Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is the causative agent of a contagious disease characterized by reproductive failure in sows and respiratory disease in piglets. This infectious disease results in significant losses in the swine industry and specific anti-PRRSV drugs are needed. In this study, we evaluated a novel class of antisense compounds, peptide-conjugated phosphorodiamidate morpholino oligomers (P-PMOs), for their ability to suppress PRRSV replication in cell culture. P-PMOs are analogs of single-stranded DNA and contain a modified backbone that confers highly specific binding to RNA and resistance to nucleases. Of six P-PMOs tested, one (‘5UP1’), with sequence complementary to the 5′-terminal 21 nucleotides of the PRRSV genome, was found to be highly effective at reducing PRRSV replication in a specific and dose-dependent manner in CRL11171 cells in culture. 5UP1 treatment generated up to a 4.5 log reduction in infectious PRRSV yield, while a control P-PMO had no effect on viral titer. Immunofluorescence assay with an anti-PRRSV monoclonal antibody confirmed the titer observations. The sequence-specificity of 5UP1 effect was confirmed in part by a cell-free luciferase reporter assay system, which showed that 5UP1-mediated inhibition of translation decreased if the target-RNA contained mispairings in relation to the 5UP1 P-PMO. Real-time RT-PCR showed that the production of PRRSV negative-sense RNA was reduced if 5UP1 was added to cells at up to 6 h post-virus inoculation. Cell viability assays detected no cytotoxicity of 5UP1 within the concentration-range of this study. These results indicate that P-PMO 5UP1 has potential as an anti-PRRSV agent.
Keywords: Porcine reproductive and respiratory syndrome virus, PRRSV, Morpholino, Antisense, Antiviral
1. Introduction
Porcine reproductive and respiratory syndrome (PRRS) is a contagious viral disease that has been causing heavy economic losses to the swine industry, even though vaccines have been commercially available for years. The causative agent, PRRSV, is an enveloped, single-stranded RNA virus in the family Arteriviridae, which includes equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV) of mice and simian hemorrhagic fever virus (SHFV) (Meulenberg et al., 1994). Arteriviruses have a genome organization and replication process similar to that of coronaviruses, and feature a positive-sense RNA genome of about 15 kb, with the coding sequence for non-structural proteins preceding that of structural proteins.
Eight open reading frames (ORFs) have been identified in the PRRSV genome. ORFs 1a and 1b comprise 80% of the viral genome sequence and encode the viral RNA replicase (Meulenberg et al., 1993). In PRRSV-infected cells, a set of six or seven nested viral subgenomic RNAs is formed (Conzelmann et al., 1993, Meng et al., 1994, Meng et al., 1996b, Meulenberg et al., 1993). All of the subgenomic RNAs have an identical 5′-leader sequence derived from the 5′-end of genomic RNA, as well as an identical 3′-terminal sequence preceding poly-(A) tails of variable length. The co-terminally identical mRNA species are generated through a process known as discontinuous subgenomic mRNA synthesis (Sawicki and Sawicki, 1995, van Marle et al., 1999). From the analysis of PRRSV genomic sequences, two genotypes of PRRSV are recognized, North American and European, which are 55–70% identical at the nucleotide level (Meng et al., 1994). Each genotype in turn contains a number of viral strains. The 5′-end of each subgenomic mRNA of North American strains contains a common leader sequence of 190 bases (Nelsen et al., 1999), whereas in the prototype strain of the European PRRSV genotype, Lelystad virus, the leader is 221 bases (Meulenberg et al., 1993). The North American and European genotypes have 3′-terminal untranslated region sequences of about 150 and 120 bases, respectively, excluding the poly-(A) tail, which is common to all of their subgenomic mRNAs. PRRSV subgenomic RNAs are polycistronic in structure, but it is believed that only the first open reading frame (ORF) of each is translated into a viral protein (Meng et al., 1996b). PRRSV can be propagated in MA-104, African green monkey kidney cells, and cells derived thereof, such as CL2621, MARC 145, and CRL11171 (Benfield et al., 1992, Horsfield et al., 2002, Kim et al., 1993, Meng et al., 1994).
Recent strategies to control PRRSV infection have not been successful. Attenuated live virus vaccine has been commercially available for years, however, outbreaks of PRRS resulting from viral strains nearly identical in sequence to the vaccine strain have been reported (Opriessnig et al., 2002). Outbreaks of atypical or acute PRRS in vaccinated pigs have raised serious concern about the efficacy of the current vaccines (Meng, 2000), and made clear the need to develop alternative strategies. Specific and efficacious antiviral compounds would be considered a welcome complement to vaccines in PRRS control. In this study, peptide-conjugated phosphorodiamidate morpholino oligomers (P-PMOs) are evaluated for their ability to block PRRSV propagation in cell culture.
Phosphorodiamidate morpholino oligomers (PMOs) are structurally similar to single-stranded DNA oligonucleotides in that they are approximately 20 bases in length and contain the same purine and pyrimidine bases as DNA. PMOs have a different backbone than DNA, however, with a morpholine ring replacing the deoxyribose sugar and a phosphorodiamidate linkage replacing the phosphodiester linkage (Fig. 1 ) (Schmajuk et al., 1999, Summerton, 1999, Summerton and Weller, 1997). PMOs are uncharged, water-soluble, and highly resistant to nuclease degradation (Hudziak et al., 1996, Nelson et al., 2005). PMOs bind to target mRNA by Watson–Crick base pairing and can exert an antisense effect by preventing access to critical segments of RNA sequence, such as a translation initiation site, through steric blockade. This is a distinctly different process from the RNase H-dependent mechanism induced by the often-used antisense structural type phosphorothioate DNA (Summerton, 1999).
Fig. 1.
