Abstract
A reverse-transcription polymerase chain reaction (RT-PCR) was developed to differentiate the bovine diarrhea virus (BVDV) from other pestiviruses, and to determine the genotype of the BVDV isolates. For this purpose, primer pairs were selected in the 5′ untranslated region (5′UTR). The primers BE and B2 were located in highly conserved regions and were pestivirus-specific. Two primer pairs named B3B4 and B5B6 were specific of BVDV genotypes I and II, respectively. With this technique, an amplification product of the expected size was obtained with either the B3B4 or the B5B6 primer pairs for the 107 BVDV isolates tested but not for BDV or CSFV. For some isolates that were grouped in the genotype II, sequence analysis of the PCR fragments confirmed their classification into this genotype.
Keywords: Diagnosis, Pestivirus, BVD, PCR, Genotyping
1. Introduction
Bovine viral diarrhea is an important disease of cattle. Acute infection is usually mild and often subclinical, and is resolved with the appearance of neutralizing antibodies. Transplacental infection of the fetus with a noncytopathic BVDV strain during the first trimester of gestation may lead to persistently infected (p.i.), immunotolerant calves. When these animals are superinfected with an antigenically similar cytopathic strain they may suffer of the fatal mucosal disease (Brownlie, 1991). Another fatal outcome of BVDV infection is associated with severe thrombocytopenia and hemorrhagic lesions. Several outbreaks of this syndrome have been reported, essentially in North America but cases have also been reported in Europe (Corapi et al., 1989; Broes et al., 1992; Pellerin et al., 1994).
BVDV belongs to the pestivirus genus that also comprises border disease virus of sheep (BDV) and classical swine fever virus (CSFV). The classification of virus types was made according to the host-species that was infected. Pestiviruses are, however, able to cross the species barrier. BVDV can cross-infect cattle, sheep, goats and pigs (Carlsson, 1991; Paton et al., 1992). BDV is an ovine pathogen that occasionally infects pigs (Roehe et al., 1992; Edwards et al., 1995; Vilcek and Belak, 1996). Differentiation between CSFV and other pestiviruses can be accomplished by the use of CSFV-specific monoclonal antibodies (Mabs) but the search for ruminant pestivirus-specific Mabs has failed due to a great antigenic diversity (Edwards et al., 1991).
RT-PCR has been proved to be a rapid and sensitive method to detect viral nucleic acids and this technique has been used by several investigators for the detection of pestiviruses using oligonucleotide primers located in conserved regions of the viral genome (Lopez et al., 1991; Katz et al., 1993; Ridpath et al., 1993; Wirz et al., 1993; Hofmann et al., 1994; Horner et al., 1995; Tajima et al., 1995; Canal et al., 1996). Most of the PCR primers that were selected in the 5′UTR recognized the greatest number of pestivirus isolates but failed to differentiate BVDV from other pestiviruses (Ridpath et al., 1993; Vilcek et al., 1994). Specific detection of CSFV by RT-PCR has been described using primers located in the coding part of the genome or by the sequence analysis of the PCR product amplified using primers located in a highly conserved region of the 5′UTR (Katz et al., 1993; Wirz et al., 1993; Hofmann et al., 1994). The gp25, gp48, p54 and p80 regions did also serve as template for BVDV amplification. However, these RT-PCR tests failed to detect all BVDV isolates (Vilcek et al., 1994; Tajima et al., 1995).
Based on the sequence comparison of 5′UTR, the BVDV isolates were subdivided in two genotypes. Sequence homology within each group was very high (>93%) while homology between group I and II droped near 74% (Pellerin et al., 1994; Ridpath et al., 1994).
An accurate and fast differentiation between BVDV and another pestivirus is of great importance for the development of control measures, particularly when an outbreak of CSFV is suspected. Sensitive detection of BVDV contamination in FCS and cell culture would help diagnostic work and would improve the safety of veterinary and human vaccines.
The aim of this study was to develop a RT-PCR test that allowed the detection of BVDV and differentiation between both genotypes. Published 5′UTR sequences were aligned and two pairs of primers were selected. Belgium field BVDV isolates were tested by amplification with both sets of primers. Sequence analysis of the PCR products confirmed the appartenance of some field isolates to the genotype II.
