Abstract
West Nile virus (WNV) is a member of the Flavivirus family and induces febrile illness, sporadic encephalitis, and paralysis. The capsid (Cp) of WNV is thought to play a role in inducing these symptoms through caspase-3- and caspase-9-dependent apoptosis. Using WNVCp as bait for a yeast two-hybrid assay, we identified that Hsp70 interacted with WNVCp. The interaction between Hsp70 and WNVCp was further substantiated using purified proteins. Deletion analysis of Hsp70 indicated that WNVCp could bind to the substrate binding domain of Hsp70. The presence of WNVCp in the Hsp70-dependent folding system inhibited the refolding of β-galactosidase (β-gal), which showed that WNVCp might function as a negative regulator of Hsp70. Finally, the cytotoxic effect of WNVCp in 293T cells was prevented by ectopic Hsp70, suggesting a negative regulatory role of Hsp70 on WNVCp. Our findings suggest a possible negative regulatory role of Hps70 in the pathway of WNV infection.
Keywords: Hsp70, WNVCp, Folding, Luciferase, β-Galactosidase, Guanidium–HCl
Eukaryotic Hsp70s are highly abundant cytosolic and nuclear molecular chaperones that play essential roles in various aspects of protein homeostasis, including protein synthesis, transportation, degradation, and folding [1]. Transient interactions between Hsp70 and unfolded polypeptides maintain its substrates in soluble, intermediate folded states and prevent protein misfolding and aggregation. Moreover, the intermediate substrates are subsequently folded into their native forms by the machinery of the Hsp70 chaperone [2], [3], [4]. This is accomplished, in part, by the help of a subclass of proteins known as co-chaperones, which modulate the chaperone function of Hsp70 and its association with other partner proteins.
Hsp70 co-chaperones can mostly be classified into three classes: the enhancers (Hsp40, GrpE, and Hip), which positively regulate Hsp70-dependent folding; the negative regulators (Bag-1, Scythe, and Chip), which inhibit the chaperone function of Hsp70; and the coadapters (Hop and Hip), which connect Hsp70 with various other co-chaperones and chaperones to form multimeric chaperone complexes [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13].
Hsp40, which couples the ATP hydrolyzing activity of Hsp70 with its substrate interaction, is required for the folding function of Hsp70 [2], [3], [4]. On the other hand, Escherichia coli DnaK, a homologue of Hsp70, requires the nucleotide exchange factor, GrpE, for full-folding activity; DnaJ, a Hsp40 orthologue, is also required [8], [14]. These are essential partner proteins of Hsp70 that enhance Hsp70-dependent folding. The tetratricopeptide repeat (TPR) domains of Hop (p60) link the multi-complex chaperone structures of Hsp70 and Hsp90; this linking helps to maintain the proper conformations of progesterone or estrogen aporeceptors prior to hormone binding [7], [11], [15]. Bag1 and Scythe are negative regulators of Hsp70-dependent folding processes [9], [10], [13]. They were initially found to interact with the anti-apoptotic protein, Bcl-2, and the apoptotic mediator, Reaper, respectively [16]. Bag1 and Scythe share common structural features, namely the carboxyl-terminal Bag-domain that interacts with Hsp70 and the amino-terminal ubiquitin-like motif, the function of which is not well characterized [17].
West Nile virus (WNV) is part of the Flaviviridae family, and it is capable of inducing febrile syndrome, meningitis, encephalitis, hepatitis, flaccid paralysis, and death [18], [19]. It contains single-stranded RNA, which encodes the capsid (Cp), envelope, premembrane proteins, and 7 non-structural proteins that have been implicated in viral replication [20], [21]. WNV has been found to induce cell death via the Bax-dependent pathway in several cancer and brain cell lines, as well as in mice [22], [23]. It was first suggested that the Cp of WNV (WNVCp) was able to induce caspase-9- and caspase-3-mediated apoptosis [24]. Later, WNV non-structural proteins 2B and 3 (NS2B-NS3) were found to induce apoptosis through the activation of caspase-8 and caspase-3 [25]. However, the detailed mechanism of the cytotoxic effects of WNV is not yet fully understood.