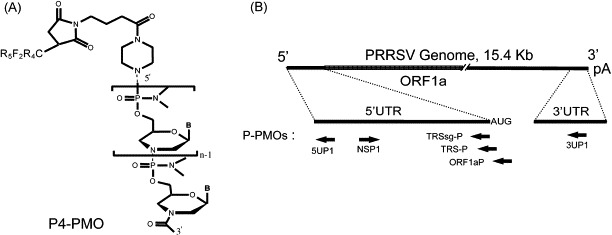
Structure of P-PMO and schematic target locations of P-PMO in the PRRSV genome. (A) P-PMO structure. A morpholine ring and a phosphorodiamidate linkage in PMO replace the deoxyribose and phosphodiester linkage, respectively, of DNA. “B” represents the bases A, G, C, or T. The peptide R5F2R4, designated P4, was covalently conjugated to the 5′-end of each PMO. (B) Positions of P-PMO target sites in PRRSV genomic RNA. The AUG translation initiation codon of ORF1a is shown after the 5′-UTR line. The arrows indicate the 5′–3′-orientation of the PMOs in relation to the PRRSV genome.
A number of studies have shown that PMOs can effectively and specifically block translation of target mRNA in vitro and in vivo (Arora et al., 2002a, Arora et al., 2002b, Brent and Drapeau, 2002, Hudziak et al., 2000, Qin et al., 2000, Stein et al., 2001, Taylor et al., 1996). It has been shown that PMOs conjugated to short arginine-rich peptides (P-PMO) have a significantly higher efficiency of delivery into culture cells than do non-conjugated PMOs (Moulton et al., 2003, Moulton and Moulton, 2004, Nelson et al., 2005). The sequence-specific antiviral efficacy of PMO compounds in vitro has been documented with caliciviruses (Stein et al., 2001), Hepatitis C virus RNA (McCaffrey et al., 2003), mouse hepatitis virus (Neuman et al., 2004), SARS coronavirus (Neuman et al., 2005), EAV (van den Born et al., 2005) and several flaviviruses (Deas et al., 2005, Kinney et al., 2005). The experiments described in our report demonstrate that a P-PMO designed to target the 5′-terminal region of the PRRSV genome can inhibit PRRSV replication in cell culture in a sequence-specific and dose-dependent manner.
2. Materials and methods
2.1. Cells and viruses
Cell line ATCC CRL11171 (Meng et al., 1996a) was maintained in DMEM medium supplemented with 10% fetal bovine serum. Cell numbers were counted with a hemocytometer and cell viability was assessed by trypan blue exclusion. PRRSV strains ATCC VR2385 (Meng et al., 1996a), Lelystad (Meulenberg et al., 1993) and NVSL (NVSL, Ames, IA) were used to inoculate CRL11171 cells at 0.5 multiplicity of infection (MOI) for PMO testing. For virus titration, 10-fold dilutions of virus were added to monolayer CRL11171 cells in a 96-well plate. The degree of cytopathic effect (CPE), characterized by cell rounding, clumping and detachment, was assessed microscopically 48 h after PRRSV inoculation, in comparison with mock-infected cells. Tissue culture infectious dose (TCID50) per milliliter was calculated based upon CPE development, according to the method of Reed and Muench, as described previously (Meng et al., 1996a).
2.2. Immunofluorescence assay (IFA)
IFA was carried out as reported previously (Zhang et al., 1998) with a nucleocapsid (N)-specific monoclonal antibody SDOW17 (Nelson et al., 1993). Specific reactions between SDOW17 and N-protein were detected with goat anti-mouse IgG-fluorescein isothiocyanate (FITC) conjugate (Sigma, St. Louis, MO) and assessed with fluorescence microscopy.
2.3. P-PMOs
Six P-PMOs complementary to specific PRRSV RNA sequences were designed. The P-PMOs used in this study and their target site locations in PRRSV are summarized in Table 1 and depicted in Fig. 1. PMOs were synthesized at AVI BioPharma Inc. (Corvallis, OR) by methods previously described (Summerton and Weller, 1997). The peptide NH2-RRRRRFFRRRRC-CONH2 (designated R5F2R4) was covalently conjugated to the 5′-end of each PMO (Fig. 1). The conjugation, purification, and analysis of R5F2R4-PMO (P4-PMO) compounds (referred to simply as ‘P-PMO’ in this report) were similar to methods described elsewhere (Moulton et al., 2004). A random sequence P-PMO (named ‘CP1’), having little agreement with PRRSV or primate mRNA sequences, was also utilized to control for non-sequence-specific activity of the P4-PMO chemistry.
Table 1.
| P-PMO name | P-PMO sequence (5′–3′) | Nucleotide position of P-PMO target in PPRSV RNA, and P-PMO orientation | P-PMO target region in PRRSV |
|---|---|---|---|
| 5UP1 | CATAGAGCCAACACCTATACG | 3–23, antisense | 5′-terminus of genomic RNA |
| NSP1 | CCCAAAAACTTGCTGCACGG | 66–85, sense | 3′-end-region of negative-sense (antigenome) RNA |
| TRS-Pc,d | GACATGGTTAAAGGGGTGGAG | 173–193, antisense | TRS-region and 5′-end of ORF1a |
| TRSsg-Pd | GGTTAAAGGGGTGGAGAGACCG | 167–188, antisense | TRS and TRS 5′-flanking sequence |
| ORF1aPc | CCGATCAAGTATCCCAGACAT | 189–209, antisense | Translation initiation region of ORF1a |
| 3UP1 | GTCGCCCTAATTGAATAGGTG | 15032–15052, antisense | 3′-end-region of genomic RNA |
| CP1 | GATATACACAACACCCAATT | None | Random sequence negative control |
The peptide R5F2R4 is conjugated to the 5′-end of all the P-PMOs in this study.
P-PMO design was based on PRRSV VR2385 genomic RNA.
The underlined nucleotides correspond to the AUG translation initiation codon of PRRSV ORF1a.
The italicized nucleotides correspond to the ‘transcription regulatory sequence’.