2. Materials and methods
2.1. Viruses and cell cultures
The BVDV reference strain NADL was received from Dr. J.M. Aynaud (INRA, Thiverval, France), the C24V strain from Dr. Straver (CDI, Amsterdam), the 3620/Han89 isolate (Hewicker-Trautwein et al., 1995) from Dr. B. Liess and the New York and BDV Aveyron (Chappuis et al., 1986) strains were received from Prof. E. Thiry (University of Liège). The viruses were propagated on bovine fetal kidney cells grown in minimum essential medium (MEM) supplemented with pestivirus free fetal bovine serum. CSFV was kindly provided by Dr. F. Koenen (CERVA).
Organs (lungs, intestines, brains) were obtained from 64 BVDV-positive cattle with respiratory, digestive or neurological symptoms. These were routine diagnostic samples obtained between 1991 and 1996 that were assessed as BVDV positive by a direct immunofluorescence test realized on frozen tissue sections. Homogenate extracts were then used for RNA isolation. Leucocytes pellets were prepared from 39 immunotolerant cattle which had previously been detected by an antigen capture ELISA kit (Rhone Mérieux).
2.2. Primer design
Oligonucleotide primers were designed from the 5′UTR. The sequence of two pairs of primers, designated B1B2 and BEB2, was located in a highly conserved region of BVDV, CSFV and BDV. These primers were tested for the amplification of pestivirus cDNAs. Their sequence, position according to the NADL genome sequence (Collett et al., 1988) and the expected size of the amplified products are presented in Table 1 .
Table 1.
Sequence of the oligonucleotide primers used in the RT-PCR reactions
| Primer | Sequence | Genome positiona | Specificity | Size of the amplified product |
| B1 | 5′AGG GTA GTC GTC AGT GGT TCG 3′ | 185–195 | Pestivirus | 210 bp (B1B2) |
| BE | 5′CAT GCC CTT AGT AGG ACT AGC 3′ | 108–127 | Pestivirus | 287 bp (BEB2) |
| B2 | 5′TCA ACT CCA TGT GCC ATG TAC 3′ | 395–375 | Pestivirus | |
| B3 | 5′GGT AGC AAC AGT GGT GAG 3′ | 139–155 | BVDV type I | 221 bp (B3B4) |
| B4 | 5′GTA GCA ATA CAG TGG GCC 3′ | 360–343 | BVDV type I | |
| B5 | 5′ACT AGC GGT AGC AGT GAG 3′ | 139–145 | BVDV type II | 221 bp (B5B6) |
| B6 | 5′CTA GCG GAA TAG CAG GTC 3′ | 360–343 | BVDV type II | |
According to the NADL genome positions.
Two pairs of primers were designed from the alignment of sequences published by others (Pellerin et al., 1994). The sequences were conserved in a BVDV genotype but not between genotype I and II. The B3B4 pair was tested for the amplification of BVDV type 1 sequences and the B5B6 pair was used for BVDV type 2 amplification.
2.3. RNA extraction and cDNA synthesis
RNA was extracted using 1 ml TRIZOL (GIBCO BRL) according to the manufacturer's instructions. The extraction was accomplished using 200 μl of organ homogenate extracts or a pellet of leucocytes corresponding to 1.5 ml of heparinized and hemolyzed blood.
The RNA was resuspended in 10 μl DEPC-treated water. The reverse transcription was carried out in a volume of 20 μl containing 50 mM Tris–HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 0.5 mM dNTP, 150 pmol of the reverse primer B2, 200 u M-MLV reverse transcriptase (GIBCO BRL) and 2 μl RNA. The cDNA was synthesized at 37°C for 15 min and the enzyme was inactivated for 5 min at 95°C.
2.4. Amplification of cDNA
The amplification of cDNA by PCR was carried out in a total volume of 50 μl containing 20 mM Tris–HCl (pH 8.4), 50 mM KCl, 3 mM MgCl2, 0.5 mM dNTP, 75 pmol of each primer, 2 μl cDNA and 2.5 u Taq DNA polymerase (GIBCO BRL). The reaction was heated in a thermocycle for 5 min at 95°C and then submitted to 35 cycles of amplification. For the B1B2 and BEB2 primer pairs, the conditions of amplification were 1 min at 94°C, 1 min at 57°C, and 1 min at 72°C. For the B3B4 and B5B6 primer pairs, the conditions were 1 min at 94°C, 1 min at 51°C, and 1 min at 72°C. Amplified products were separated by electrophoresis in 1.5% agarose gel in Tris–borate EDTA buffer.
