 0.01; **p < 0.05).
4. Discussion
Eukaryotic translation initiation factor 3, subunit i (eIF3i) is a subunit of the multi-subunit eIF3 translation factor, which is essential for translation in vivo. EIF3i not only regulates translation but is also involved in cellular signaling transduction. For instance, eIF3i was shown to interact with Akt1 and prevent PP2A dephosphorylation of Akt1 to activate Akt1 oncogenic signaling (Wang et al., 2013). By selectively increasing VEGFR2 and ERK translation, eIF3i promotes vascular endothelial cell proliferation and migration (Zhang et al., 2017). To date, the role of eukaryotic translation initiation factor in the replication of VSV remains unknown.
In this study, the matrix protein of VSV was shown to interact with the eukaryotic initiation factor eIF3i. By using GST pull-down and laser confocal assays, the M-eIF3i interaction was validated. We also demonstrated that the globular domain of M was indispensable for the M–eIF3i interaction. Through a mutagenesis analysis, we found that some mutations in M between amino acids 122 and 181 impaired but did not completely abolish the M–eIF3i interaction. To investigate the role of the M-eif3i interaction in VSV propagation, we silenced the eIF3i gene by using siRNA. The results showed that silencing the eIF3i gene decreased the replication of VSV in the early stages but promoted transcription of VSV in the late stages. Over-expression of eIF3i promoted transcription of VSV in the early stages but had no effect on the transcription of VSV in the late stages.
Many viruses have been reported to inhibit host translation by interacting with the eukaryotic translation initiation factor complex. Rabies virus (RV) matrix protein can interact with eIF3 h to inhibit the expression of cellular genes (Komarova et al., 2007). In addition, the spike (S) protein of SARS-CoV and infectious bronchitis virus (IBV) can interact with eIF3f to inhibit host translation. This mechanism has been exploited by coronaviruses to counteract the host-antiviral response and to modulate coronavirus pathogenesis (Xiao et al., 2008). Wataru Kamitani had reported that the nsp1 protein of SARS-Cov utilizes a two-pronged strategy to inhibit translation of host proteins: nsp1 blocked the formation of 80S by binding to the 40S ribosomal subunit and facilitated the degradation of the 5′-end of mRNA degradation, rendering the mRNA translationally inactive (Kamitani et al., 2009). However, many reports showed that VSV M did not inhibit host protein translation (Black et al., 1994, Connor and Lyles, 2005, Redondo et al., 2015). However, Zackary demonstrated that VSV could inhibit the translation of mRNAs transfected prior to infection (Whitlow et al., 2008). To further confirm whether M played a key role in inhibiting the translation of mRNAs during VSV infection, HeLa cells were transfected with in vitro transcribed luciferase and M mRNAs. Our result showed that M had no effect on inhibiting the translation of luciferase mRNAs transfected prior to the M mRNA. Thus, there might be some other viral components involved in the inhibition of host translation.
A previous study has demonstrated that VSV matrix protein could induce Akt dephosphorylation and inhibit the AKT/mTOR signaling pathway in the absence of other viral components (Dunn and Connor, 2011). Furthermore, Ezell et al. demonstrated that Akt1 could modulate antiviral immunity through phosphorylation of the transcriptional repressor EMSY (Ezell et al., 2012). To determine whether eIF3i exploited these mechanisms to affect VSV growth in HeLa cells, we have determined the effect of VSV infection on the phosphorylation of Akt1 in HeLa cells. The results showed that Akt1 phosphorylation was inhibited in cells infected with VSV. To further confirm this hypothesis, we investigated the effect of eIF3i on the expression of Akt/EMSY-regulated ISGs after infection with VSV, and the results showed that the knockdown of eIF3i inhibited the expression of these ISGs. We also demonstrated that neither silencing the eIF3i gene nor over-expression of eIF3i could promote VSV growth in type-I IFN system-deficient Vero cells (data not shown). These results indicated that eIF3i might affect VSV growth by modulating host antiviral response in HeLa cells. In this study, we also found that knockdown of eIF3i by RNA interference decreased replication and transcription and over-expression of eIF3i increased VSV transcription in the early stages. Thus, we speculated that eIF3i might exploit other mechanisms to affect VSV replication in HeLa cells.
Competing interests
The authors declare that they have no conflict of interest.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (No. 31572473).