In these studies, we have shown that WNVCp was able to bind to Hps70 and to function as a negative regulator of Hsp70. Furthermore, WNVCp was able to induce cell cycle arrest in 293T cells. This cytotoxic effect was prevented when Hsp70 was overexpressed, which suggests that, as a molecular chaperone, Hsp70 might have a protective effect against WNV infection. These findings are expected to contribute to the identification of the infectious pathway induced by WNVCp.
Materials and methods
Plasmids. pGBK-T7-WNVCp, a bait plasmid for yeast two-hybrid screening, was created by cloning an EcoRI–XhoI fragment from pcDNA3-His-WNVCp into pGBK-T7 (Clontech). pcDNA3-HA-WNVCp and pET28a-His-WNVCp were prepared by subcloning an EcoRI–XhoI fragment from pcDNA3-his-WNVCp into pcDNA3-HA and pET28a-His (Invitrogen). The bacterial expression plasmids for Hsp70 and Hsp70AAAA were used as described previously [2]. Hsp70 mammalian expression vector, pcDNA/HA-Hsp70, was used as described previously (Nollen et al. [10]). pcDNA3.1/Luciferase was obtained from pGL3 (Promega) using XbaI and HindIII restriction, and was subcloned into plasmid pcDNA3.1 (Invitrogen). All DNA constructs were produced by PCR and sequenced.
Yeast two-hybrid screening. Yeast two-hybrid screening was performed according to the instructions of the manufacturer using the pretransformed MATCHMAKER human brain cDNA library (Clontech). In brief, pGBK-T7-WNVCp was transformed into yeast strain AH109. The transformants containing bait plasmid were mated with the pre-transformed human brain cDNA library. Candidates for two-hybrid interaction were initially selected on SD (-His, -Leu, -Trp) medium and further confirmed on SD (-Ade, -His, -Leu, -Trp) medium containing X-gal. The plasmid DNA was isolated from the positive clones and sequenced according to the instructions of the manufacturer. To further confirm the interaction between WNVCp and Jab1, Y187 and AH109, each of which contained pGAD-T7-Jab1 and pGBK-T7-WNVCp, respectively, were mated and tested using SD (-Ade, -His, -Leu, -Trp) medium containing X-gal.
Cell biology. A human cancer cell line, 293T (kidney carcinoma), was maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone) and 1% penicillin/streptomycin (Invitrogen). Transient transfections were performed using Lipofectamine plus reagent (Invitrogen) or Welfect (Wellgene) according to the recommendations of the manufacturers. Immunoprecipitation (IP) and Western blot analysis were carried out as described previously [17].
Biochemical analyses. Recombinant Hsp40, Hsp70, ATPase domain, SBD, and Hsp70AAAA were expressed and purified as described previously [2], [26]. His-WNVCp was prepared according to the instructions of the manufacturer (Invitrogen). In brief, BL21 cells, which were transformed with pET28a-His-wnvcp or pET28a-His-P5/8A, were treated with IPTG (1 mM) at 25 °C. The crude extract was prepared using lysozyme (Sigma). The extract was resolved over the Resource Q (35 ml, Pharmacia-LKB), His-bind resin (20 ml, Novagen), and DEAE column (50 ml, Pharmacia-LKB). Fractions containing His-WNVCp was concentrated using a Centriprep-30 (Millipore). Concentrations of proteins were determined using the bicinchoninic acid (BCA) protein assay (Pierce, IL) relative to standard BSA (Pierce, IL). Folding assays of guanidine–HCl denatured β-galactosidase and luciferase (Sigma, MO) followed existing protocols [2], [26]. The pull-down experiment was carried out as described previously [17]. Immunofluorescence staining was carried out as follows. Cells were plated in six-well plates with coverslips; transfections were performed using lipofectamine reagent. After 24 h, cells were fixed with 4% paraformaldehyde solution for 15 min at room temperature, washed with PBS (Invitrogen), and permeabilized with 0.5% Triton X-100 in PBS for 15 min. The cells were then blocked with 5% bovine serum albumin (Santa Cruz) in PBS for 30 min and incubated overnight with specific primary antibody at room temperature. The samples were incubated with Alexa Fluor 488 anti-mouse or Alexa 594 anti-rabbit antibodies (each diluted at 1:400) for 1 h at room temperature. The cells were stained with DAPI (4,6-diamidino-2-phenylindole, Sigma) for 5 min. The slides were analyzed using confocal or immunofluorescent microscopes (Carl Zeiss Vision, LSM510 and 5203 Axiophot, respectively, Oberkochen).