2.5. Southern blot hybridization
After electrophoresis of the BEB2 amplification products, the agarose gel was soaked two times for 15 min in denaturation solution (1.5 M NaCl, 0.5 M NaOH), then in neutralization solution (0.5 M Tris–HCl (pH 7.5), 1.5 M NaCl). The DNA was then transfered to two positively charged nylon membranes (Boehringer) and the DNA was cross-linked by UV irradiation. The filter was incubated for 30 min at 44°C in DIG Easy Hyb solution (Boehringer). Then 100 pmol of 3′ DIG labeled B4 or B6 oligonucleotide (Eurogentec) was added and the incubation was continued overnight at 44°C.
Filters were washed and the hybridized probes were detected using DIG luminescent detection kit (Boehringer) according to the manufacturer's instructions.
2.6. Sequence analysis
The PCR products obtained with the B1B2 or BEB2 primers were purified on Centricon 100 filters and were cloned in a PGEM-T vector (Promega). Plasmid DNA was purified using Qiagen tip 20 columns. A minimum of two clones were sequenced with the sequencing primer M13–21, dye terminator cycle sequencing kit (Perkin-Elmer) and ABI PRISM 310 automated cycle sequencer according to the manufacturer's recommendations.
Alignment of the sequences was realized using the PILEUP program included in the GCG software package (Devereux et al., 1984). Distances were calculated by the Kimura 2-parameter method (Kimura, 1980) and used to construct trees using the GROWTREE program of the GCG. Sequence analysis was achieved using the Belgian EMBnet Node facility.
3. Results
3.1. Genotyping of BVDV reference strains by RT-PCR and Southern blot hybridization
Total RNA extracted from infected cell cultures was submitted to RT-PCR by using the BEB2 pestivirus-specific primers. A PCR product of the expected size (287 bp) was amplified from BVDV reference strains NADL, New York, C24V, 3620/Han89, BDV Aveyron and CSFV, and from the BVDV field isolate UVR420.
The results obtained for BVDV NADL, New York, UVR420, BDV Aveyron and CSFV are shown in Fig. 1 (A). In Fig. 1(B), the same cDNAs were amplified with BVDV type I and type II-specific primer pairs B3B4 and B5B6. A fragment of 221 bp was obtained with the former for BVDV NADL and New York, and with the latter for BVDV UVR420. This isolate was then used as positive control for the type II amplifications. No amplification product was obtained for BDV Aveyron and CSFV. The BVDV strains C24V and 3620/Han89 were classified in the genotype I (data not shown).
Fig. 1.
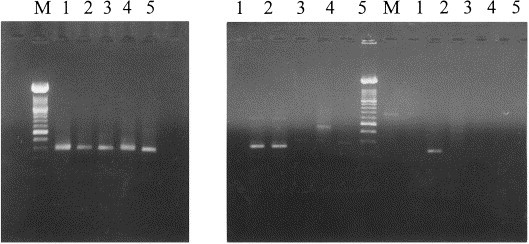
Agarose gel electrophoresis of RT-PCR amplification products derived from BVDV NADL (1), New York (2), UVR420 (3), BDV (4), CSFV (5). M: Molecular weight marker XIV (Boehringer). (A) Primers BEB2, (B) Primers B3B4 and B5B6.
The same results were obtained after Southern blot hybridization performed on the BEB2 amplification products by using the DIG-labeled genotype-specific probes. The BEB2 amplification products shown in Fig. 1(A) were hybridized with B4DIG probe in Fig. 2 (A) and with B6DIG probe in Fig. 2(B). With the BVDV type I-specific probe, the amplification products from BVDV NADL and New York were detected. The BVDV type II-specific probe reacted only with the BVDV UVR420 isolate. The BDV and CSFV amplification fragments did not react with either BVDV-specific probe.
Fig. 2.
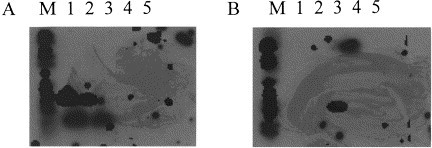
Southern blot hybridization of pestivirus-specific amplification DNA fragments from BVDV NADL (1), New York (2), UVR420 (3), BDV (4), CSFV (5). M: Molecular weight marker 6 DIG (Boehringer). (A) B4DIG Probe, (B) B6DIG Probe.
3.2. Genotyping of BVDV field isolates
A total of 107 BVDV positive samples were tested. RNA was extracted from organs or from leukocytes of infected animals. All of the samples contained a pestivirus as demonstrated by a RT-PCR test using the pestivirus-specific primers B1B2 or BEB2. The samples were also tested by both genotype-specific PCR and hybridization. The results are presented in Table 2 . Of a total of 64 organs (lungs, intestines, brains), 55 were assessed as genotype I and nine as genotype II. Two out of 39 immunotolerant cattle were infected with a genotype II isolate and two out of four laboratory viruses were also classified as genotype II. As a total, 13 of the 107 isolates tested belonged to the genotype II. Organs positive for infectious bovine rhinotracheitis, bovine parainfluenza type 3, coronavirus and rotavirus were used as controls. No amplification product was obtained (data not shown).
