Abstract
Infectious bronchitis virus (IBV) causes avian infectious bronchitis, an important disease that produces severe economic losses in the poultry industry worldwide. Recent IBV infections in Sweden have been associated with poor growth in broilers, drop in egg production and thin egg shells in layers. The complete spike gene of selected isolates from IBV cases was amplified and sequenced using conventional RT-PCR. Nucleotide and amino acid sequence comparisons have shown that the recent isolates bear 98.97% genetic similarity with strains of the QX-like genotype. The phylogenetic analysis revealed that strains predominant in the nineties, which were of the Massachusetts type, have been replaced by D388/QX-like strains, however the evolutionary link could not be established. The homology between the two genotypes was 79 and 81%. Remarkably, a strong positive selection pressure was determined, mostly involving the S1 subunit of the S gene. This strong selective pressure resulted in recombination events, insertions and deletions in the S gene. Two new isolates generated from recombination were found with nucleotide sequence diverging 1.7–2.4% from the D388/QX-like branch, indicating the emergence of a new lineage. The study demonstrates a constant evolution of IBV that might be in relation to increased poultry farming, trade and vaccine pressure. The findings underscore the importance of continuous monitoring to control spread of infections, as well as to timely adjust diagnostic methods, molecular epidemiological studies, development and use of vaccines that are adapted to the changing disease scenario.
Keywords: IBV, Avian infectious bronchitis, S gene, Phylogeny, Evolution
1. Introduction
Infectious bronchitis virus (IBV) causes avian infectious bronchitis (IB), a highly contagious disease that affects poultry worldwide and produces severe economic losses. The disease is clinically manifested by respiratory distress, drop in egg production, poor egg quality in layers and some strains causes nephritis (Cavanagh, 2003, Cavanagh, 2005, Mahmood et al., 2011). The disease may lead to loss of weight and predisposes to secondary bacterial infections that may be fatal (Cavanagh, 2003). Initially IBV infects the respiratory tract, however, infections of kidneys and oviduct may follow (Cavanagh and Naqi, 2003). Also a shift of tissue tropism in the virus has been reported (Liu et al., 2006, Zhou et al., 2004), resulting in infection of a wide range of avian host species especially those reared close to domestic fowl. For example, IBV cases were found in Chinese peafowl (Pavo), guinea fowl (Numida meleagris), partridge (Alectoris) and teal (Anas) (Cavanagh, 2005).
IBV belongs to the order of Nidovirales, family Coronaviridae and to genera of Gamma-coronavirus group 3 (Gonzalez et al., 2003). The genome is positive-sense single stranded RNA of about 27.6 kb and contains 5′ and 3′ untranslated regions (UTRs). The proximal two-thirds of the genome encode two overlapping open reading frames (ORFs) 1a and 1b that are translated into large polyproteins. These are then cleaved to generate 15 non-structural proteins (Nsp2-16), comprising a main protease, an RNA-dependent RNA-polymerase and other non-structural proteins that carry different functions in transcription and replication. The remaining third of the genome encodes four main structural proteins: the envelope protein (E), the spike glycoprotein (S), the nucleocapsid protein (N) and the membrane protein (M). IBV has two accessory genes 3 and 5 that express accessory proteins 3a, 3b and 5a, 5b, respectively. (Cavanagh, 2007, Pasternak et al., 2006).
Generally, the spike glycoprotein of the virus is translated as a pre-cursor protein (SO) and later cleaved into the S1 and S2 subunits. However, this cleavage of the spike glycoprotein is not observed in all coronaviruses (Holmes, 1990). The spike glycoprotein of IBV consists of 1145 amino acids and its functions include attachment to the host cell, neutralization and induction of protective immunity (Ignjatovic and Galli, 1994, Johnson et al., 2003). It is the most variable of the IBV proteins, with most of the variability mapping to the S1 part (Adzhar et al., 1997, Farsang et al., 2002, Keeler et al., 1998, Kingham et al., 2000) due to mutations or recombination in this segment.
The sequence variability in the S1 coding gene determines serotype specificity of IBV. More than 25 serotypes have been recognized to date, differing by 20–25% and sometimes 50% at amino acid level (Adzhar et al., 1997, Gelb et al., 2005). As a result, cross protection between serotypes is poor (Kuo et al., 2010) and changes as small as 5% in the amino-terminal half (S1) of the S protein have been shown to alter the protection ability of a vaccine (Cavanagh, 2003). The several different IBV variants are present around the globe some of them exist in particular region, while others are generally distributed (de Wit et al., 2011). The increasing number of new serotypes and genotypes of IBV is a major challenge for the prevention and control of the disease.
IB had never been reported in Sweden until the first outbreak of IB in commercial poultry farms in 1970s. From 1994 a series of outbreaks have been reported in layer flocks and later in parent flocks for broilers in the South of the country. Due to this situation a decision was made in 1997 to vaccinate with a live attenuated vaccine of Massachusetts (Ma) type IB Ma5. Vaccination reduced the frequency and seriousness of the outbreaks. Subsequent to introduction of vaccination, our study has found indications of outbreaks initiated by the vaccine virus (Farsang et al., 2002). Investigations as to the origins of IB cases during 2006 have shown presence of antibodies reacting with the 4/91 variant. In 2007 and 2008, the D388 variant of IBV was identified in chickens.