FACS analysis. For FACS analysis, 293T cells were transfected with pcDNA3-HA-WNVCp, with or without pcDNA/HA-Hsp70. At 48 h after transfection, the cells were harvested using trypsin and were subsequently fixed with 70% EtOH. After washing three times with PBS, the fixed cells were incubated with RNase A (1 μg/ml) at 37 °C for 1 h, followed by treatment with propidium iodide (PI) (50 μg/ml) for 30 min. Stained cells were detected by flow cytometric analysis (Becton–Dickinson). Data were analyzed using CellQuest Pro software (Becton–Dickinson).
Antibodies and chemicals. Hsp70 (SPA810) was purchased from Stressgen. Luciferase (L0159) and Flag (M2) antibodies were purchased from Sigma. Alexa Fluor 488 anti-mouse and Alexa 594 anti-rabbit antibodies were obtained from Alexa.
Results and discussion
WNVCp could bind to Hsp70 independently of ATP
WNVCp is known to induce apoptosis by inducing mitochondrial dysfunction and caspase-9 activation in tissue culture (24). Furthermore, when its gene is delivered to mouse brain or skeletal muscle, it can cause cell death and inflammation in vivo (24). To further identify the functional role of WNVCp, we carried out a yeast two-hybrid assay using the entire region of WNVCp as bait. The result showed that several chaperones, including Hsp70, were able to bind to WNVCp. To confirm the binding ability of Hsp70 to WNVCp, recombinant Hsp70 and His-WNVCp were purified (Fig. 1 A). The interaction between purified recombinant proteins was tested using a Ni+–agarose pull-down assay (Fig. 1B). The results indicated that Hsp70 was able to bind to WNVCp (Fig. 1B, lanes 1 and 2). ATP was added to the reaction system to exclude the possibility of denatured WNVCp binding to Hsp70 as an unfolded substrate (Fig. 1B, lanes 3 and 4). As a control, recombinant Hsp70 was shown not to bind to Ni+–agarose non-specifically (Fig. 1B, lanes 5 and 6). It is known that the presence of ATP facilitates the release of unfolded substrate from Hsp70 [1]. As a control, we carried out an interaction between Hsp70 and chemically denatured luciferase in the presence or absence of ATP (Fig. 1C). The data indicated that, in the presence of ATP, the denatured substrates did not bind to Hsp70 (Fig. 1C, lanes 3 and 4). The immunoprecipitation assays of ectopic WNVCp overexpressed in a human kidney tumor cell line, 293T, with endogenous Hsp70 showed that the WNVCp and Hsp70 bind in cells, indicating physiologically viable interaction between the two proteins (Fig. 1C). Overall, the interaction between Hsp70 and WNVCp was not affected, which suggests that WNVCp could bind to Hsp70 as a binding partner rather than binding to unfolded substrates. Furthermore, Hsp70 could directly bind to WNVCp independently of ATP.
Fig. 1.