2.4. P-PMO treatment of cells
CRL1171 cells were seeded in 12-well plates at 5 × 105 cells per well and grown overnight. For experiments that employed virus inoculation before P-PMO treatment, VR2385 virus stock was diluted and the cells were inoculated at 0.5 MOI. After 2 h incubation at 37 °C, the virus inoculum was removed. P-PMOs were diluted in DMEM and added to the cell monolayers. P-PMO CP1 was included as a negative control, and DMEM without P-PMO was included as a mock treatment control. After gentle shaking, the cells were incubated for 4 h at 37 °C. The PMO solution was removed and maintenance medium (DMEM supplemented with 2% FBS) was then added. The cells were then incubated for 48 h at 37 °C, after which cell culture supernatants and cells were harvested for further analysis.
For experiments that employed virus inoculation post P-PMO treatment, cell monolayers were first treated with P-PMOs in the same manner as described above, for 4 h at 37 °C. The P-PMO solution was then removed, and cells were inoculated with virus at MOI of 0.5. After 2 h incubation at 37 °C, the virus inoculum was removed and maintenance medium was added. Cell culture supernatants and cells were harvested for further analysis at 48 h post-inoculation.
2.5. Plasmid construction and cell-free luciferase reporter assay
Complementary DNA oligonucleotides were synthesized so as to include the first 26 nucleotides (nt) of the 5′-untranslated region (UTR) of PRRSV VR2385 genome sequence (CTAGA CGTAT AGGTG TTGGC TCTAT G). The oligomers were subcloned upstream of a luciferase coding sequence in a T7 promoter-containing reporter plasmid, as described previously (Neuman et al., 2004). The ‘pre-luciferase’ leader sequence included the 21 nt target sequence of 5UP1 P-PMO. Five variants of the 5UP1 P-PMO target sequence, containing one to four mismatches, were also synthesized and subcloned, as above, into the luciferase reporter vector. DNA sequencing was conducted to confirm all ‘leader’ sequences in this series of six plasmids. Each plasmid DNA was linearized with NotI downstream of the PRRSV sequence-luciferase fusion region, and in vitro transcription was conducted with the MegaScript T7 Kit following the manufacturer's instructions (Ambion, Austin, TX). In vitro translations were carried out using the transcribed RNA with nuclease-treated rabbit reticulocyte lysate (Promega, Madison, WI). Each translation reaction was mixed with luciferase assay reagent (Promega) according to the manufacturer's instructions, and data was collected using an FL×800 microplate luminometer (Bio-Tek Instruments Inc., Winooski, VT). The relative light units produced by each reaction were normalized to the mean of all control reactions and expressed as percent inhibition of luciferase translation.
2.6. RNA isolation and real-time RT-PCR
Total RNA was isolated from PRRSV-infected CRL11171 cells using Tri-Zol reagent following the manufacturer's instructions (Invitrogen, Carlsbad, CA). For quantitative RT-PCR analysis, the RNA was treated with RNase-free DNase to remove DNA carryover during RNA isolation, and then reverse transcribed. For detection of negative-sense RNA, reverse transcription was initiated by a forward primer, 5UF1 (5′-GACGT ATAGG TGTTG GCTC-3′), which consisted of a sequence identical to the 5′-terminal region of PRRSV genomic RNA. PRRSV genomic RNA isolated from virions was quantified and used to generate standard curves for the real-time RT-PCR. Real-time PCR was conducted on an iCycler system (Bio-Rad Laboratories, Hercules, CA) with cDNAs generated from negative-sense RNA, a primer set (5URR-F1, 5′-GTCTG TCCCT AGCAC CTTG-3′ and 1aRR-R1, 5′-GCCCT CCGCC ATAAA CAC-3′) designed to amplify sequence in the 3′-end of the full-length negative-sense PRRSV antigenomic RNA, and iQ SYBR Green Supermix (Bio-Rad). Transcripts of β-actin were also amplified from the samples in order to assure normalized quantitative RT-PCR detection of PRRSV RNA from the cells.
2.7. Cell viability assay
The viability of CRL11171 cells after P-PMO treatment was determined with CellTiter-Blue (Promega) following the manufacturer's instructions. Briefly, CRL11171 cells were treated with 5UP1 or CP1 under conditions identical to those described above in “P-PMO treatment of cells”. Mock-treated cells were included for comparison. CellTiter-Blue reagent was added and incubated for 1 h at 37 °C. The fluorescence signal was measured with a Synergy HT Multi-Detection Microplate Reader (Bio-Tek). In this assay, fluorescence intensity reflects the number of viable cells. Relative percentages of fluorescence intensity were calculated by comparison to mock-treated controls.
3. Results
3.1. Design of anti-PRRSV P-PMO compounds
Parallel sequence alignments of each of the two major genotypes of PRRSV show that parts of the 5′- and 3′-UTRs, and the 5′-region of ORF1a, within each genotype, are highly conserved, and likely have essential functions in molecular events of the viral life cycle (Tan et al., 2001, Verheije et al., 2002), making them rational targets for antisense PMO design. For this study, P-PMOs were designed against the sequence of PRRSV VR2385, a virulent strain of the North American genotype.
P-PMO target sites were selected from the common 5′-leader and 3′-terminal sequences found in PRRSV genomic and subgenomic RNAs, the transcription regulatory sequence (TRS) region, the translation initiation region of ORF1a, and 3′-end of negative-sense anti-genomic RNA (Fig. 1). Six P-PMOs were synthesized; their exact base-sequences and PRRSV RNA target locations are listed in Table 1. P-PMO 5UP1 was designed to target the 5′-terminal 21 nt of the PRRSV genome, in an attempt to block translation. 3UP1 was designed to target the genomic 3′-end-region, in an attempt to interfere with RNA synthesis. Two P-PMOs were designed against the TRS-region, in an effort to interfere with PRRSV subgenomic RNA synthesis and/or translation. TRSsg-P is complementary to sequence immediately upstream of, and including, the TRS. The target of TRSsg-P is present in the leader of genomic and all subgenomic RNAs (Meng et al., 1994, Nelsen et al., 1999). The target of TRS-P includes the TRS and the first 5 nt of ORF1a. NSP1 is complementary to the sequence in the 3′-end of negative-sense PRRSV RNA, and was designed with the intention of interfering with the synthesis of positive-sense PRRSV RNA. ORF1aP is complementary to the sequence at the 5′-end of ORF1a, and was designed to inhibit translation of ORF1a, which encodes the proteins of the PRRSV replicase complex, including the RNA-dependent RNA polymerase.