Despite the immunization of a number of Swedish poultry flocks with live attenuated vaccines, cases of infections were detected again from 2006 to 2010 in broilers. Reports of similar situations abound also in other countries (Beato et al., 2005, Handberg et al., 2009, Worthington et al., 2008, Worthington and Jones, 2006) indicating a sub-optimal protection provided by the vaccines. The appearance of two new economically important field strains of IBV QX-like and Italy-02 in commercial poultry flocks in Western Europe (Beato et al., 2005, Worthington et al., 2008) has further raised the need for a better understanding of the epidemiology of IB all over the world. Clinically, the D388/QX-like and ITA-02 genotypes are characterized by nephritis, production of severe disease symptoms and mortality (Ammayappan and Vakharia, 2009). Such symptoms have not been observed in Sweden, where signs of infection are limited to poor growth in broilers.
The objective of the present study was to perform molecular characterization of IBV isolates in Sweden in order to better understand the epidemiology and the factors behind the occurrence of new infections.
2. Materials and methods
2.1. Virus isolation
Samples from IB cases (trachea, bronchi, ceaca), collected in Sweden from the different outbreaks between 1995 and 2010, were obtained from The National Veterinary Institute (SVA) sample bank. Available information of the isolates is summarized in Table 1 . All the IBV strains have been propagated in specific pathogen-free (SPF) embryonated chicken eggs (Lohmann Tierzucht, Cuxhaven, Germany) to maintain the stock. Virus isolation was performed by inoculation of 9–11 day old embryonated hen's eggs with 200 μl of 10% tissue homogenates. The eggs were incubated at 37 °C for 72 h. At the end of the incubation the eggs were chilled at 4 °C for 18 h and the allantoic fluid was harvested and centrifuged at 2500 rpm for 15 min.
Table 1.
List of selected Swedish IBV strains isolated during 1995–2010 used in this study.
| Strain | Year | Country | NCBI accession number |
|---|---|---|---|
| 242 | 1995 | Sweden | JN022536 |
| 261 | 1995 | Sweden | JN022537 |
| 381 | 1995 | Sweden | JN022538 |
| 397 | 1995 | Sweden | JN022539 |
| 423 | 1997 | Sweden | JN022540 |
| 748 | 1995 | Sweden | JN022541 |
| 1096 | 1997 | Sweden | JN022542 |
| 1489 | 1999 | Sweden | JN022543 |
| 6904 | 1999 | Sweden | JN022544 |
| 09620 | 2010 | Sweden | JN022545 |
| 09621 | 2010 | Sweden | JN022546 |
| 09622 | 2010 | Sweden | JN022547 |
| 062545 | 2009 | Sweden | JN022548 |
| 062561 | 2009 | Sweden | JN022549 |
| 065846 | 2010 | Sweden | JN022550 |
| 079663 | 2010 | Sweden | JN022551 |
| 079692 | 2010 | Sweden | JN022552 |
| 082066 | 2010 | Sweden | JN022553 |
| 900419 | 1997 | Sweden | JN022554 |
| A889 | 1999 | Sweden | JN022555 |
2.2. RNA extraction and synthesis of cDNA
The RNA was extracted using TRIzol reagent as recommended by the manufacturer (Invitrogen, Carlsbad, USA). Synthesis of cDNA was performed with a primer complementary to the IBV spike gene and using SuperScript II reverse transcriptase (Invitrogen™ Life technologies, Carlsbad, USA) according to the manufacturer's instructions, in a total reaction volume of 20 μl.
2.3. PCR amplification of the spike gene (S1 and S2 units)
Two sets of primers were used to amplify the spike gene by RT PCR using the Phusion high-fidelity PCR enzyme (Finnzymes, Espoo, Finland). The PCR mix contained 31 μl of distilled water, 10 μl of 5× Phusion GC buffer, 1 μl of dNTP mix, 1 μl of each primer, 1 μl of Phusion™ High-Fidelity DNA polymerase and 5 μl of cDNA. The thermo-profile was initial denaturation at 98 °C/30 s; and 35 cycles of 98 °C/10 s, 55 °C/10 s, and 72 °C/1 min. A final extension at 72 °C/7 min was included. PCR products of were run on 0.8% agarose gel containing ethidium bromide. The specific bands were excised from gels and purified using Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA).
2.4. Sequencing reactions
Sequencing reactions were carried out using Big Dye terminator sequencing kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. A set of internal primers was used to generate overlapping sequences in both directions. Due to sequence diversity, primers have been designed as new sequence data were generated. The final sequence data enables the selection of consensus primers for sequencing of similar strains. The sequences of the primers are available on request. The thermal profile was setup at 96 °C/4 min; and 25 cycles at 96 °C/15 s, 50 °C/10 s and 60 °C/4 min. The reactions were precipitated in sodium acetate and 95% ethanol. Thereafter, they were centrifuged. The pellets were washed with 75% ethanol and dried at room temperature. Sequencing was carried out on a 3100 DNA analyzer (Applied Biosystems, Foster City, CA).
The sequences of S1 and S2 obtained were edited and assembled to the 3540 bp of the S gene. For sequence analysis, the sequences data set was pair wise aligned and edited using software Lasergene DNASTAR (Madison, USA).
2.5. Selection pressure in the spike glycoprotein
Selective pressure of the spike glycoprotein was determined by using SNAP (Korber, 2000) services available at http://hcv.lanl.gov/content/sequence/SNAP/SNAP.html. The difference between Synonymous (dS) and non-synonymous (dNS) substitutions in the codons was calculated to evaluate the substitution rate in the spike gene of the virus.
