Abstract
The purpose of this study was to describe clinical, hematological and fecal PCR results from 161 horses involved in outbreaks associated with ECoV. The outbreaks happened at four separate boarding facilities between November 2011 and April 2012 in the States of CA, TX, WI and MA. Following the molecular detection of ECoV in the feces from the initial index cases, the remaining herdmates were closely observed for the development of clinical signs. Fecal samples were collected from sick and healthy horses for the PCR detection of ECoV. All four outbreaks involved primarily adult horses. Fifty-nine horses developed clinical signs with 12–16 sick horses per outbreak. The main clinical signs reported were anorexia, lethargy and fever. Four horses from 3 different outbreaks were euthanized or died due to rapid progression of clinical signs. The cause of death could not be determined with necropsy evaluation in 2 horses, while septicemia secondary to gastrointestinal translocation was suspected in 2 horses. Blood work was available from 10 horses with clinical disease and common hematological abnormalities were leucopenia due to neutropenia and/or lymphopenia. Feces were available for ECoV testing by real-time PCR from 44 and 96 sick and healthy horses, respectively. 38/44 (86%) horses with abnormal clinical signs tested PCR positive for ECoV, while 89/96 (93%) healthy horses tested PCR negative for ECoV. The overall agreement between clinical status and PCR detection of ECoV was 91%. The study results suggest that ECoV is associated with self-limiting clinical and hematological abnormalities in adult horses.
Keywords: Equine coronavirus, Horse, Outbreak
1. Introduction
Coronaviruses are a large group of enveloped, positive-stranded RNA viruses that can cause respiratory and enteric diseases in a variety of avian and mammalian species (Wege et al., 1982). On the basis of serological cross-reactivity and genetic differences, these viruses are grouped into three different genera: Alphacoronavirus, Betacoronavirus and Gammacoronavirus (Woo et al., 2009). Equine coronavirus (ECoV) shares the Betacoronavirus-1 species together with human coronavirus OC43 and HKU1, murine hepatitis virus, bovine coronavirus, porcine hemagglutinating encephalomyelitis virus and rat coronavirus (Zhang et al., 2007). Equine coronavirus has been identified by electron microscopy, culture and more recently by PCR in feces of foals with and without enteric disease (Davis et al., 2000, Guy et al., 2000, Slovis et al., 2010). A recent study reported on the isolation of ECoV from the feces of 2- to 4-year-old horses with pyrogenic and enteric disease living in stables of a racetrack in Japan (Oue et al., 2011). However, little is known about ECoV, especially with regard to molecular diagnostics of field samples and the clinical significance of ECoV PCR positive fecal results. The purpose of this study was to describe clinical, hematological and fecal PCR results from 161 horses involved in four outbreaks associated with ECoV.
2. Materials and methods
2.1. Study population
The outbreaks happened at four separate boarding facilities between November 2011 and April 2012 in the States of TX, WI, CA, and MA (Table 1 ). ECoV was determined to be the etiological agent associated with the different outbreaks, based on the clinical presentation of the diseased horses and the molecular exclusion of common respiratory (equine herpesvirus-1/-4, equine influenza virus, Streptococcus equi subsp. equi, equine arteritis virus, equine rhinitis A and B virus) and enteric pathogens (Salmonella spp., Clostridium difficile toxin A and B, Neorickettsia risticii) associated with outbreaks. The population of horses per stable ranged from 28 to 65 horses. Following the molecular detection of ECoV in the feces from the initial index cases, the remaining herdmates were closely observed for the development of clinical signs. Fecal samples were collected from sick and healthy horses for the PCR detection of ECoV. Clinical pathology from sick horses was evaluated when available.
Table 1.
Demographic, clinical and molecular results from 161 horses involved in four separate ECoV outbreaks.