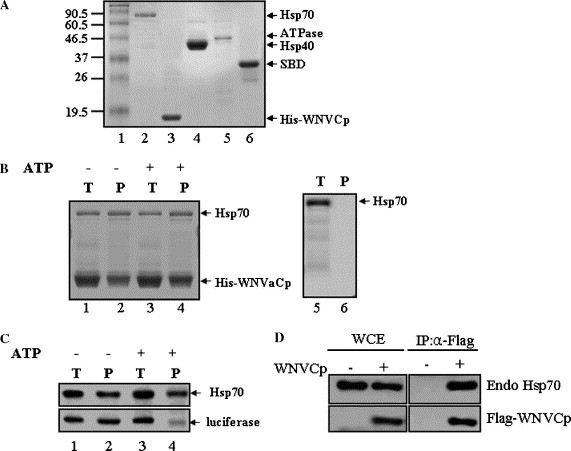
WNVCp forms a complex with Hsp70. (A) Recombinant proteins, Hsp70, His-WNVCp, Hsp40, ATPase domain, and SBD, were purified and analyzed by SDS–PAGE and visualized with Coomassie blue. (B) Reaction mixtures containing Hsp70 (0.5 μM) + His-WNVCp (4 μM) (lanes 1–4) or Hsp70 (1 μM) alone (lanes 5 and 6) were incubated (T: total, lanes 1, 3, and 5) in the absence (lanes 1, 2, 5, and 6) or presence (lanes 3 and 4) of ATP, and protein complexes were precipitated (P: precipitated, lanes 2, 4, and 6) by Ni2+–agarose. Precipitated proteins were analyzed by SDS–PAGE followed by Coomassie blue staining. (C) Reaction mixtures containing Hsp70 (0.5 μM) and luciferase that had been chemically denatured by guanidium–HCl (0.5 μM) (lanes 1–4) were incubated (T: total, lanes 1 and 3) in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of ATP, and protein complexes were precipitated (P: precipitated, lanes 2, 4, and 6) by Hsp70 antibody (5a5). Precipitated proteins were analyzed by SDS–PAGE followed by Western blotting using mono-Hsp70 mouse (5a5) and poly-luciferase rabbit (L0159) antibodies. (D) The plasmid expressing HA-WNVCp was transfected into 293T cells. The cell lysates were immunoprecipitated using anti-HA mouse antibodies. The whole cell extract (WCE) and immunoprecipitates (IP) were detected using anti-HA rabbit and anti-Hsp70 mouse antibodies.
WNVCp could function as a negative regulator of Hsp70 by binding to its substrate binding domain (SBD)
To further confirm the interaction between Hsp70 and WNVCp, we carried out refolding assays using purified Hsp70 and Hdj1 as folding mediators and β-gal and luciferase that had been chemically denatured by guanidium–HCl as substrates. This assay allows us to examine whether WNVCp could still bind to Hsp70 in the presence of its co-chaperone, Hsp40, and whether it could still function as a regulator of Hsp70. The data indicated that WNVCp was able to suppress 50% of the β-gal folding activities of Hsp70 when the ratio of WNVCp and Hsp70 was 1:1. When the ratio of Hsp70 and WNVCp was 1:2, the folding of β-gal was almost completely suppressed, indicating that WNVCp could function as a potent inhibitor of Hsp70 inhibitor (Fig. 2 A). Similar results were obtained using luciferase as unfolded substrate (Fig. 2B).
Fig. 2.