3.2. Initial evaluation of P-PMO compounds against PRRSV
A virulent PRRSV strain, VR2385, was used to evaluate the ability of the various P-PMOs to inhibit PRRSV replication in CRL11171 cells. At the MOI used in this study, VR2385 reached peak replication in CRL11171 cells at around 48 h post-inoculation (pi). In initial experiments, P-PMOs were applied to cell monolayers at a final concentration of 16 μM in DMEM and incubated for 4 h immediately before the virus inoculation period. The cells were then observed daily for CPE development, and supernatants were collected 2 days pi for virus titration in CRL11171 cells to determine PRRSV levels.
Of the six P-PMOs tested, only 5UP1 was found to be highly effective at inhibiting PRRSV production. Cells treated with 5UP1 showed far less CPE development than cells treated with CP1 or mock-treated (Fig. 2A). Cells treated with NSP1 exhibited somewhat less severe CPE than the control. The other four P-PMOs did not have an observable effect on CPE development. Titrations of PRRSV from cell culture supernatants showed that treatment with 5UP1 reduced virus yield 214-fold compared to mock treatment (Fig. 2B), while treatments with other P-PMOs generated titers similar to CP1 or mock treatment, confirming the above CPE observations. Further confirmation of these CPE observations was obtained by conducting IFA on CRL11171 cells after P-PMO treatment and subsequent virus inoculation. IFA with a monoclonal antibody against the PRRSV N-protein showed that treatment with 5UP1 led to far fewer fluorescent-positive cells than treatment with CP1 (Fig. 2C), indicating that 5UP1 interfered with PRRSV propagation. There were a few fluorescent foci in the wells treated with 16 μM 5UP1, but only a few individual positive cells in the wells treated with 32 μM 5UP1. CP1 treatment resulted in nearly all cells supporting PRRSV replication, which was similar to mock treatment control.
Fig. 2.

P-PMO 5UP1-mediated inhibition of PRRSV replication in infected CRL11171 cells. (A) Cytopathic effect (CPE) is clearly visible at 48 h post PRRSV (image with label of ‘PRRSV VR2385, no PMO’) infection, while uninfected control cells (image with label of ‘CRL11171, no virus’) retain an intact monolayer. Cells treated with 16 μM 5UP1 for 4 h immediately before virus inoculation display reduced CPE development (image with label of ‘5UP1, VR2385’), while treatment with CP1 or other P-PMOs did not result in reduced CPE under identical conditions. (B) Virus yield titration shown as log TCID50/ml from similar samples as pictured in (A). The experiment was repeated three times and error bars are shown. (C) Immunofluorescence assay with a monoclonal antibody against PRRSV shows green fluorescence, indicating detection of PRRSV nucleocapsid protein in infected cells. P-PMO 5UP1 suppressed PRRSV replication (5UP1 16 and 32 μM), while CP1 P-PMO had no effect.
P-PMO treatment was also conducted post-virus inoculation, and a similar inhibitory effect on CPE development was observed. To avoid the possibility that 5UP1 P-PMO was affecting virus entry, cells underwent a virus-adsorption period before the beginning of P-PMO treatment in the remaining experiments, unless otherwise specified.
3.3. Dose-responsive inhibition of viral replication by 5UP1 P-PMO
Having found 5UP1 to be an effective inhibitor of PRRSV replication, we further characterized this P-PMO through a dose–response challenge. CRL11171 cells were inoculated with VR2385 and, after inoculum removal, 5UP1 P-PMO was added to the cells in incremental concentrations of 2, 4, 8, 16, and 32 μM. Cell culture supernatants were harvested 48 h later and titrated for PRRSV yield. PRRSV titer was reduced concomitantly with increasing 5UP1 concentration (Fig. 3 ), demonstrating that 5UP1 was effective in a dose–responsive manner. CP1 did not have any effect on virus replication in comparison with the mock-treatment control, indicating that the antiviral action of 5UP1 was a sequence-specific effect.
Fig. 3.
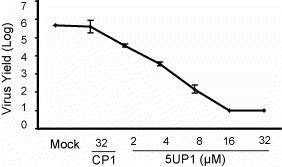
P-PMO 5UP1 reduces virus yield in cell culture in a dose-responsive manner, as measured by log TCID50/ml. Cells treated with 16 or 32 μM 5UP1 for 4 h post-virus inoculation had virus yields below the level of detection (approximately 1 log) of this assay. This experiment was repeated three times and the error bars indicate the standard deviations of those replicates.
We next tested the effect of the duration of P-PMO treatment on viral propagation. Cell monolayers were inoculated with virus for 2 h, and then treated with 16 μM 5UP1 for 0.5, 1, 2 or 4 h, after which maintenance medium replaced the P-PMO solution. At 48 h post-inoculation, cell culture supernatants were harvested for virus titration. Virus yields from VR2385-inoculated cells were 1.5-, 22-, 22- and 4677-fold less for cells treated with P-PMO for 0.5, 1, 2 and 4 h, respectively, than mock-treatment controls. Extension of 5UP1 treatment time past 4 h did not lead to a further reduction of VR2385 yield. The experiment was repeated three times, and the same pattern of virus yield reduction was observed each time. The incremental increase in P-PMO treatment times in this experiment resulted in a corresponding incremental reduction in viral titer, indicating 5UP1 uptake was time-dependent. The results showed that a 4 h incubation was required to achieve a high reduction in PRRSV replication. Consequently, for the remainder of the experiments reported herein, a post-inoculation P-PMO treatment period of 4 h was employed, unless otherwise specified.