| Outbreak | TX | WI | CA | MA |
|---|---|---|---|---|
| Date | November 2011 | February 2012 | April 2012 | April 2012 |
| Affected horses/total horses (%) | 16/28 (57) | 16/33 (48) | 13/65 (20) | 14/35 (40) |
| Clinical signs | ||||
| Anorexia | 16 | 14 | 10 | 12 |
| Lethargy | 10 | 13 | 12 | 11 |
| Fever (T > 101.5 °F) | 10 | 14 | 11 | 8 |
| Colic | 0 | 0 | 2 | 2 |
| Soft formed feces/diarrhea | 0 | 2 | 5 | 5 |
| No clinical signs | 12 | 17 | 52 | 21 |
| Fatality | 1 | 2 | 0 | 1 |
| PCR results for ECoV | ||||
| Positive sick horses | 8 | 11 | 14 | 5 |
| Negative sick horses | 0 | 3 | 1 | 2 |
| Positive healthy horses | 0 | 6 | 0 | 1 |
| Negative healthy horses | 0 | 17 | 17 | 55 |
2.2. PCR analysis
Fecal samples were processed on the day of submission to the laboratory. Two milliliters of PBS were added to 200 μg of feces in a conical tube. Thereafter, each sample was vortexed for 10 s and centrifuged at 13,000 × g for 10 s. Nucleic acid purification from 200 μl of supernatant was performed using an automated nucleic acid extraction system (CAS-1820 X-tractor Gene, Corbett Life Science, Sydney, Australia) according to the manufacturer's recommendations. Total RNA was converted to complementary DNA (cDNA) as previously reported (Pusterla et al., 2011). All samples were assayed for the presence of ECoV by real-time PCR. Briefly, the PCR assay used was based on the detection of a specific 142 base-pair product of the N gene of ECoV (GenBank accession number EF446615; oligonucleotides: forward primer ECoV-380f TGGGAACAGGCCCGC, reverse primer ECoV-522r CCTAGTCGGAATAGCCTCATCAC, probe ECoV-436p 6FAM-TGGGTCGCTAACAAG-TAMRA). The samples were amplified in a combined thermocycler/fluorometer (7900 HT Fast, Applied Biosystems, Foster City, CA, USA) with the standard thermal cycling protocol: 2 min at 50 °C, 10 min at 95 °C, and 40 cycles of 15 s at 95 °C and 60 s at 60 °C. Furthermore, a real-time PCR assay targeting a universal sequence of the bacterial 16S rRNA gene was used as quality control (i.e. efficiency of DNA purification and amplification) and as an indicator of fecal inhibition (Mapes et al., 2007). A standard curve was run for the assay using ECoV plasmids and the amplification efficiency calculated from the slope using the formula E = 101/−s − 1. The amplification efficiency was 98.3% for the N gene of ECoV indicating a very high analytical sensitivity. To test for analytical specificity, 4 TaqMan PCR products from each of the 4 outbreaks were sequenced using standard sequencing procedures (BigDye Terminator chemistry, ABI 3730, Applied Biosystems, Foster City, CA, USA) and 100% homology was determined with the N gene of previously deposited sequences of ECoV. Plate to plate variability of the assay was 0.15 CT standard variations while the reproducibility in triplicate was 0.08 CT standard variation. Detection limit of the assay was determined using 10-fold dilutions of the ECoV plasmid added to PBS or to ECoV free equine feces. The detection limit for the assay was 5 and 20 ECoV genome equivalents when the cDNA was purified from samples diluted from PBS and feces, respectively. Real-time PCR results were reported quantitatively following absolute quantitation using a standard curve and expressed as ECoV genome equivalents per gram of feces.
A 435-nucleotide segment of the N gene of ECoV was generated for each of the outbreak strains in order to determine phylogenetic relationship according to Zhang et al. (2007). Sequence of both the 5′ and 3′ ends of a partial segment of the N gene of ECoV were determined using standard sequencing procedures (BigDye Terminator chemistry, ABI 3730, Applied Biosystems, Foster City, CA, USA).
3. Results
All four outbreaks involved primarily adult horses ranging in age from 1 to 29 years (median 15 years). Fifty-nine horses developed clinical signs with 12–16 sick horses per outbreak (Table 1). The main clinical signs reported were anorexia (52 horses), lethargy (46) and fever (43). The rectal temperature of febrile horses ranged from 101.5 to 105.8 °F (median 104.1 °F). Changes in fecal character, ranging from soft-formed to watery consistency, and colic were observed in 12 and 4 horses, respectively. Clinical signs generally resolved within 1–4 days with supportive care. Four horses from 3 different outbreaks were euthanized or died due to rapid progression of clinical signs. Two horses showed acute onset of neurological disease including depression, ataxia and recumbency. Two additional horses were euthanized due to severe endotoxemia. The cause of death could not be determined with necropsy evaluation in the two horses with neurological signs, while bacteremia secondary to gastrointestinal translocation was suspected in the additional 2 horses. The outbreak period lasted approximately 3 weeks for each of the stables.
Blood work was available from 10 horses with clinical disease, and common hematological abnormalities were leucopenia (<4600/μl; 7 horses) due to neutropenia (<2260/μl; 6 horses) and/or lymphopenia (<1500/μl; 9 horses).