References
- Barr J.N., Whelan S.P.J., Wertz G.W. Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus. Bba-Gene Struct. Expr. 2002;1577:337–353. doi: 10.1016/s0167-4781(02)00462-1. [DOI] [PubMed] [Google Scholar]
- Beilstein F., Obiang L., Raux H., Gaudin Y. Characterization of the interaction between the matrix protein of vesicular stomatitis virus and the immunoproteasome subunit LMP2. J. Virol. 2015;89:11019–11029. doi: 10.1128/JVI.01753-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.L., Lyles D.S. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J. Virol. 1992;66:4058–4064. doi: 10.1128/jvi.66.7.4058-4064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.L., Brewer G., Lyles D.S. Effect of vesicular stomatitis virus matrix protein on host-directed translation in vivo. J. Virol. 1994;68:555–560. doi: 10.1128/jvi.68.1.555-560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel D., Harmison G.G., Schubert M. Role of matrix protein in cytopathogenesis of vesicular stomatitis virus. J. Virol. 1990;64:1716–1725. doi: 10.1128/jvi.64.4.1716-1725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.-H., Miettinen P.J., Maruoka E.M., Choy L., Derynck R. A WD-domain protein that is associated with and phosphorylated by the type II TGF-β receptor. Nature. 1995;377:548–552. doi: 10.1038/377548a0. [DOI] [PubMed] [Google Scholar]
- Connor J.H., Lyles D.S. Inhibition of host and viral translation during vesicular stomatitis virus infection. eIF2 is responsible for the inhibition of viral but not host translation. J. Biol. Chem. 2005;280:13512–13519. doi: 10.1074/jbc.M501156200. [DOI] [PubMed] [Google Scholar]
- Das A., Dinh P.X., Panda D., Pattnaik A.K. Interferon-inducible protein IFI35 negatively regulates RIG-I antiviral signaling and supports vesicular stomatitis virus replication. J. Virol. 2014;88:3103–3113. doi: 10.1128/JVI.03202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn E.F., Connor J.H. Dominant inhibition of Akt/protein kinase B signaling by the matrix protein of a negative-strand RNA virus. J. Virol. 2011;85:422–431. doi: 10.1128/JVI.01671-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezell S.A., Polytarchou C., Hatziapostolou M., Guo A., Sanidas I., Bihani T., Comb M.J., Sourvinos G., Tsichlis P.N. The protein kinase Akt1 regulates the interferon response through phosphorylation of the transcriptional repressor EMSY. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E613–621. doi: 10.1073/pnas.1115029109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudier M., Gaudin Y., Knossow M. Crystal structure of vesicular stomatitis virus matrix protein. EMBO J. 2002;21:2886–2892. doi: 10.1093/emboj/cdf284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin Y., Barge A., Ebel C., Ruigrok R.W.H. Aggregation of VSV M protein is reversible and mediated by nucleation sites: implications for viral assembly. Virology. 1995;206:28–37. doi: 10.1016/s0042-6822(95)80016-6. [DOI] [PubMed] [Google Scholar]
- Ge P., Tsao J., Schein S., Green T.J., Luo M., Zhou Z.H. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science. 2010;327:689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty R.N., Paragas J., Sudol M., Palese P. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty R.N., Brown M.E., McGettigan J.P., Wang G., Jayakar H.R., Huibregtse J.M., Whitt M.A., Schnell M.J. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 2001;75:10623–10629. doi: 10.1128/JVI.75.22.10623-10629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her L.S., Lund E., Dahlberg J.E. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol.r Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova A.V., Real E., Borman A.M., Brocard M., England P., Tordo N., Hershey J.W., Kean K.M., Jacob Y. Rabies virus matrix protein interplay with eIF3, new insights into rabies virus pathogenesis. Nucleic Acids Res. 2007;35:1522–1532. doi: 10.1093/nar/gkl1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky S.A., Willingham M.C., Lyles D.S. Matrix protein and another viral component contribute to induction of apoptosis in cells infected with vesicular stomatitis virus. J. Virol. 2001;75:12169–12181. doi: 10.1128/JVI.75.24.12169-12181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letchworth G.J., Rodriguez L.L., Del cbarrera J. Vesicular stomatitis. Vet. J. 1999;157:239–260. doi: 10.1053/tvjl.1998.0303. [DOI] [PubMed] [Google Scholar]
- Rajani K.R., Pettit Kneller E.L., McKenzie M.O., Horita D.A., Chou J.W., Lyles D.S. Complexes of vesicular stomatitis virus matrix protein with host Rae1 and Nup98 involved in inhibition of host transcription. PLoS Pathog. 2012;8:e1002929. doi: 10.1371/journal.ppat.1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raux H., Obiang L., Richard N., Harper F., Blondel D., Gaudin Y. The matrix protein of vesicular stomatitis virus binds dynamin for efficient viral assembly. J. Virol. 2010;84:12609–12618. doi: 10.1128/JVI.01400-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo N., Madan V., Alvarez E., Carrasco L. Impact of vesicular stomatitis virus M proteins on different cellular functions. PLoS One. 2015;10:e0131137. doi: 10.1371/journal.pone.0131137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente S.T., Gilmartin G.M., Mott C., Falkard B., Goff S.P. Inhibition of HIV-1 replication by eIF3f. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4071–4078. doi: 10.1073/pnas.0900557106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kobbe C., van Deursen J.M., Rodrigues J.P., Sitterlin D., Bachi A., Wu X., Wilm M., Carmo-Fonseca M., Izaurralde E. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell. 2000;6:1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- Wang Y.W., Lin K.T., Chen S.C., Gu D.L., Chen C.F., Tu P.H., Jou Y.S. Overexpressed-eIF3I interacted and activated oncogenic Akt1 is a theranostic target in human hepatocellular carcinoma. Hepatology. 2013;58:239–250. doi: 10.1002/hep.26352. [DOI] [PubMed] [Google Scholar]
- Whitlow Z.W., Connor J.H., Lyles D.S. New mRNAs are preferentially translated during vesicular stomatitis virus infection. J. Virol. 2008;82:2286–2294. doi: 10.1128/JVI.01761-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Xu L.H., Yamada Y., Liu D.X. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS One. 2008;3:e1494. doi: 10.1371/journal.pone.0001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang P., Zhang Q., Yao X., Zhao L., Liu Y., Liu X., Tao R., Yu C., Li Y., Song X., Yao S. eIF3i activity is critical for endothelial cells in tumor induced angiogenesis through regulating VEGFR and ERK translation. Oncotarget. 2017;8:19968–19979. doi: 10.18632/oncotarget.15274. [DOI] [PMC free article] [PubMed] [Google Scholar]