To appraise the persistence of active P-PMO in CRL11171 cells over time, VR2385 inoculum was added to cells and incubated for 2 h at 24, 48 or 72 h after the commencement of a 4 h P-PMO treatment. Noticeably less CPE developed in cells receiving virus inoculation at 24 h after initiation of P-PMO treatment than in cells that were not inoculated until 72 h after P-PMO treatment began. Virus titrations showed that virus yields for samples from inoculations at 24, 48, and 72 h post P-PMO treatment-commencement were 35-, 5- and 0.5-fold less, respectively, than mock-treatment controls. This result suggests that the effect of P-PMO is diluted as cell metabolism and growth proceed, and that periodic addition of fresh P-PMO is likely required to maintain high antiviral activity.
To test whether 5UP1 could effectively inhibit a preexisting PRRSV infection, we commenced P-PMO treatment at 2, 4, 8, 10, and 24 h pi, and then fixed the cells at 48 h pi for further analysis. The application of 5UP1 led to reduced PRRSV propagation for all time points compared to controls, as evidenced by IFA analysis showing fewer fluorescent-positive cells (data not shown). In this time-course, the number of PRRSV-positive cells increased in concert with the delay in initiation of 5UP1 treatment. The result demonstrates that P-PMO treatment commencing at any time earlier than 24 h in the PRRSV infection cycle was effective at significantly reducing virus amplification to some degree, but the earlier the better.
3.4. Sequence specificity of 5UP1 inhibition
Having shown that 5UP1 inhibits VR2385 replication in a dose-responsive manner, we further scrutinized the sequence specificity of 5UP1 with a cell-free reporter assay system. A P-PMO binding to its RNA target in translation assays should result in translation inhibition of downstream luciferase coding sequence. Incremental concentrations of P-PMO were added to in vitro translation reactions to determine the degree of the translation inhibition. Extinction concentration 50 [EC50], which refers to the amount of PMO compound required to reduce luciferase signal by 50% compared to untreated controls, was obtained. The EC50s of 5UP1 P-PMO for RNA targets with two-, three-, or four-mismatches were 12-, 58-, and 167-fold higher, respectively, compared to the wild-type (e.g. zero mismatch) target (EC50 of 87, 404, 1170 and 7 nM, respectively) (Fig. 4 ). This result demonstrated that 5UP1 exhibited less inhibition when RNA targets contained more mismatches. Among the two RNA target sequences containing one mismatch, 5UP1 was nearly two-fold less active against the target that contained a mismatch on its 5′-side, compared to a mismatch on the 3′-side (EC50 of 21 and 12 nM, respectively) (Fig. 4B). This series of reporter translation assays further confirmed the sequence-specific inhibition of target-RNA translation by 5UP1, and showed that three or four mispairings between a P-PMO and its RNA target markedly reduces duplexing.
Fig. 4.
In vitro translations with luciferase reporter assay to assess P-PMO 5UP1's effect on variable target sequences. (A) DNA versions of the complementary target sequence of 5UP1 and five variants of the target sequence with one to four mismatches (MM) each were cloned upstream of luciferase in reporter plasmids. RNA was in vitro transcribed and in vitro translation reactions were conducted to measure luciferase translation by the six RNAs in the presence of various concentrations of P-PMO 5UP1. EC50s are shown and represent the effective concentration of a compound resulting in a 50% reduction in luciferase signal compared with mock treatment controls. (B) Relative percentages of inhibition were calculated in comparison with mock treatment controls. Note the decreasing level of inhibition of luciferase production caused by P-PMO 5UP1 against RNAs with which it has an increasing number of mispairs.
Sequence alignment analysis shows that PRRSV strains of the North American genotype (to which VR2385, used in the above experiments, belongs) exhibit high sequence conservation in the 5′-terminal region of the genome. Strains of the European PRRSV genotype, however, differ from the North American strains in this region. The Lelystad strain, a prototypic European PRRSV, and NVSL strain, a second North American PRRSV, were selected to test 5UP1's comparative effect on viral replication in cell culture. CRL11171 cells were inoculated with Lelystad and NVSL strains, and P-PMO treatment was then conducted for 4 h at 37 °C. As expected, treatment with 16 μM 5UP1 did not have an inhibitory impact on replication of the Lelystad strain, but did on NVSL, in a manner similar to that on VR2385 (data not shown). This result further confirms 5UP1's sequence-specific inhibition of PRRSV propagation.
3.5. Effect of 5UP1 post-infection treatment timing on PRRSV RNA synthesis
P-PMO 5UP1 was designed to be complementary to the sequence of the 5′-terminal region of the PRRSV genome. This region is known to be critical in the pre-initiation of translation (van den Born et al., 2004). We suspected that this P-PMO exerted its antiviral effect by interfering with the translation of the product of the first two open reading frames, the PRRSV replicase complex. We expected that as a result of lowered levels of replicase proteins, PRRSV RNA synthesis would ultimately be reduced. To investigate this hypothesis, we commenced P-PMO treatment at multiple time points pi, and harvested the cells at 24 h pi for RNA isolation. Strand-specific real-time RT-PCR was performed to measure the levels of PRRSV negative-sense RNA (an indicator of PRRSV RNA synthesis). The PCR results showed that application of 16 μM 5UP1 at 2, 4, and 6 h pi led to a significant reduction of negative-sense PRRSV RNA in comparison with 16 μM CP1 and mock treatments (P < 0.001) (Fig. 5 ). Application of 5UP1 at 10 h pi did not result in any differences when compared to CP1 or mock treatments. The results indicate that 5UP1 treatment led to a decrease in PRRSV minus-strand RNA synthesis when treatment commenced before or at 6 h pi.
Fig. 5.
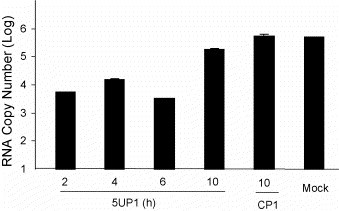
Real-time RT-PCR to assess negative-sense PRRSV RNA level. P-PMOs 5UP1 or CP1 were applied to CRL11171 cells at various time points post-virus inoculation (pi) and the cells then harvested at 24 h pi. Total RNA was isolated and strand-specific real-time RT-PCR was performed as described in Section 2.