2.6. Recombination analysis
The sequences were analysed for recombination events, as confirmed by analysis with a recombination detection program, RDP V.3.44. The window size was adjusted to 30 with the highest P value 0.05. The detection of recombination events were applied between sequences sharing 0 and 100% identity.
2.7. Phylogenetic analysis
Phylogenetic analysis was performed with 20 sequences generated in the present study and 95 sequences of the partial S1 and complete spike gene downloaded from the GenBank (accession numbers are presented in Table 1, Table 2 ). All the gaps in the sequences created by insertions were deleted by using software Mega4 version 4 (Tamura et al., 2007). Phylogenic trees were constructed by the Neighbour-joining method with the nucleotide substitution model of Kimura-2 parameter model. The tree reliability was evaluated by bootstrap 1000 replicates. The data set was kept similar for the analysis. The tree topology was prepared in the FigTree_v1.3.1.
Table 2.
List of reference strains of IBV of which sequences were downloaded from GenBank for the phylogenetic analysis in this study.
| Strain | Year | Country | NCBI accession number |
|---|---|---|---|
| LSD | 2008 | China | GQ258333 |
| 2886 | China | AY846750 | |
| IBVX8 | 2009 | China | FJ793940 |
| IBVX9 | 2009 | China | GQ853589 |
| IBVX16 | 2009 | China | FJ793941 |
| LSD | 2007 | China | FJ345390 |
| LSD03 | 2007 | China | FJ345383 |
| LHLJ | 2007 | China | FJ345366 |
| LSDV | 2007 | China | FJ345388 |
| LHLJ | 2009 | China | HM194662 |
| QX99 | 1999 | China | AF193423 |
| JS | 2006 | China | EU031525 |
| PSH050513 | 2005 | China | DQ160004 |
| SAIBK | 2005 | China | DQ288927 |
| YZ07 | 2008 | China | FJ807653 |
| LDT3 | 2005 | China | AY702975 |
| A2 | 2008 | China | EU526388 |
| DY07 | 2007 | China | HM245923 |
| LX4 | 2002 | China | AY189157 |
| LX4 | 2002 | China | AY338732 |
| LH2 | 2002 | China | AY180958 |
| LDL97I | 1997 | China | DQ068701 |
| W | 1996 | China | GQ149080 |
| ZJ971 | 2005 | China | AF352311 |
| H52 | 2005 | China | AF352315 |
| LKQ3 | 2005 | China | AY702085 |
| IBN | 2006 | China | AY856348 |
| HK | 2006 | China | AY761141 |
| 1450L | 2005 | France | EF079117 |
| 1450T | 2005 | France | EF079118 |
| 1061-PH | 2004 | Iraq | AY544778 |
| 1062 | 2004 | Iraq | AY544777 |
| 1201 | 2004 | Israel | DQ400359 |
| 236 | 2002 | Israel | AY135205 |
| 90254 | 2005 | Italy | FN430414 |
| 497 | 2002 | Italy | DQ901377 |
| 283 | 2004 | Korea | FJ807923 |
| 463 | 2004 | Korea | FJ807924 |
| 154 | 2005 | Korea | FJ807922 |
| 33 | 2005 | Korea | AY790367 |
| 1019 | 2003 | Korea | FJ807927 |
| 630 | 2002 | Korea | FJ807925 |
| 1583 | 2004 | Korea | FJ807931 |
| 1148 | 2004 | The Netherlands | DQ431199 |
| 1449K | 2004 | The Netherlands | EF079115 |
| 1449T | 2004 | The Netherlands | EF079116 |
| 18 | 2008 | Spain | GQ253483 |
| 17 | 2008 | Spain | GQ253482 |
| 79 | 2008 | Spain | GQ253484 |
| 116 | 2009 | Spain | GQ253485 |
| 170 | 2009 | Spain | GQ253486 |
| 308 | 1998 | Spain | DQ064807 |
| 339 | 2000 | Spain | DQ064815 |
| Beaudette | 2005 | Singapore | DQ001337 |
| P36 | 2005 | Singapore | DQ001342 |
| Beaudette | 2005 | Singapore | DQ001334 |
| Beaudette | 2005 | Singapore | DQ001335 |
| Beaudette | 2005 | Singapore | DQ001336 |
| P20C222 | 2005 | Singapore | DQ001340 |
| P36C12 | 2005 | Singapore | DQ001341 |
| 3468 | 2007 | Taiwan | EU822336 |
| 3382 | 2006 | Taiwan | GQ229232 |
| 1171 | 1992 | Taiwan | DQ646406 |
| 2296 | 1995 | Taiwan | DQ646404 |
| 3263 | 2004 | Taiwan | EU822338 |
| 2992 | 2002 | Taiwan | EU822340 |
| 3374 | 2005 | Taiwan | EU822337 |
| H120 | 2005 | Taiwan | EU822341 |
| 2994 | 2002 | Taiwan | GU386375 |
| 340552 | 2009 | Thailand | GQ885140 |
| 320352 | 2009 | Thailand | GQ885138 |
| 241251 | 2008 | Thailand | GQ885131 |
| KKU1 | 2008 | Thialand | GQ906706 |
| 2150 | 2007 | UK | EU914939 |
| 633 | 2004 | UK | DQ901376 |
| Ma5 | USA | AY561713 | |
| MassAreiso | USA | EU283075 | |
| MassAvial1 | USA | EU283073 | |
| MassAvial2 | USA | EU283074 | |
| MassDC6dpvc | USA | EU283088 | |
| Mass41 | 2006 | USA | DQ664534 |
| Beaudette42 | 2007 | USA | DQ830981 |
| Beaudette42 | 1995 | USA | X02342 |
| Massachusetts 41 | 2006 | USA | DQ830980 |
| Ark99 | 1995 | USA | L10384 |
| AL5364 | 2000 | USA | EU359656 |
| AL7149 | 2000 | USA | DQ458218 |
| AL0803 | 2001 | USA | EU359653 |
| ArkDPI | 2007 | USA | EU359652 |
| ArkDPI | USA | EU359649 | |
| ArkDPI | USA | EU359651 | |
| MassDCvial | USA | EU283085 | |
| 4/91 | AF093794 | ||
| Massachusetts 41 | 1995 | M21883 | |
| Massachusetts 41 | 1993 | X04722 |
3. Results
3.1. Virus isolation
Between 1995 and 2010, the SVA received specimens from different outbreaks of avian infectious bronchitis that had been submitted by commercial poultry farms for diagnosis. Initially outbreaks were confirmed as IBV positive by real-time PCR. Twenty Swedish IBV isolates from different outbreaks were selected for comprehensive study of the complete spike gene of the virus. The isolates were assigned into two categories: one comprising isolates from nineties 1995 to 1999 and the other containing isolates from 2009 to 2010.