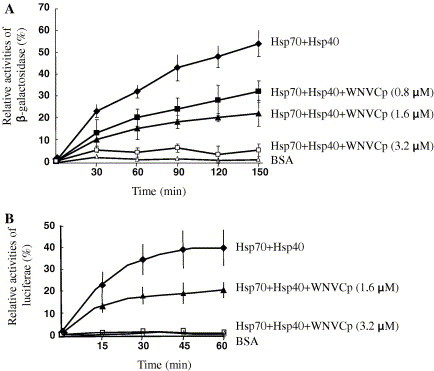
WNVCp inhibits Hsp70-dependent folding. (A) The folding of β-gal (40 nM) that had been chemically denatured by guanidium–HCl was measured in the presence of BSA (3.2 μM) (▵), Hsp70 (1.6 μM) + Hsp40 (3.2 μM) (♦), Hsp70 (1.6 μM) + Hsp40 (3.2 μM) + His-WNVCp (0.8 μM) (■), Hsp70 (1.6 μM) + Hsp40 (3.2 μM) + His-WNVCp (1.6 μM) (▴), and Hsp70 (1.6 μM) + Hsp40 (3.2 μM) + His-WNVCp (3.2 μM) (□). (B) The folding of chemically denatured luciferase (2 μM) was measured in the presence of BSA (3.2 μM) (▵), Hsp70 (1.6 μM) + Hsp40 (3.2 μM) (♦), Hsp70 (1.6 μM) + Hsp40 (3.2 μM) + His-WNVCp (1.6 μM) (▴), and Hsp70 (1.6 μM) + Hsp40 (3.2 μM) + His-WNVCp (3.2 μM) (□).
The regulation of Hsp70 by the Hsp70 partner proteins could take place through their binding to ATPase domain or SBD of Hsp70. For example, Hop, SGT, and TPR are individually known to interact with Hsp70 through its conserved carboxyl-terminal EEVD regulatory motif, while Bag1 and Hip interact with the ATPase domain [5], [6], [7], [8], [9], [11], [18], [26]. To examine the domain of Hsp70 responsible for binding with WNVCp, we purified the recombinant ATPase domain or SBD of Hsp70 (Fig. 1A). The data indicated that the SBD was responsible for the interaction between Hsp70 and WNVCp (Fig. 3 A). Because WNVCp binds to the SBD, we further tested whether the carboxyl-terminal EEVD motif was required for the interaction between the two proteins. The EEVD motif is considered to be important for its role in linking the SBD and ATPase domain of Hsp70, which enables the crosstalk between the two domains [2]. Furthermore, this motif is considered important for the refolding activities of Hsp70, since the lack of this motif terminates the refolding activities of Hsp70 [2]. To identify the role of EEVD motif in the interaction between Hsp70 and WNVCp, we employed the HSP70AAAA mutant. In this mutant, the four amino acids, EEVD, of the C-terminus are changed to AAAA [2]. When this mutant was purified as a recombinant protein, it was easily degraded to the ATPase domain and SBD [2]. Thus, the purified proteins displayed both full-length Hsp70AAAA and ATPase domain (Fig. 3B). The data showed that this mutant was not able to bind to WNVCp, which suggested that the C-terminus might function as a motif for the interaction with WNVCp (Fig. 3B).
Fig. 3.
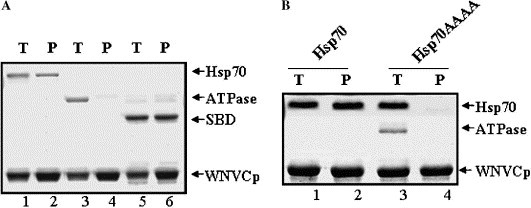
WNVCp binds to the substrate binding domain of Hsp70. (A) Reaction mixtures containing Hsp70 (0.5 μM) + His-WNVCp (4 μM) (lanes 1 and 2), ATPase (1 μM) + His-WNVCp (4 μM) (lanes 3 and 4), and SBD (1 μM) + His-WNVCp (4 μM) (lanes 5 and 6) were incubated (T: lanes 1, 3, and 5), and protein complexes were precipitated (P: lanes 2, 4, and 6) by Ni2+–agarose. (B) Reaction mixtures containing Hsp70 (1 μM) + His-WNVCp (5 μM) (lanes 1 and 2) and Hsp70AAAA (1 μM) + His-WNVCp (5 μM) (lanes 3 and 4) were incubated (T: lanes 1 and 3), and protein complexes were precipitated (P: lanes 2 and 4) by Ni2+–agarose. Precipitated proteins were analyzed as described in Fig. 1B.