3.6. Cell viability assay
Cell viability assays were conducted in order to assess the cytotoxicity of P-PMO 5UP1 and exclude the possibility that a reduction in virus yield was due to 5UP1-induced reduction in cell viability. CRL11171 cells were treated with 5UP1 or CP1 at 16 μM under conditions identical to the dose–response experiment above. The experiment was repeated twice, each with four replicates. The average of the relative effect, when compared to controls, was 103% for cells treated with 16 μM 5UP1, and 103% for cells treated with 16 μM CP1. The percentage of viable cells was nearly identical between 5UP1-, CP1-, and mock-treated cells, demonstrating that the P-PMOs were not cytotoxic at 16 μM under these conditions.
4. Discussion
Most of the P-PMOs in this study were designed to target highly conserved sequences in the 5′- and 3′-UTRs of the genomic and subgenomic RNAs of the North American PRRSV genotype. Of a total of six antisense P-PMOs tested, only one, 5UP1, was found to be highly effective at inhibiting PRRSV replication in CRL11171 cell culture. Virus titration and IFA results demonstrated that the 5UP1 P-PMO was effective in a dose-responsive and sequence-specific manner. The reasons for the ineffectiveness of the other five P-PMOs are unclear, but could include the inability of certain P-PMO to access complementary PRRSV target sequence, or, that successful P-PMO/target-RNA duplexing did not ultimately affect PRRSV replication. The former speculation is supported by secondary structural analysis with RNA folding programs, as extensive stem-loop structures in the PRRSV 5′-UTR are predicted (van den Born et al., 2004). The latter speculation is suggested if successful PMO duplexing to particular PRRSV target-RNA sites simply could not cause significant interference with critical events in the viral life cycle, such as translation or RNA synthesis. Antisense P-PMOs against EAV (a closely related arterivirus) (van den Born et al., 2005) and SARS (a fellow Nidovirus) (Neuman et al., 2005), which were designed to target their respective TRS-regions, showed substantial antiviral activity. In our study, the two P-PMOs targeting the PRRSV TRS-region were ineffective, indicating that these particular compounds were unable to significantly interfere with the processes of PRRSV RNA synthesis or translation. van den Born et al. also found that several other antisense P-PMOs, each targeting a different location in the 5′-UTR of EAV, had high antiviral activity in cell culture (van den Born et al., 2005). In our study, only the 5′-terminus targeted 5UP1 P-PMO showed antiviral activity, and further work was conducted to characterize this particular P-PMO.
To determine whether 5UP1 P-PMO was capable of acting as an inhibitor of translation, and also to characterize the effect of mispairs between 5UP1 and its target-RNA, a cell-free luciferase reporter assay study employing in vitro transcription/translation was designed. When the 5UP1 RNA target-sequence was modified to contain one- to four-base mismatches in relation to 5UP1 P-PMO, incrementally less inhibitory effect by 5UP1 P-PMO was evident, thus providing a demonstration that this P-PMO could interfere with the process of translation, and that the inhibition exerted by this compound was sequence-specific. The result clearly indicates that a reduction in the level of agreement between a P-PMO and its RNA target-sequence diminishes their hybridization affinity.
Treatment of PRRSV-infected CRL11171 cells with 16 μM 5UP1 generated low, but detectable, virus yield. This could be due to some cells having little or no uptake of 5UP1, or that an insufficient quantity of 5UP1 made its way to the subcellular locations where PRRSV replication occurs. This speculation may provide a potential explanation as to why higher P-PMO concentrations were required in cultured cells than those needed in a cell-free reporter assay to achieve an equivalent level of inhibition of target-RNA expression.
In our initial evaluation of the P-PMOs, we employed a ‘pre-treatment’ protocol, i.e., the cells were treated with P-PMO compounds before virus inoculation. The results indicated that 5UP1 P-PMO was effective at reducing PRRSV replication. In the remainder of the experiments, a post-treatment procedure was performed. Although one might expect a higher PRSSV titer reduction when cells are ‘pre-loaded’ with P-PMO, in comparison to post-treated cells, we did not find that to be the case. Several experiments showed that there was a stronger PRRSV titer reduction in post-treated cells (of about 4.5 logs), in comparison to pretreated (about 2 logs). We speculate that the peptide portion of the P-PMO compound could assist virus entry into pretreated cells, as was likewise suggested by van den Born et al. in their P-PMO experiments with EAV (van den Born et al., 2005). More virus particles from the inoculum may adhere to the surface of cells pretreated with P-PMO than to the surface of non-pretreated cells. Therefore, cells pretreated with this P-PMO may engender more virus entry and replication than cells post-treated. van den Born et al. also noticed that high concentrations of certain P-PMO appeared sometimes to be associated with increased virus titers (van den Born et al., 2005).
This study, in cultured CRL11171 cells, showed that if P-PMO treatment commenced at 2 h pi, at least 4 h of subsequent incubation with 16 μM 5UP1 was necessary to significantly lower VR2385 replication when compared to controls. Furthermore, substantially less inhibition by 5UP1 was observed if P-PMO-treatment was begun after 24 h pi than was the case at earlier commencement times. This observation is consistent with the expected mechanism of action of 5UP1, i.e., blocking the translation of RNA into protein. IFA detection of PRRSV N-protein indicated that addition of 5UP1 at up to 24 h pi resulted in some reduction of PRRSV-positive cells at 48 h pi. The reduced N-protein synthesis in IFA observations may be due to 5UP1 interfering with either the synthesis or translation of N subgenomic RNA. The latter speculation seems more likely. N subgenomic RNA possesses the 5′-leader sequence common to all PRRSV subgenomic RNAs, which includes the 5UP1 target site and has been implicated in the events of the pre-initiation of translation in arteriviruses (van den Born et al., 2004).