3.2. Analysis of the nucleotide sequences
The complete spike gene sequences were subject to BLAST searches in GenBank and the results confirmed the isolates as IBV. Comparative analysis of the nucleotide sequences of the complete spike gene was carried out to determine the relatedness among 20 Swedish isolates (Table 3 ). The isolates from 1995 to 1999 showed 92.10–99.90% nucleotide sequence homology among themselves. The higher nucleotide homology 99.90% was shared by isolates 423 and 1096. The isolates from 2009 to 2010 showed 97.20–100% nucleotide sequence homology among themselves. Isolates 079663 and 079692 have shown 1.7–2.4% nucleotide sequence divergence than other isolates from 2009 to 2010. The isolates 1995–1999 exhibited 79.0–81.30% homology with the isolates 2009–2010.
Table 3.
The nucleotide sequence pairwise comparison of percentage identity and divergence based on complete spike gene between Swedish IBV isolates.
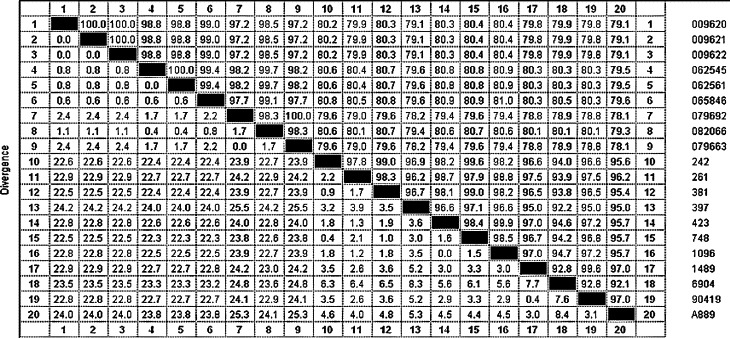
The sequences were also analysed for the presence of nucleotide insertions or deletions in the spike gene. Nucleotide insertions were found at position 69–71 (TCT) and 361–366 (GGGTCT) in the spike gene of all the recent isolates (2009–2010) analysed. Also nucleotide deletions were found at positions 2483, 2536–2537, 2568–2569 and 2614–2615 in the Swedish IBV isolates from 2009 to 2010. There were no insertions or deletions of nucleotides observed in the sequences of the Swedish IBV isolates from 1995 to 1999.
3.3. Analysis of the deduced amino acid sequences
The deduced amino acid sequences of the spike glycoprotein of the Swedish IBV isolates from 2009 to 2010 exhibited 95.30–100% homology. The isolates from 2009 to 2010 contained three amino acid insertions, aspartic acid, glycine and serine at positions 24, 121 and 122, respectively. The new isolates 079663 and 079692 had a deletion of arginine at position 411, whereas isolates 09620, 09621 and 09622 have lost threonine, asparagine, glutamine and arginine at positions 838, 845, 859 and 871, respectively, consequence to the nucleotide deletions. The Swedish IBV isolates from 1995 to 1999 had shared homology of 89.20–99.10%. Hypervariable regions were found encompassing positions 53–80, 270–296 and 521–547 within the S1 in all the analysed Swedish isolates. Positions 538 and 566 were highly subjected to amino acid substitutions. Overall, high rates of amino acids substitution were observed in the sequences of spike glycoprotein of the virus.
3.4. Selective evolutionary pressure in the spike glycoprotein
A bioinformatics and pair wise comparison approach was applied to estimate the synonymous and non-synonymous substitution rates and selective evolutionary pressure in the spike glycoprotein. The analyses revealed that most of the S1 subunit was under strong positive selective pressure, showing positive codon specific differences and a high level of genetic diversity across the S1 gene (Fig. 1 ). In particular, codon regions 360–384 and 598–619 contained a high number of non-synonymous substitutions. Overall, the S2 gene showed negative selective pressure, however, the codon regions encompassing positions 693–711 and 790–801 were found to contain non-synonymous substitutions, thus contributing to overall variation of the S gene.
Fig. 1.