WNVCp has been known to localize to the nucleoli when expressed alone [24]. Thus, we next tested whether the expression of Hsp70 could affect the localization of WNVCp. When both proteins were expressed, we found that WNVCp, usually localized to the nucleoli, was dispersed into the cytoplasm (Fig. 4 ). These results indicate that WNVCp could interact with Hsp70 in the cells. Furthermore, the interaction seems to affect the cellular localization of WNVCp from the nucleoli to the cytoplasm. It is not certain, however, whether Hsp70 participates in the nuclear export of WNVCp or inhibits its localization of WNVCp into the nucleoli. Taken together, WNVCp was able to bind to the substrate domain of Hsp70 and inhibited its folding activities. Conversely, Hsp70 seems to be able to induce the cytoplasmic localization of WNVCp through direct interaction.
Fig. 4.
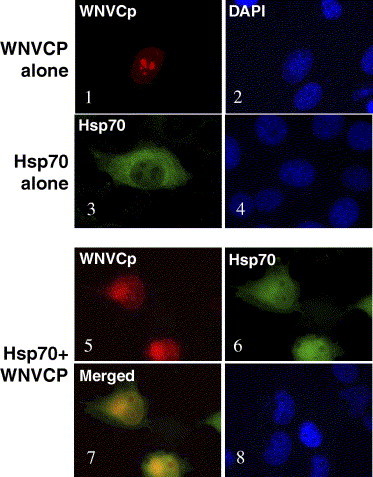
Hsp70 prevents localization of WNVCp to the nucleoli. H1299 cells were transfected with plasmids expressing HA-WNVCp, Hsp70, or both proteins. Twenty-four after transfection, cells were fixed and stained with polyclonal anti-HA and Alexa Fluor 594 anti-rabbit antibodies, or with monoclonal anti-Hsp70 and Alexa 488 anti-mouse antibodies. The images were captured by immunofluorescence microscopy. The cells were counterstained with DAPI to visualize the nuclei.
Hsp70 has a protective role against the cytotoxic effect of WNVCp
Various viral proteins have been known to localize to the nucleoli, which could strategically aid in the propagation of many viruses. These proteins usually participate in the disruption of cytokinesis of mammalian cells, as seen in the N protein of the coronavirus and the capsid of human hepatitis B virus [27], [28]. It is also likely that the WNVCp might have disruptive activities against mammalian cells by localizing to the nucleoli. Moreover, the prevention of the nucleolus localization of WNVCp by Hsp70 might protect cells from the cytotoxic effect of WNVCp (Fig. 4). To examine the cellular effect of WNVCp, we transfected the plasmid expressing WNVCp into the 293T cells and observed the cell cycle in the 293T cells. The data showed that WNVCp-induced G2 arrest in 293T cells (Fig. 5 A and B, lane 2). When Hsp70 was co-expressed, the cytotoxic effect of WNVCp was extinguished, which indicates that Hsp70 has a protective role against WNVCp (Fig. 5A and B, lane 4). It is assumed that, by forming a complex, Hsp70 inhibited the localization of WNVCp to the nucleoli and prevented the cytotoxic effect of the viral protein. We cannot, however, exclude the possibility that Hsp70 is able to indirectly protect the cells from cytotoxic effects imposed by WNVCp, since Hsp70 generally functions against various stresses exerted on cells [1].
Fig. 5.
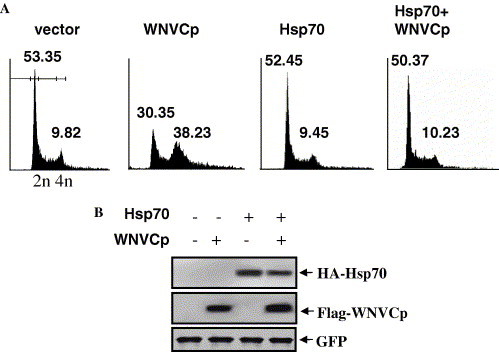
In 293T cells, Hsp70 negatively regulates WNVCp-induced cell cycle arrest at G2 phase. (A) 293T cells were transfected with the plasmid expressing HA-WNVCp, HA-Hsp70, or both proteins. After harvesting, cells were fixed and stained with EtOH and PI, respectively. Stained cells were detected by flow cytometric analysis and analyzed using CellQuest Pro software. (B) The plasmids expressing HA-WNVCp, HA-Hsp70, or both proteins were transfected into 293T cells. The cell lysates were detected using anti-HA rabbit antibodies.