Table 2.
Number of BVDV isolates classified in each genotype
| Source | Genotype I | Genotype II | Total |
|---|---|---|---|
| Organs | 55 | 9 | 64 |
| Leucocytes (PI) | 37 | 2 | 39 |
| Others | 2 | 2 | 4 |
| Total | 94 | 13 | 107 |
Type II viruses were isolated from 1991 to 1996 in Belgium (Table 3 ). Four isolates came from calves with respiratory disease, one from the intestine of a calf. Three viruses were isolated from the brain of animals showing neurological symptoms. Two genotype II viruses were isolated from the blood of immunotolerant animals and only one virus was isolated from a case of haemorrhagic syndrome (Broes et al., 1992).
Table 3.
Characterization of the genotype II isolates
| Strain | Year of isolation | Organ | Characteristics |
|---|---|---|---|
| UVR420 | 1991 | Lung (calf) | Respiratory symptoms |
| BSE341 | 1991 | Brain | Neurologic symptoms |
| UVD493 | 1992 | Intestine (calf) | Hemorrhagic syndrome |
| WVD829 | 1994 | Intestine (calf) | |
| XVR2518 | 1995 | Lung (calf) | Respiratory symptoms |
| XVR2154 | 1995 | Lung | Respiratory symptoms |
| XVR2790 | 1995 | Lung | Respiratory symptoms |
| BSE921 | 1995 | Brain | Neurologic symptoms |
| YVR1298 | 1995 | Blood | Persistently infected |
| 1877 | 1995 | Blood | Persistently infected |
| BSE1239 | 1996 | Brain | Neurologic symptoms |
3.3. Sequencing of the genotype II isolates
The alignment of nucleotide sequence of a 168 bp DNA fragment, flanked by primers B1 and B2 is presented in Fig. 3 . The sequences of 10 Belgian type II isolates were compared to other genotype II sequences extracted from a genebank: 890, isolated from a haemorrhagic syndrome outbreak (Pellerin et al., 1994), CPA virus isolated from cell culture (Harasawa, 1996), the ovine pestivirus 175 375 isolated in the UK (Vilcek et al., 1997). Reference BVDV type I strains NADL and New York and three Belgian virus isolated from organs (ZVD278, ZVR711) and from a p.i. calf were also included in the comparison.
Fig. 3.

Alignment of nucleotide sequences from the 5′UTR of BVDV types I and II isolates. The sequences are flanked by primers B1 and B2. Genotype II sequences are presented above the horizontal line and genotype I below. Sequence of the B4 and B6 oligonucleotides is boxed.
The sequence alignment confirmed the classification of all the isolates. The sequence of the region corresponding to the oligonucleotides B4 and B6, represented in Fig. 3 by a box, showed a high degree of conservation inside both genotypes, and were divergent between the genotypes.
A phylogenetic tree was constructed and is presented in Fig. 4 . Genotype II viruses were segregated into two groups. The first englobed eight Belgian isolates which showed very little genetic divergence. In the other group were classified viruses isolated from cell culture (CPA), from the USA, the UK and two Belgian isolates. Interestingly, one of them (BSE921) was nearly identical to the CPA virus.
Fig. 4.
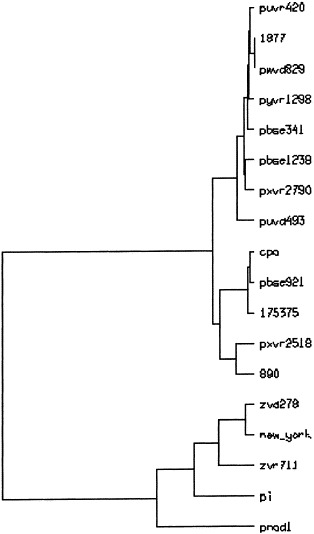
Phylogenetic tree of selected BVDV field isolates. A portion of the 5′UTR corresponding to BVDV-NADL base positions 206–374 was used. The horizontal branch lenghts are proportional to the similarity between the sequences.
The ZVD278, ZVR711 and p.i. viruses were classified in genotype I. The ZVD278 isolate showed a strong homology to the New York strain.