Feces were available for ECoV testing by real-time PCR from 44 to 96 sick and healthy horses, respectively. 38/44 (86%) horses with abnormal clinical signs tested PCR positive for ECoV with absolute genome equivalents ranging from 7887 to 7.2 × 107 equivalents/g of feces (median 538,410 genome equivalents/g of feces). 89/96 (93%) healthy horses tested PCR negative for ECoV. The seven healthy horses had viral loads ranging from 55,683 to 3.9 × 106 equivalents/g of feces (median 228,520 genome equivalents/g of feces). There was no statistical difference in absolute ECoV quantitation between PCR positive sick and healthy horses (Fig. 1 ; Mann–Whitney test, P = 0.71). The overall agreement between clinical status and PCR detection of ECoV was 91%. Follow-up samples were available from 7 sick horses. PCR detection of ECoV persisted for 3–9 days (median 4 days) in these 7 horses. Sequence comparison of a 435-nucleotide segment of the N gene of ECoV showed between 99.5 and 100% homology amongst the 4 study strains and between 97.9 and 99.0% homology with the NC99 and the Tokachi09 strain (Fig. 2 ; Zhang et al., 2007).
Fig. 1.
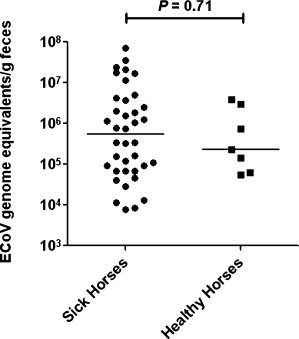
Results of ECoV detection in feces from symptomatic and asymptomatic horses involved in ECoV outbreaks. The results are expressed as number of ECoV genome equivalents/g of feces, horizontal bars represent median.
Fig. 2.
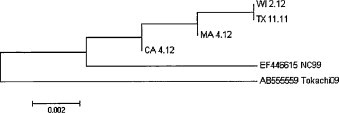
Phylogenetic relationship of 4 field strains of ECoV and the NC99 and the Tokachi09 strain based on a comparison of a partial sequence of the N gene. The field isolates are labeled with the state of origin and the month and year of the outbreak. Analyses were done using the MEGA 5.10 version Beta #4 software package (www.megasoftware.net).
4. Discussion
Despite the reports of probable coronavirus infections in foals, there has been no definitive confirmation of ECoV involved with diarrhea in foals before 2000. Guy et al. (2000) and Davis et al. (2000) each reported on one diarrheic foal with isolation and characterization and histological detection of ECoV, respectively. Further, ECoV has recently been associated with fever and diarrhea in adult horses (Oue et al., 2011). During this outbreak 132 of approximately 600 horses developed disease with most horses recovering in 2–4 days.
Coronavirus infection typically begins in the proximal small intestine and subsequently spreads to the colonic crypt cells, leading to blunting of the villi and subsequent villous atrophy. It is the loss of epithelial cells that results in malabsorption and maldigestion of nutrients and acute diarrhea in foals (Gonzales Arguedas, 2007). Although diarrhea is a consistent hallmark of ECoV infection in foals, diarrhea was only seen in 12/59 (20%) of affected study horses. The main clinical signs observed in the 4 separate outbreaks were anorexia (88%), lethargy (78%) and fever (73%). Overall, the clinical presentations in the study horses were consistent with a previous report (Oue et al., 2011). The morbidity for the 4 outbreaks ranged from 20 to 57% with 4 deaths occurring at the same time. ECoV infections are generally self-limiting, however, secondary complications, including inadequate perfusion due to dehydration and gastrointestinal translocation, have been previously reported (Davis et al., 2000). In two study horses with signs of diarrhea and endotoxemia, bacteremia due to gastrointestinal translocation was suspected based on the culture of enteric microorganisms from the liver. Both horses with acute onset of neurological disease had pathological changes consistent with moderate encephalopathy. It is possible that metabolic derangements, such as hyperammonemia or uremia, could have contributed to the clinical presentation and pathological findings in these horses. There was no evidence of a non-suppurative encephalomyelitis and edema in these 2 horses, which would have been suggestive of a viral infection, as reported for other coronaviruses, such as porcine hemagglutinating encephalomyelitis virus and murine hepatitis virus (Lane and Hosking, 2010, Gao et al., 2011).
Hematological changes observed in 10 horses were characterized by leucopenia due to neutropenia and/or lymphopenia. These changes are non-specific and reflect an acute inflammatory disease process (Morris, 2009) and do not allow differentiation between bacterial and viral diseases.