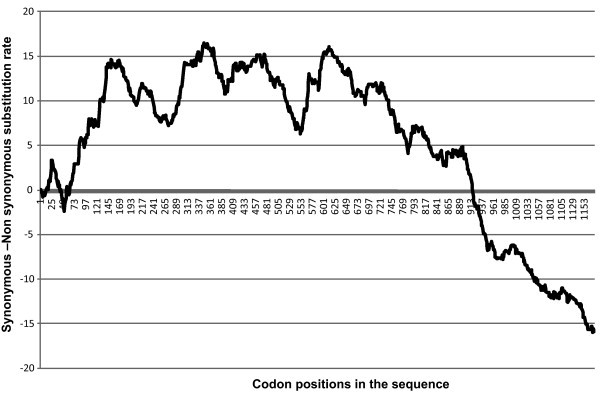
The selective pressure analysis in the spike glycoprotein of IBV. The graph illustrates the S1 part of the spike glycoprotein was mostly subject to positive selective pressure. In S2, the selective pressure was mostly negative.
3.5. Recombination analysis
There was evidence of recombination events in the S1 subunit of the Swedish isolates from 2009 to 2010. Recombination events were mapped to begin with break point position at amino acid 536 in isolates 079663 and 079692. The ending break point strong signals were observed at amino acid position 834.
3.6. Phylogenetic analysis
Phylogenetic analyses of the Swedish IBV isolates were carried out based on the partial S1 and complete spike gene. Published IBV sequences of Massachusetts type, M41, H120, Baudette, Italy02, QX-like and 4/91 available in the GenBank were used to determine likely origin, genetic characteristics and molecular epidemiology of the Swedish isolates.
The phylogenetic analysis based on the partial S1 gene revealed the sequences distinguished into four main groups (Fig. 2 ). One group was composed of Massachusetts-type, M41, H120 and Beaudette strains, where the Swedish isolates from 1995 to 1999 clustered, together with strains from Spain, Korea Thailand, China and other vaccine strains. This group was classified as Massachusetts genotype. The second group was divided into two sub-groups: one sub-group comprising sequences from Sweden, France, The Netherlands, Spain, Italy, UK, Israel, Korea and China known to belong QX-like genotype. This sub-group was further composed of three clusters from Sweden, Spain and mixed strains from France and the Netherlands. Three isolates from Israel, Italy and UK occupied a separate position near to these subgroups. The recent Swedish isolates from 2009 to 2010 were found distributed in this sub-group as a separate cluster and closely related to the Dutch strain NL1148. Two new Swedish isolates 079663 and 079692 branched out separately from the other Swedish isolates. The remaining two sub-groups in this group consisted mostly of Korean and Chinese QX strains. The third group consisted of two isolates from UK and Italy related to ITA-02 and belonging to the ITA-02 genotype. The fourth group has shown relation to the 4/91 genotype and consisted of reference strain 4/91 and two isolates from Iraq.
Fig. 2.
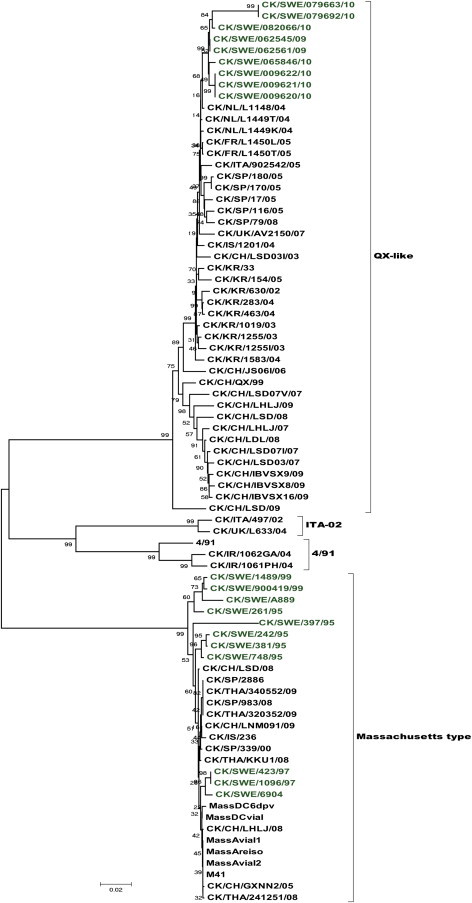
Phylogenetic tree based on partial S1 gene showing the relationship of different IBV isolates. The isolates sequenced in this study are indicated in green and grouped with Mass and the QX-like genotypes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
There were no complete spike gene sequences of the European D388/QX-like genotype available in the GenBank. Thus the comparison was limited to available sequences in the databases. The phylogenetic relationship based on the complete spike gene of IBV revealed that these sequences were separated into two main groups (Fig. 3 ). The first group contained sequences of the strains from USA, Sweden, China and Singapore related to Baudette, H120, Ark 99 and Massachusetts genotype. This group was further divided into subgroups. Swedish isolates from 1999 to 1995 were found in this group. The Swedish isolates 1489, 900419 and A889 have fallen in the subgroup which is closely related to Mass 41 isolates from USA. The Swedish isolates 242, 748, 381, 397, 261, 423 and 1096 clustered together and showed identity as a separate subgroup of Massachusetts type. One Swedish isolate 6904 occupied a distinct place in this group.
Fig. 3.