4. Discussion
RT-PCR tests and sequence analysis of the p20 and E2 regions were used by several authors to classify the pestiviruses. Using these techniques, four genotypes were identified. The CSFV group was restricted to swine, whereas BVDV types I and II included strains isolated from cattle, sheep, goats and wild ruminants. The BDV genotype comprised strains that were isolated from sheep and pigs (Becher et al., 1995, Becher et al., 1997; van Rijn et al., 1997; Vilcek et al., 1997). This classification did not correlate with the host-species origin but was supported by serological investigations (Dekker et al., 1995; Paton et al., 1995). This accurate method cannot however be used for diagnostic work.
Specific primers located in the 5′UTR region were used by others for the diagnosis of BVDV infection by RT-PCR. Canal et al. (1996)reported the use of a conserved antisense oligonucleotide and two sense primers, CSFV and ruminant-pestivirus-specific, respectively, to distinguish these viruses. Another set of primers was designed by Sandvik et al. (1997). PCR fragments from BVDV types I and II originating from different host-species were successfully amplified.
Here, we describe a differential system to discriminate between BVDV and other pestiviruses and to determine the genotype of the BVDV isolates, using conserved regions located in the 5′UTR. The sequence of the primer used for the cDNA synthesis was conserved between the pestiviruses. This cDNA could thus be used in pestivirus-specific but also BVDV-specific PCR. The primers BE and B2 have already been used by others and were designed to detect all the pestiviruses (Vilcek et al., 1994). Based on the search for genotype-conserved regions, two primer pairs were selected. This allowed us to determine the genotype of the BVDV isolates.
The RT-PCR tests were validated with reference BVDV, BDV and CSFV strains. With a combination of PCR reactions, the BVDV strains could be differentiated from BDV and CSFV and the genotype of BVD viruses could be determined. No cross-reactivity was detected between the type-specific primers. Then, the PCR assays were used to characterise field isolates that were previously shown BVDV-positive by other techniques. The RNA extraction was performed on organ homogenate extracts or on leukocytes. This excluded a BVDV contamination during cell culture multiplication. Among the 107 samples examined, 94 were classified as genotype I and 13 as genotype II.
From the 13 genotype II viruses that were obtained, five were isolated from organs, two from the blood of p.i. animals and for two other isolates, we were not able to trace the origin of the virus. Only one virus was isolated from a case of hemorrhagic syndrome. Eight BVDV-positive brains were also tested. Three of them harboured genotype II viruses. Our hypothesis is that these were p.i. animals. Indeed, the presence of BVDV in the central nervous system (CNS) has been described and this seems to be an important location for the persistence of BVDV. Primary infection of the CNS most likely occurs across the blood–brain barrier (Fernandez et al., 1989; Hewicker et al., 1990; Hewicker-Trautwein et al., 1992). Other experiments showed that BVDV was found in the CNS of all p.i. animal tested, without causing obvious cellular destruction (Wöhrmann et al., 1992).
Ten genotype II and three genotype I isolates were further analyzed by sequencing the amplification DNA fragment. Based on pairwise alignment, conservation of the sequences inside the genotype II group was at least 92%. A minimum of 88% homology was found inside the genotype I group. When NADL strain was excluded from the comparison, the homology rose to 95%, suggesting that the Belgian isolates tested were closer to genotype Ib than Ia strains. The homology between genotype I and II was approximately 75%.
The greatest divergence between genotype II isolates was between UVD493 and 890, which were both isolated from a haemorrhagic syndrome.
Our results demonstrate that RT-PCR could be performed on RNA extracted from organ samples. Furthermore, the BVDV genotype could be determined. The same approach has been used by Sullivan and Akkina (1995). Consensus primers located in the gp48 region were tested on bovine and ovine isolates, whereas combination of specific primers allowed the differentiation between BVDV type I, type II and BDV.
BVDV has been known not only as a veterinary pathogen but also as a common contaminant in cell culture work and in live vaccines for human, bovine and swine use (Harasawa and Tomiyama, 1994; Harasawa, 1995; Harasawa and Mizusawa, 1995). Genotype II BVDV has been described as a contaminant of FBS (Ridpath et al., 1994), of cell lines (Harasawa, 1996), IFN for human use (Harasawa and Sasaki, 1995) and live virus vaccine for human use (Harasawa, 1994).
The origin of the Belgian type II isolates is not known. The RNA was extracted from organs or leukocytes avoiding a possible contamination of the samples by the cell line or FBS. However, we cannot exclude the introduction of these strains via live vaccine for bovine use.