Historically, the detection of ECoV has relied on either electron microscopy, antigen-capture ELISA or viral isolation from the feces. All these detection modalities lack sensitivity, especially if viral particles are not present in sufficient numbers. Real-time PCR for the detection of coronaviruses has supplanted many conventional virological assays, mainly due to its short turn-around-time, high throughput capability and increased analytical sensitivity and specificity (Bustin and Mueller, 2005). The molecular detection of ECoV from the feces of horses does not necessarily mean that the virus is involved in the disease process. This is especially true for foals, since a similar percentage of healthy foals and foals with diarrhea tested PCR positive for ECoV in a recent report (Slovis et al., 2010). The present study results showed a strong association between clinical status and PCR detection of ECoV. ECoV was the likely putative virus linked to these four outbreaks based on: (i) detection of ECoV in feces from the majority of sick horses (86%), (ii) lack of detection of ECoV in feces from the majority of horses without clinical signs (93%), (iii) temporal association between PCR detection of ECoV and clinical status, and (iv) short detection period of ECoV for clinically affected horses. PCR disagreement with clinical status in a few study horses may have been related to subclinical shedding in PCR positive healthy horses and disease stage and amount of viral shedding in PCR negative sick horses. Sequence analysis for the different ECoV strains showed high level of homology for the partially sequenced N gene amongst the different study strains and a previously isolated strain (NC99) from a diarrheic foal from NC (Zhang et al., 2007).
In conclusion, the study results suggest that ECoV is associated with self-limiting clinical and hematological abnormalities in adult horses. Real-time PCR is a sensitive and fast diagnostic tool to document the presence of ECoV in feces from horses with unspecific clinical signs such as lethargy, anorexia, fever and changes in fecal character.
Acknowledgement
This study was supported by Boehringer Ingelheim Vetmedica.
References
- Bustin S.A., Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin. Sci. 2005;109:365–379. doi: 10.1042/CS20050086. [DOI] [PubMed] [Google Scholar]
- Davis E., Rush B.R., Cox J., DeBey B., Kapil S. Neonatal enterocolitis associated with coronavirus infection in a foal: a case report. J. Vet. Diagn. Invest. 2000;12:153–156. doi: 10.1177/104063870001200210. [DOI] [PubMed] [Google Scholar]
- Gao W., Zhao K., Zhao C., Du C., Ren W., Song D., Lu H., Chen K., Li Z., Lan Y., Xie S., He W., Gao F. Vomiting and wasting disease associated with hemagglutinating encephalomyelitis viruses infection in piglets in Jilin, China. Virol. J. 2011;21:130–136. doi: 10.1186/1743-422X-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales Arguedas M. Coronavirus infections. In: Sellon D., Long M., editors. Equine Infectious Diseases. Saunders Elsevier; St. Louis: 2007. pp. 184–185. [Google Scholar]
- Guy J.S., Breslin J.J., Breuhaus B., Vivrette S., Smith L.G. Characterization of a coronavirus isolated from a diarrheic foal. J. Clin. Microbiol. 2000;38:4523–4526. doi: 10.1128/jcm.38.12.4523-4526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T.E., Hosking M.P. The pathogenesis of murine coronavirus infection of the central nervous system. Crit. Rev. Immunol. 2010;30:119–130. doi: 10.1615/critrevimmunol.v30.i2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapes S., Rhodes D.M., Wilson W.D., Leutenegger C.M., Pusterla N. Comparison of five real-time PCR assays for detecting virulence genes in isolates of Escherichia coli from septicaemic neonatal foals. Vet. Rec. 2007;161:716–718. doi: 10.1136/vr.161.21.716. [DOI] [PubMed] [Google Scholar]
- Morris D. Alterations I the leukogram. In: Smith B.P., editor. Large Animal Internal Medicine. Mosby Elsevier; St. Louis: 2009. pp. 405–410. [Google Scholar]
- Oue Y., Ishihara R., Edamatsu H., Morita Y., Yoshida M., Yoshima M., Hatama S., Murakami K., Kanno T. Isolation of an equine coronavirus from adult horses with pyrogenic and enteric disease and its antigenic and genomic characterization in comparison with the NC99 strain. Vet. Microbiol. 2011;150:41–48. doi: 10.1016/j.vetmic.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusterla N., Kass P.H., Mapes S., Johnson C., Barnett D.C., Vaala W., Gutierrez C., McDaniel R., Whitehead B., Manning J. Surveillance programme for important equine infectious respiratory pathogens in the USA. Vet. Rec. 2011;169:12. doi: 10.1136/vr.d2157. [DOI] [PubMed] [Google Scholar]
- Slovis N.M., Elam J., Estrada M., Ferrero Thao M., Leutenegger C.M. Comprehensive analysis of infectious agents associated with diarrhea in foals in Central Kentucky. Proceedings of the 56th Annual Convention of the American Association of Equine Practitioners; Baltimore, MD, USA, December 4–8.; 2010. p. 262. [Google Scholar]
- Wege H., Siddell S., Ter Meulen V. The biology and pathogenesis of coronavirus. Curr. Top. Microbiol. Immunol. 1982;99:165–200. doi: 10.1007/978-3-642-68528-6_5. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Zhang J., Guy J.S., Snijder E.J., Denniston D.A., Timoney P.J., Balasuriya U.B. Genomic characterization of equine coronavirus. Virology. 2007;369:92–104. doi: 10.1016/j.virol.2007.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]