Real-time RT-PCR of samples taken at 24 h pi showed that application of 5UP1 at or before 6 h pi led to a reduction of viral negative-sense RNA production, which suggest that 5UP1 can suppress translation of PRRSV replicase. Addition of 5UP1 at 10 h pi failed to reduce the level of negative-sense viral RNA, which suggests that by this time point PRRSV replicase production had reached a level that was sufficient to maintain PRRSV RNA synthesis.
A cell viability assay of CRL11171 cells treated with 5UP1, at concentrations up to and including 16 μM, detected no cytotoxicity, and indicates that suppression of PRRSV replication was due to the 5UP1-specific inhibition of PRRSV molecular events. The absence of 5UP1-induced cytotoxicity at effective antiviral concentrations is an important characteristic of this compound for potential in vivo applications.
The prevalence of PRRSV infection in swine herds is high, and current strategies to control infection, including vaccines, are less than adequately effective (Meng, 2000, Opriessnig et al., 2002). Specific anti-PRRSV drugs are needed to complement other strategies in PRRSV prevention and control. Considering the feasibility of P-PMO synthesis, further investigation into the pharmacokinetic, toxicological and antiviral properties of 5UP1 P-PMO in vivo is desirable.
Acknowledgements
The authors wish to express their gratitude to the Chemistry Dept. at AVI Biopharma for the expert production and quality control of all PMO compounds used in this study.
References
- Arora V., Cate M.L., Ghosh C., Iversen P.L. Phosphorodiamidate morpholino antisense oligomers inhibit expression of human cytochrome P450 3A4 and alter selected drug metabolism. Drug Metabol. Dispos. 2002;30:757–762. doi: 10.1124/dmd.30.7.757. [DOI] [PubMed] [Google Scholar]
- Arora V., Knapp D.C., Reddy M.T., Weller D.D., Iversen P.L. Bioavailability and efficacy of antisense morpholino oligomers targeted to c-myc and cytochrome P-450 3A2 following oral administration in rats. J. Pharm. Sci. 2002;91:1009–1018. doi: 10.1002/jps.10088. [DOI] [PubMed] [Google Scholar]
- Benfield D.A., Nelson E., Collins J.E., Harris L., Goyal S.M., Bobinson D., Christianson W.T., Morrison R.B., Gorcyca D., Chladek D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332) J. Vet. Diagn. Invest. 1992;4:127–133. doi: 10.1177/104063879200400202. [DOI] [PubMed] [Google Scholar]
- Brent L., Drapeau P. Targeted ‘knockdown’ of channel expression in vivo with an antisense morpholino oligonucleotide. Neuroscience. 2002;114:275. doi: 10.1016/s0306-4522(02)00270-1. [DOI] [PubMed] [Google Scholar]
- Conzelmann K.K., Visser N., Van Woensel P., Thiel H.J. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology. 1993;193:329–339. doi: 10.1006/viro.1993.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas T.S., Binduga-Gajewska I., Tilgner M., Ren P., Stein D.A., Moulton H.M., Iversen P.L., Kauffman E.B., Kramer L.D., Shi P.Y. Inhibition of Flavivirus Infections by Antisense Oligomers Specifically Suppressing Viral Translation and RNA Replication. J. Virol. 2005;79:4599–4609. doi: 10.1128/JVI.79.8.4599-4609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield J., Ramachandran A., Reuter K., LaVallie E., Collins-Racie L., Crosier K., Crosier P. Cadherin-17 is required to maintain pronephric duct integrity during zebrafish development. Mech. Dev. 2002;115:15–26. doi: 10.1016/s0925-4773(02)00094-1. [DOI] [PubMed] [Google Scholar]
- Hudziak R.M., Barofsky E., Barofsky D.F., Weller D.L., Huang S.B., Weller D.D. Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucl. Acid Drug Dev. 1996;6:267–272. doi: 10.1089/oli.1.1996.6.267. [DOI] [PubMed] [Google Scholar]
- Hudziak R.M., Summerton J., Weller D.D., Iversen P.L. Antiproliferative effects of steric blocking phosphorodiamidate morpholino antisense agents directed against c-myc. Antisense Nucl. Acid Drug Dev. 2000;10:163–176. doi: 10.1089/oli.1.2000.10.163. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Kwang J., Yoon I.J., Joo H.S., Frey M.L. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 1993;133:477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- Kinney R.M., Huang C.Y., Rose B.C., Kroeker A.D., Dreher T.W., Iversen P.L., Stein D.A. Inhibition of Dengue Virus Serotypes 1 to 4 in Vero Cell Cultures with Morpholino Oligomers. J Virol. 2005;79:5116–5128. doi: 10.1128/JVI.79.8.5116-5128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey A.P., Meuse L., Karimi M., Contag C.H., Kay M.A. A potent and specific morpholino antisense inhibitor of hepatitis C translation in mice. Hepatology. 2003;38:503–508. doi: 10.1053/jhep.2003.50330. [DOI] [PubMed] [Google Scholar]
- Meng X.J. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet. Microbiol. 2000;74:309–329. doi: 10.1016/S0378-1135(00)00196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.J., Paul P.S., Halbur P.G. Molecular cloning and nucleotide sequencing of the 3′ terminal genomic RNA of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 1994;75:1795–1801. doi: 10.1099/0022-1317-75-7-1795. [DOI] [PubMed] [Google Scholar]
- Meng X.J., Paul P.S., Halbur P.G., Lum M.A. Characterization of a high-virulence US isolate of porcine reproductive and respiratory syndrome virus in a continuous cell line, ATCC CRL11171. J. Vet. Diagn. Invest. 1996;8:374–381. doi: 10.1177/104063879600800317. [DOI] [PubMed] [Google Scholar]
- Meng X.J., Paul P.S., Morozov I., Halbur P.G. A nested set of six or seven subgenomic mRNAs is formed in cells infected with different isolates of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 1996;77:1265–1270. doi: 10.1099/0022-1317-77-6-1265. [DOI] [PubMed] [Google Scholar]