Epizootical problems due to the circulation of genotype II viruses in Belgium have not yet been encountered. However, these viruses are antigenically different from classical BVDV viruses (Ridpath et al., 1994). The complete sequence of a genotype II virus has been published showing that the weak homology with type I strains extended through the complete genome. The gp53 protein had less than 70% identity with the type I counterpart (Ridpath and Bolin, 1995). Recently, genotype II strains have been isolated in Japan. No cross-neutralization was observed between these type II and type I isolates (Nagai et al., 1998). One should thus take account of these viruses for the development of control programs and vaccines against BVDV.
Acknowledgements
The authors thank Hans Vanderhallen for performing the automated sequencing.
References
- Becher P., König M., Paton D.J., Thiel H.-J. Further characterization of border disease virus isolates: Evidence for the presence of more than three species within the genus pestivirus. Virology. 1995;209:200–206. doi: 10.1006/viro.1995.1243. [DOI] [PubMed] [Google Scholar]
- Becher P., Orlich M., Shannon A.D., Horner G., König M., Thiel H.-J. Phylogenic analysis of pestiviruses from domestic and wild ruminants. J. Gen. Virol. 1997;78:1357–1366. doi: 10.1099/0022-1317-78-6-1357. [DOI] [PubMed] [Google Scholar]
- Broes A., Wellemans G., Dheedene J. Syndrome hémorragique chez des bovins infectés par le virus de la diarrhée bovine (BVD/MD) Ann. Méd. Vét. 1992;137:33–38. [Google Scholar]
- Brownlie, J., 1991. The pathways for bovine virus diarrhoea virus biotypes in the pathogenesis of disease. Arch. Virol. (Suppl 3), 79–96 [DOI] [PubMed]
- Canal C.W., Hotzel I., de Almeida L.L., Roehe P.M., Masuda A. Differentiation of classical swine fever virus from ruminant pestiviruses by reverse transcription and polymerase chain reaction. Vet. Microbiol. 1996;48:373–379. doi: 10.1016/0378-1135(95)00156-5. [DOI] [PubMed] [Google Scholar]
- Carlsson U. Border disease in sheep caused by transmission of virus from cattle persistently infected with BVDV. Vet. Rec. 1991;128:145–147. doi: 10.1136/vr.128.7.145. [DOI] [PubMed] [Google Scholar]
- Chappuis, G., Brun, A., Kato, F., Dauvergne, M., Reynaud G., Duret, C., 1986. Etudes sérologiques et immunologiques réalisées à la suite de l'isolement d'un pestivirus dans un foyer ovina chez des moutons. In: Pestiviroses des ovins et des bovins. J. Espinasse, M. Savey, Société française de buiatrie, pp. 55–66
- Collett M.S., Larson R., Gold C., Strick D., Anderson D.K., Purchio A.F. Molecular cloning and nucleotide sequence of the pestivirus bovine diarrhea virus. Virology. 1988;165:191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- Corapi W.V., French T.W., Dubovi E.J. Severe thrombocytopenia in young calves experimentally infected with noncytopathic bovine diarrhea virus. J. Virol. 1989;63:3934–3943. doi: 10.1128/jvi.63.9.3934-3943.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker A., Wensvoort G., Terpstra C. Six antigenic groups within the genus pestivirus as identified by cross neutralization assays. Vet. Microbiol. 1995;47:317–329. doi: 10.1016/0378-1135(95)00116-6. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S., Moennig V., Wensvoort G. The development of an international reference panel of monoclonal antibodies for the differentiation of hog cholera virus from other pestiviruses. Vet. Microbiol. 1991;29:101–108. doi: 10.1016/0378-1135(91)90118-y. [DOI] [PubMed] [Google Scholar]
- Edwards S., Roehe P.M., Ibata G. Comparative studies of Border disease and closely related virus infections in experimental pigs and sheep. Br. Vet. J. 1995;151:181–187. doi: 10.1016/s0007-1935(95)80008-5. [DOI] [PubMed] [Google Scholar]