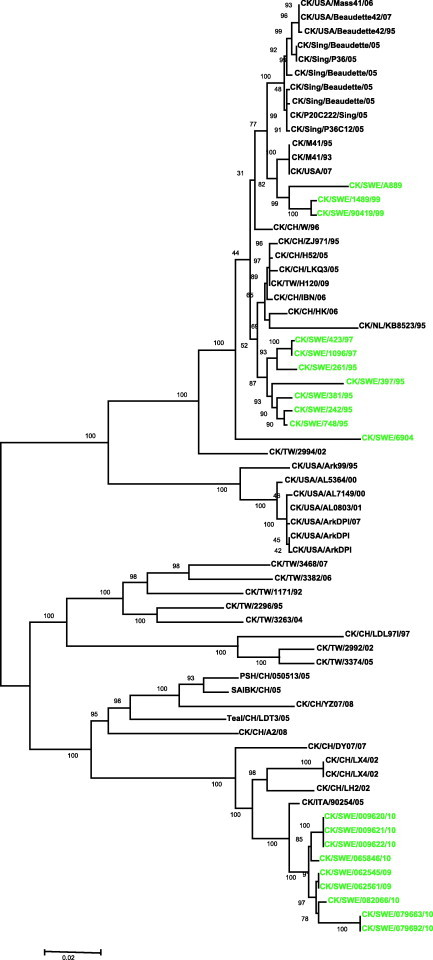
Phylogenetic tree based on the complete spike gene showing the relationship between different IBV strains.
The second group consisted of two subgroups. One of the subgroup mostly contained isolates from Taiwan. The second sub-group consisted of two main clusters. The Swedish isolates from 2009 to 2010 were found forming a separate cluster in this sub-group of the sequences. The closest relationship of these Swedish isolates shared with the CK/ITA/90254/05 strain. CK/ITA/90254/05 has occupied a distinct place within this subgroup. The other cluster consisted of different Chinese isolates.
4. Discussion
IBV has been detected in the poultry population in Sweden in the nineties and then in more recent years cases were reported. Despite current immunization with live attenuated vaccines, occasional cases of IB have occurred in poultry flocks in 2007–2010. Therefore, this study was designed to characterize strains of IBV associated with outbreaks in the nineties and in recent years in order to understand the molecular epidemiology of this disease in Sweden.
Analysis of the nucleotide sequences of spike glycoprotein of Swedish isolates from 1995 to 2010 showed that they comprise distinct sets of IBV variants; differing among themselves along that timeline (Table 3). It is obvious that the outbreaks in the nineties and those in 2000s were caused by different IB viruses. The isolates 079663 and 079692, standing alone had the genetic signature of a new variant, as they have shown unique substitutions compared to the remaining sequences in this cluster. The nucleotide sequence data showed different regions of hypervariability in the S1 subunit of the spike glycoprotein. This is consistent with the notion that the S1 is prone to mutations, as demonstrated in different studies (Adzhar et al., 1997, Farsang et al., 2002, Keeler et al., 1998, Kingham et al., 2000).
Previously it has been reported that Ark-like strains have a deletion of three nucleotides in the S1 gene at nucleotides positions 69–71 (Ammayappan and Vakharia, 2009) but in this context, we have observed insertion rather than deletion in S1 unit of the sequences of D388/QX-like strains. These insertions and deletions possibly have some significance in the properties related to S1 functions. Further studies are needed to determine the role of these insertions and deletions in the spike glycoprotein for example in neutralization phenotype or virus tropism, which could explain the reported change in the clinical manifestation of infectious bronchitis.
The S1 gene of the isolates showed much higher variation as compared to S2 gene that was mostly conserved, as previously reported (Adzhar et al., 1997, Keeler et al., 1998, Kingham et al., 2000). However, certain regions in the S2 were determined to be also variable, and, to an extent, contributing to sequence discrimination based on analysis of the complete S gene. Therefore, it is plausible that the S2 subunit may have some role in development of new field variants. It has been reported that the evolutionary rates in the S1 of vaccinated birds were 1.5–2.5% per year. Nucleotide point mutations, deletions, insertions, and RNA recombination in the sequences of spike gene lead to generation of different IBV serotypes (Wang and Huang, 2000).
The deduced amino acid sequence data revealed different patterns of substitutions resulting from point mutations and insertions in the Swedish IBV isolates especially in S1 subunit. The insertions of aspartic acid, glycine and serine in the sequences of the isolates may affect antigenicity of the virus, and this will be a question for further studies on serotyping of the isolates. Our results are consistent to those were obtained by previous studies (Ammayappan and Vakharia, 2009, Liu et al., 2009).
The strong positive selective pressure resulting from mutations in S1 gene is likely responsible for the genetic diversity and in the appearance of new phenotypic and antigenic variants. Recombination events were found in the spike gene of recent Swedish IBV isolates. In the particular case of strains 079663 and 079692 the recombination event was mapped to amino acid 536 and is believed to have determined a new lineage within Swedish D388/QX-like cluster.
In order to obtain more information concerning to molecular epidemiology of IB, We have carried out a comprehensive phylogenetic study based on partial S1 and complete spike gene of the selected Swedish isolates. The phylogenetic studies, based on the partial S1 gene regions, showed that Swedish IBV isolates from 1995 to 1999 were related to the Massachusetts genotype. The findings confirmed our previous observation (Farsang et al., 2002). It is hypothesized that these genotypic variants have been reduced by vaccine and did not appear through diagnosis after 1999. The Swedish isolates from 2009 to 2010 were found to be closely related to other Western European isolates from comparison of sequences available in the GenBank especially to one isolate from The Netherlands (Fig. 2).