- Meulenberg J.J., Hulst M.M., de Meijer E.J., Moonen P.L., Den Besten A., de Kluyver E.P., Wensvoort G., Moormann R.J. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenberg J.J., Hulst M.M., de Meijer E.J., Moonen P.L., den Besten A., de Kluyver E.P., Wensvoort G., Moormann R.J. Lelystad virus belongs to a new virus family, comprising lactate dehydrogenase-elevating virus, equine arteritis virus, and simian hemorrhagic fever virus. Arch. Virol. Suppl. 1994;9:441–448. doi: 10.1007/978-3-7091-9326-6_43. [DOI] [PubMed] [Google Scholar]
- Moulton H.M., Hase M.C., Smith K.M., Iversen P.L. HIV Tat peptide enhances cellular delivery of antisense morpholino oligomers. Antisense Nucl. Acid Drug Dev. 2003;13:31–43. doi: 10.1089/108729003764097322. [DOI] [PubMed] [Google Scholar]
- Moulton H.M., Moulton J.D. Arginine-rich cell-penetrating peptides with uncharged antisense oligomers. Drug Discov. Today. 2004;9:870. doi: 10.1016/S1359-6446(04)03226-X. [DOI] [PubMed] [Google Scholar]
- Moulton H.M., Nelson M.H., Hatlevig S.A., Reddy M.T., Iversen P.L. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug. Chem. 2004;15:290–299. doi: 10.1021/bc034221g. [DOI] [PubMed] [Google Scholar]
- Nelsen C.J., Murtaugh M.P., Faaberg K.S. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 1999;73:270–280. doi: 10.1128/jvi.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson E.A., Christopher Hennings J., Drew T., Wensvoort G., Collins J.E., Benfield D.A. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 1993;31:3184–3189. doi: 10.1128/jcm.31.12.3184-3189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M.H., Stein D.A., Kroeker A.D., Hatlevig S.A., Iversen P.L., Moulton H.M. Arginine-rich peptide conjugation to morpholino oligomers: effects on antisense activity and specificity. Bioconjug. Chem. 2005;16:959–966. doi: 10.1021/bc0501045. [DOI] [PubMed] [Google Scholar]
- Neuman B.W., Stein D.A., Kroeker A.D., Churchill M.J., Kim A.M., Kuhn P., Dawson P., Moulton H.M., Bestwick R.K., Iversen P.L., Buchmeier M.J. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 2005;79:9665–9676. doi: 10.1128/JVI.79.15.9665-9676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Stein D.A., Kroeker A.D., Paulino A.D., Moulton H.M., Iversen P.L., Buchmeier M.J. Antisense morpholino-oligomers directed against the 5′ end of the genome inhibit coronavirus proliferation and growth. J. Virol. 2004;78:5891–5899. doi: 10.1128/JVI.78.11.5891-5899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opriessnig T., Halbur P.G., Yoon K.J., Pogranichniy R.M., Harmon K.M., Evans R., Key K.F., Pallares F.J., Thomas P., Meng X.J. Comparison of molecular and biological characteristics of a modified live porcine reproductive and respiratory syndrome virus (PRRSV) vaccine (ingelvac PRRS MLV), the parent strain of the vaccine (ATCC VR2332), ATCC VR2385, and two recent field isolates of PRRSV. J. Virol. 2002;76:11837–11844. doi: 10.1128/JVI.76.23.11837-11844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Taylor M., Ning Y.Y., Iversen P., Kobzik L. In vivo evaluation of a morpholino antisense oligomer directed against tumor necrosis factor-alpha. Antisense Nucl. Acid Drug Dev. 2000;10:11–16. doi: 10.1089/oli.1.2000.10.11. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- Schmajuk G., Sierakowska H., Kole R. Antisense oligonucleotides with different backbones. Modification of splicing pathways and efficacy of uptake. J. Biol. Chem. 1999;274:21783–21789. doi: 10.1074/jbc.274.31.21783. [DOI] [PubMed] [Google Scholar]
- Stein D.A., Skilling D.E., Iversen P.L., Smith A.W. Inhibition of Vesivirus infections in mammalian tissue culture with antisense morpholino oligomers. Antisense Nucl. Acid Drug Dev. 2001;11:317–325. doi: 10.1089/108729001753231696. [DOI] [PubMed] [Google Scholar]
- Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- Summerton J., Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucl. Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- Tan C., Chang L., Shen S., Liu D.X., Kwang J. Comparison of the 5′ leader sequences of North American isolates of reference and field strains of porcine reproductive and respiratory syndrome virus (PRRSV) Virus Genes. 2001;22:209–217. doi: 10.1023/A:1008179726163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.F., Paulauskis J.D., Weller D.D., Kobzik L. In vitro efficacy of morpholino-modified antisense oligomers directed against tumor necrosis factor-alpha mRNA. J. Biol. Chem. 1996;271:17445–17452. doi: 10.1074/jbc.271.29.17445. [DOI] [PubMed] [Google Scholar]
- van den Born E., Gultyaev A.P., Snijder E.J. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA. 2004;10:424–437. doi: 10.1261/rna.5174804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E., Stein D.A., Iversen P.L., Snijder E.J. Antiviral activity of morpholino oligomers designed to block various aspects of equine arteritis virus amplification in cell culture. J. Gen. Virol. 2005;86:3081–3090. doi: 10.1099/vir.0.81158-0. [DOI] [PubMed] [Google Scholar]
- van Marle G., Dobbe J.C., Gultyaev A.P., Luytjes W., Spaan W.J., Snijder E.J. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Olsthoorn R.C., Kroese M.V., Rottier P.J., Meulenberg J.J. Kissing interaction between 3′ noncoding and coding sequences is essential for porcine arterivirus RNA replication. J. Virol. 2002;76:1521–1526. doi: 10.1128/JVI.76.3.1521-1526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sharma R.D., Paul P.S. Monoclonal antibodies against conformationally dependent epitopes on porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 1998;63:125–136. doi: 10.1016/s0378-1135(98)00231-4. [DOI] [PubMed] [Google Scholar]