- Fernandez A., Hewicker M., Trautwein G., Pohlenz J., Liess B. Viral antigen distribution in the central nervous system of cattle persistently infected with bovine diarrhea virus. Vet. Pathol. 1989;26:26–32. doi: 10.1177/030098588902600105. [DOI] [PubMed] [Google Scholar]
- Harasawa R. Comparative analysis of the 5′non-coding region of pestivirus RNA detected from live virus vaccines. J. Vet. Med. Sci. 1994;56:961–964. doi: 10.1292/jvms.56.961. [DOI] [PubMed] [Google Scholar]
- Harasawa R., Tomiyama T. Evidence of pestivirus RNA in human virus vaccine. J. Clin. Microbiol. 1994;32:1604–1605. doi: 10.1128/jcm.32.6.1604-1605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harasawa R. Adventitious pestivirus RNA in live virus vaccines against bovine and swine diseases. Vaccine. 1995;13:100–103. doi: 10.1016/0264-410x(95)80018-9. [DOI] [PubMed] [Google Scholar]
- Harasawa R., Mizusawa H. Demonstration and genotyping of pestivirus RNA from mammalian cell lines. Microbiol. Immunol. 1995;39:975–985. doi: 10.1111/j.1348-0421.1995.tb03301.x. [DOI] [PubMed] [Google Scholar]
- Harasawa R., Sasaki T. Sequence analysis of the 5′UTR of pestivirus RNA demonstrated in interferons for human use. Biologicals. 1995;23:263–269. doi: 10.1006/biol.1995.0044. [DOI] [PubMed] [Google Scholar]
- Harasawa R. Phylogenetic analysis of pestivirus based on the 5′UTR. Acta Virologica. 1996;40:49–54. [PubMed] [Google Scholar]
- Hewicker M., Wöhrmann T., Fernandez A., Trautwein G., Liess B., Moennig V. Immunohistological detection of bovine viral diarrhoea virus antigen in the central nervous system of persistently infected cattle using monoclonal antibodies. Vet. Microbiol. 1990;23:203–210. doi: 10.1016/0378-1135(90)90150-t. [DOI] [PubMed] [Google Scholar]
- Hewicker-Trautwein M., Trautwein G., Moennig V., Liess B. Infection of ovine fetal brain cell cultures with cytopathogenic and non-cytopathogenic bovine viral diarrhoea virus. Vet. Microbiol. 1992;33:239–248. doi: 10.1016/0378-1135(92)90052-u. [DOI] [PubMed] [Google Scholar]
- Hewicker-Trautwein M., Trautwein G., Frey H.-R., Liess B. Variation in neuropathogenicity in sheep fetuses transplacentally infected with non-cytopathogenic and cytopathogenic biotypes of BVDV. J. Vet. Med. 1995;42:557–567. doi: 10.1111/j.1439-0450.1995.tb00748.x. [DOI] [PubMed] [Google Scholar]
- Hofmann M.A., Brechtbühl K., Stäuber N. Rapid characterization of new pestivirus strains by direct sequencing of PCR-amplified cDNA from the 5′ noncoding region. Arch. Virol. 1994;139:217–229. doi: 10.1007/BF01309467. [DOI] [PubMed] [Google Scholar]
- Horner G.W., Tham K.-M., Orr D., Ralston J., Rowe S., Houghton T. Comparison of an antigen capture enzyme-linked assay with reverse transcription-polymerase chain reaction and cell culture immunoperoxidase tests for the diagnosis of ruminant pestivirus infections. Vet. Microbiol. 1995;43:75–84. doi: 10.1016/0378-1135(94)00080-g. [DOI] [PubMed] [Google Scholar]
- Katz J.B., Ridpath J.F., Bolin S.R. Presumptive diagnostic differentiation of hog cholera virus from bovine viral diarrhea and border disease viruses by using a cDNA nested-amplification approach. J. Clin. Microbiol. 1993;31:565–568. doi: 10.1128/jcm.31.3.565-568.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Lopez O.J., Osorio F.A., Donis R.O. Rapid detection of bovine viral diarrhea virus by polymerase chain reaction. J. Clin. Microbiol. 1991;29:578–582. doi: 10.1128/jcm.29.3.578-582.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M., Sato M., Nagano H., Pang H., Kong X., Murakami T., Ozawa T., Akashi H. Nucleotide sequence homology to BVDV 2 in the untranslated region of BVDVs from cattle with mucosal disease or persistent infection in Japan. Vet. Microbiol. 1998;60:271–276. doi: 10.1016/s0378-1135(98)00158-8. [DOI] [PubMed] [Google Scholar]
- Paton D.J., Simpson V., Done S.H. Infection of pigs and cattle with bovine viral diarrhoea virus on a farm in England. Vet. Rec. 1992;131:185–188. doi: 10.1136/vr.131.9.185. [DOI] [PubMed] [Google Scholar]