The QX strains were first isolated in China in 1998 and were found associated mostly with proventriculitis, diarrhea and loss of body weight in 25–70 day old chickens (Wang et al., 1998). It was interesting that the Chinese QX-like genotype of IBV was isolated from a backyard flock in a Vladimir Oblast (Russia) where the prevalence of Chinese QX-like genotype was common and this region is close to Europe (Bochkov et al., 2006, Gough et al., 2008). It is unclear how the QX viruses spread to Europe. There were more cases of QX-like virus infection with presentation of nephritis rather than proventriculitis, and false layers in mature hens reported from Belgium, China, France, Germany, The Netherlands, Russia, Taiwan and UK (Ammayappan and Vakharia, 2009, Beato et al., 2005, Bochkov et al., 2006, Worthington et al., 2008, Zhou et al., 2004). Similar type of IBV isolates has been reported in Israel and Korea (Meir et al., 2004, Park et al., 2005). A QX-like strain in chicken associated with nephritis, rare proventriculitis and mortality was reported in Poland in 2006 (Domanska-Blicharz et al., 2006). These results demonstrated that QX-like strains comprise new emerging IBVs and presented evidence of its involvement in epidemics in many countries.
It is noteworthy that despite a number of outbreaks in Europe caused by QX-like strains, only a few sequences are available in the GenBank. The lack of sufficient information in this aspect is limiting the comprehensible tracing of routes of infection in the continent. For example in Denmark QX-like IBV variants have been reported (Handberg et al., 2009) but the sequences of these viruses are not available in GenBank. There is a possibility that trade and import of day old chicks and poultry products from other European countries or indirect routes may have contributed to the spread of the QX-like strains in Sweden. The role of wild or migratory birds in dissemination of the IBV new variants still remained unclear. However, previous studies on other coronaviruses and IBV in wild birds have indicated a potential role of wild birds in dissemination of various coronaviruses, including IBV in Europe (Gough et al., 2008, Muradrasoli et al., 2010).
Taken together, the complete spike sequence data revealed different isolates of IBV of the Massachusetts type and European D388/QX-like strains circulating over a time-line in Sweden. So far, there was no evidence of presence of the ITA-02 and 4/91 genotypes in Sweden.
5. Conclusions
The data suggest that the Massachusetts type strains have been replaced recently with European D388/QX-like strains in Sweden. Sequence diversity and different molecular characteristics were observed among the Swedish isolates from 1995 to 2010. The investigations revealed that presumably, D388/QX-like viruses were introduced from other European countries through imports. Considering the number of investigated viruses, the present study provides the most comprehensive sequence data on IBV variants related to the D388/QX-like genotype, detected in Europe to date. Thus, the sequence data of this article will provide valuable reference for future studies on the evolution of genotypes, molecular epidemiology, improved diagnosis and on the virus-evolution in relation to vaccine development. Ultimately, the information provided in these studies, will contribute to a more effective control of IB, this viral disease in poultry of global importance.
Acknowledgements
This study was funded by Grant 70030/2009 of SVA Research Fund and grant from the Swedish Poultry Meat Association (Svenskfågel).
References
- Adzhar A., Gough R.E., Haydon D., Shaw K., Britton P., Cavanagh D. Molecular analysis of the 793/B serotype of infectious bronchitis virus in Great Britain. Avian Pathol. 1997;26:625–640. doi: 10.1080/03079459708419239. [DOI] [PubMed] [Google Scholar]
- Ammayappan A., Vakharia V.N. Complete nucleotide analysis of the structural genome of the infectious bronchitis virus strain Md27 reveals its mosaic nature. Viruses-Basel. 2009;1:1166–1177. doi: 10.3390/v1031166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M.S., De Battisti C., Terregino C., Drago A., Capua I., Ortali G. Evidence of circulation of a Chinese strain of infectious bronchitis virus (QXIBV) in Italy. Vet. Rec. 2005;156:720. doi: 10.1136/vr.156.22.720. [DOI] [PubMed] [Google Scholar]
- Bochkov Y.A., Batchenko G.V., Shcherbakova L.O., Borisov A.V., Drygin V.V. Molecular epizootiology of avian infectious bronchitis in Russia. Avian Pathol. J. W.V.P.A. 2006;35:379–393. doi: 10.1080/03079450600921008. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol. 2003;32:567–582. doi: 10.1080/03079450310001621198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Naqi S. Infectious bronchitis. In: Barnes H.J., Fadly A.M., Glisson J.R., McDougald L.R., Swayne D.E., editors. Diseases of Poultry. 11th ed. Iowa State University Press; Ames: 2003. pp. 101–119. [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- de Wit J.J., Cook J.K.A., van der Heijden H.M.J.F. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40:223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanska-Blicharz K., Minta Z., Smietanka K., Porwan T. New variant of IBV in Poland. Vet. Rec. 2006;158:808. doi: 10.1136/vr.158.23.808-c. [DOI] [PubMed] [Google Scholar]