- Paton D.J., Sands J.J., Lowings J.P., Smith J.E., Ibata G., Edwards S. A proposed division of the pestivirus genus using monoclonal antibodies, supported by cross-neutralisation assays and genetic sequencing. Vet. Res. 1995;26:92–109. [PubMed] [Google Scholar]
- Pellerin C., Van den Hurk J., Lecomte J., Tijssen P. Identification of a new group of bovine viral diarrhea virus strains associated with severe outbreaks and high mortalities. Virology. 1994;203:260–268. doi: 10.1006/viro.1994.1483. [DOI] [PubMed] [Google Scholar]
- Ridpath J.F., Bolin S.R., Katz J. Comparison of nucleic acid hybridization and nucleic acid amplification using conserved sequences from the 5′ noncoding region for detection of bovine viral diarrhea virus. J. Clin. Microbiol. 1993;31:986–989. doi: 10.1128/jcm.31.4.986-989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridpath J.F., Bolin S.R., Dubovi E.J. Segregation of bovine viral diarrhea virus into genotypes. Virology. 1994;205:66–74. doi: 10.1006/viro.1994.1620. [DOI] [PubMed] [Google Scholar]
- Ridpath J.F., Bolin S.R. The genomic sequence of a virulent bovine viral diarrhea virus (BVDV) from the type 2 genotype: Detection of a large genomic insertion in a noncytopathic BVDV. Virology. 1995;212:39–46. doi: 10.1006/viro.1995.1451. [DOI] [PubMed] [Google Scholar]
- Roehe P.M., Woodward M.J., Edwards S. Characterisation of p20 sequences from a border disease-like pestivirus isolated from pigs. Veterinary Microbiol. 1992;33:231–238. doi: 10.1016/0378-1135(92)90051-t. [DOI] [PubMed] [Google Scholar]
- Sandvik T., Paton D.J., Lowings P.J. Detection and identification of ruminant and porcine pestiviruses by nested amplification of 5′ untranslated cDNA regions. J. Virol. Methods. 1997;64:43–56. doi: 10.1016/s0166-0934(96)02136-2. [DOI] [PubMed] [Google Scholar]
- Sullivan D.G., Akkina R.K. A nested polymerase chain reaction assay to differentiate pestiviruses. Virus Res. 1995;38:231–239. doi: 10.1016/0168-1702(95)00065-x. [DOI] [PubMed] [Google Scholar]
- Tajima M., Kirisawa R., Taguchi M., Iwai H., Kawakami Y., Hagiwara K., Ohtsuka H., Sentsui H., Takahashi K. Attempt to discriminate between bovine viral-diarrhea virus strains using polymerase chain reaction. J. Vet. Med. 1995;42:257–265. doi: 10.1111/j.1439-0450.1995.tb00709.x. [DOI] [PubMed] [Google Scholar]
- van Rijn P.A., van Gennip H.G.P., Leendertse C.H., Bruschke C.J.M., Paton D.J., Moormann R.J.M., van Oirschot J.T. Subdivision of the pestivirus genus based on envelope glycoprotein E2. Virology. 1997;237:337–348. doi: 10.1006/viro.1997.8792. [DOI] [PubMed] [Google Scholar]
- Vilcek S., Belak S. Genetic identification of pestivirus strain Frijters as a border disease virus from pigs. J. Virol. Methods. 1996;60:103–108. doi: 10.1016/0166-0934(96)02031-9. [DOI] [PubMed] [Google Scholar]
- Vilcek S., Herring A.J., Herring J.A., Nettleton P.F., Lowings J.P., Paton D.J. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch. Virol. 1994;136:309–323. doi: 10.1007/BF01321060. [DOI] [PubMed] [Google Scholar]
- Vilcek S., Nettleton P.F., Paton D.J., Belak S. Molecular characterization of ovine pestiviruses. J. Gen. Virol. 1997;78:725–735. doi: 10.1099/0022-1317-78-4-725. [DOI] [PubMed] [Google Scholar]
- Wöhrmann T., Hewicker-trautwein M., Fernandez A., Moennig V., Liess B., Trautwein G. Distribution of bovine diarrhea viral antigens in the central nervous system of cattle with various congenital manifestations. J. Vet. Med. 1992;39:599–609. doi: 10.1111/j.1439-0450.1992.tb01211.x. [DOI] [PubMed] [Google Scholar]
- Wirz B., Tratschin J.-D., Müller H.K., Mitchell D.B. Detection of hog cholera virus and differentiation from other pestiviruses by polymerase chain reaction. J. Clin. Microbiol. 1993;31:1148–1154. doi: 10.1128/jcm.31.5.1148-1154.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]