- Farsang A., Ros C., Renstrom L.H.M., Baule C., Soos T., Belak S. Molecular epizootiology of infectious bronchitis virus in Sweden indicating the involvement of a vaccine strain. Avian Pathol. 2002;31:229–236. doi: 10.1080/03079450220136530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb J., Weisman Y., Ladman B.S., Meir R. Gene characteristics and efficacy of vaccination against infectious bronchitis virus field isolates from the United States and Israel (1996–2000) Avian Pathol. 2005;34:194–203. doi: 10.1080/03079450500096539. [DOI] [PubMed] [Google Scholar]
- Gonzalez J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough R.E., Cox W.J., Welehman D.D., Worthington K.J., Jones R.C. Chinese QX strain of infectious bronchitis virus isolated in the UK. Vet. Rec. 2008;162:99–100. doi: 10.1136/vr.162.3.99. [DOI] [PubMed] [Google Scholar]
- Handberg K.J., Kabell S., Olesen L., Jrgensen P.H. Rauischholzhausen; Germany: 2009. VI International Symposium on Avian Corona- and Pneumoviruses and Complicating Pathogens. pp. 14–17. [Google Scholar]
- Holmes K.V. Coronaviridae and their replication. In: Fields B.N., Knipe D.M., editors. Virology. 2nd ed. Raven Press, Ltd.; New York: 1990. pp. 841–856. [Google Scholar]
- Ignjatovic J., Galli L. The S1 glycoprotein but not the N-protein or M-protein of avian infectious-bronchitis virus induces protection in vaccinated chickens. Arch. Virol. 1994;138:117–134. doi: 10.1007/BF01310043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.A., Pooley C., Ignjatovic J., Tyack S.G. A recombinant fowl adenovirus expressing the S1 gene of infectious bronchitis virus protects against challenge with infectious bronchitis virus. Vaccine. 2003;21:2730–2736. doi: 10.1016/s0264-410x(03)00227-5. [DOI] [PubMed] [Google Scholar]
- Keeler C.L., Reed K.L., Nix W.A., Gelb J. Serotype identification of avian infectious bronchitis virus by RT-PCR of the peplomer (S1) gene. Avian Dis. 1998;42:275–284. [PubMed] [Google Scholar]
- Kingham B.F., Keeler C.L., Nix W.A., Ladman B.S., Gelb J. Identification of avian infectious bronchitis virus by direct automated cycle sequencing of the S1 gene. Avian Dis. 2000;44:325–335. [PubMed] [Google Scholar]
- Korber B. In: Gerald and Learn, HIV Signature and Sequence Variation Analysis. Computational Analysis of HIV Molecular Sequences. Allen G., Rodrigo H., editors. Kluwer Academic Publishers; Dordrecht, Netherlands: 2000. pp. 55–72. [Google Scholar]
- Kuo S.M., Wang C.H., Hou M.H., Huang Y.P., Kao H.W., Su H.L. Evolution of infectious bronchitis virus in Taiwan: characterisation of RNA recombination in the nucleocapsid gene. Vet. Microbiol. 2010;144:293–302. doi: 10.1016/j.vetmic.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.W., Zhang Q.X., Chen J.D., Han Z.X., Liu X., Feng L., Shao Y.H., Rong J.G., Kong X.G., Tong G.Z. Genetic diversity of avian infectious bronchitis coronavirus strains isolated in China between 1995 and 2004. Arch. Virol. 2006;151:1133–1148. doi: 10.1007/s00705-005-0695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.L., Su J.L., Zhao J.X., Zhang G.Z. Complete genome sequence analysis of a predominant infectious bronchitis virus (IBV) strain in China. Virus Genes. 2009;38:56–65. doi: 10.1007/s11262-008-0282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood Z.H., Sleman R.R., Uthman A.U. Isolation and molecular characterization of Sul/01/09 avian infectious bronchitis virus, indicates the emergence of a new genotype in the Middle East. Vet. Microbiol. 2011;150:21–27. doi: 10.1016/j.vetmic.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir R., Rosenblut E., Perl S., Kass N., Ayali G., Hemsani E., Perk S. Identification of a novel nephropathogenic infectious bronchitis virus in Israel. Avian Dis. 2004;48:635–641. doi: 10.1637/7107. [DOI] [PubMed] [Google Scholar]
- Muradrasoli S., Balint A., Wahlgren J., Waldenstrom J., Belak S., Blomberg J., Olsen B. Prevalence and phylogeny of coronaviruses in wild birds from the bering strait area (Beringia) Plos One. 2010:5. doi: 10.1371/journal.pone.0013640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.Y., Pak S.I., Sung H.W., Kim J.H., Song C.S., Lee C.W., Kwon H.M. Variations in the nucleocapsid protein gene of infectious bronchitis viruses isolated in Korea. Virus Genes. 2005;31:153–162. doi: 10.1007/s11262-005-1788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Spaan W.J.M., Snijder E.J. Nidovirus transcription: how to make sense…? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. Molecular evolutionary genetic ananlysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wang C.H., Huang Y.C. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch. Virol. 2000;145:291–300. doi: 10.1007/s007050050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.D., Wang Y.L., Zhang Z., Fan G., Jiang Y., Liu X., Ding J., Wang S. Isolation and identification of glandular stomach type IBV (QX IBV) in chickens. Chin. J. Anim. Quarantine. 1998;15:1–3. [Google Scholar]
- Worthington K.J., Currie R.J., Jones R.C. A reverse transcriptase–polymerase chain reaction survey of infectious bronchitis virus genotypes in Western Europe from 2002 to 2006. Avian Pathol. 2008;37:247–257. doi: 10.1080/03079450801986529. [DOI] [PubMed] [Google Scholar]
- Worthington K.J., Jones R.C. New genotype of infectious bronchitis virus in chickens in Scotland. Vet. Rec. 2006;159:291–292. doi: 10.1136/vr.159.9.291-b. [DOI] [PubMed] [Google Scholar]
- Zhou J.Y., Zhang D.Y., Ye J.X., Cheng L.Q. Characterization of an avian infectious bronchitis virus isolated in China from chickens with nephritis. J. Vet. Med. 2004;B51:147–152. doi: 10.1111/j.1439-0450.2004.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