Our findings suggest that WNVCp could function as a negative regulator of Hsp70 chaperone activities by binding to the substrate binding domain of Hsp70 (Fig. 2). At the same time, the cellular stresses imposed by WNVCp could be protected against by the ectopic expression of Hsp70 (Fig. 5). WNVCp does not seem to bind to Hsp70 as an unfolded substrate, which could prevent its chaperoning of other substrates, as ATP did not have any effects on the binding abilities of Hsp70 and WNVCp (Fig. 1). Thus, it is possible that Hsp70 might function as a negative regulatory protein of WNVCp, which weakened the cytotoxic effects of WNVCp. However, it is still not clear from these studies whether the toxic effect of WNV itself could be prevented by the overexpression of Hsp70. Furthermore, it has not yet been determined whether infection with WNV induced the expression of Hsp70, which, in turn, might function to protect cells from WNV-induced death. However, our studies suggest the possible role of Hsp70 as a protector of the cell against infection with WNV through the suppression of one of the toxic proteins of WNV. Although further studies are required to determine the detailed mechanistic pathways involved in the relationship between Hsp70 and WNV infection, this is a novel finding that suggests how WNV protein could be negatively controlled by the proteins of host cells. We expect that this finding will aid investigators in identifying the mechanistic pathway of how mammalian cells adjust to the WNV infection.
Acknowledgments
We thank Prof. Young-Ki Paik (Yonsei University) and Jae Kyoon Shin (Sungkyunkwan University) for their kind advice and assistance. This project was supported by SeokChon Grants 2002-0706-000 from Sungkyunkwan University and R01-2002-000-00445-0 from the Korea Science and Engineering Foundation.
References
- 1.Morimoto R.I., Tissieres A., Georgopoulos C. Cold Spring Harbor Laboratory; Cold Spring Harbor NY: 1994. The Biology of Heat Shock Proteins and Molecular Chaperones. [Google Scholar]
- 2.Freeman B.C., Morimoto R.I. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- 3.Levy E.J., McCarty J., Bukau B., Chirico W.J. Conserved ATPase and luciferase refolding activities between bacteria and yeast Hsp70 chaperones and modulators. FEBS Lett. 1995;368:435–440. doi: 10.1016/0014-5793(95)00704-d. [DOI] [PubMed] [Google Scholar]
- 4.Minami Y., Hohfeld J., Ohtsuka K., Hartl F.U. Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog, Hsp40. J. Biol. Chem. 1996;271:19617–19624. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- 5.Hohfeld J., Minami Y., Hartl F.U. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- 6.Prapapanich V., Chen S., Smith D.F. Mutation of Hip’s carboxy-terminal region inhibits a transitional stage of progesterone receptor assembly. Mol. Cell. Biol. 1998;18:944–952. doi: 10.1128/mcb.18.2.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F.U., Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell. 2000;101:99–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 8.Langer T., Lu C., Echols H., Flanagan J., Hayer M.K., Hartl F.U. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 9.Bimston D., Song J., Winchester D., Takayama S., Reed J.C., Morimoto R.I. BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J. 1998;17:6871–6878. doi: 10.1093/emboj/17.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nollen E.A., Brunsting J.F., Song J., Kampinga H.H., Morimoto R.I. Bag1 functions in vivo as a negative regulator of Hsp70 chaperone activity. Mol. Cell. Biol. 2000;20:1083–1088. doi: 10.1128/mcb.20.3.1083-1088.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S., Smith D.F. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J. Biol. Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 12.Ballinger C.A., Connell P., Wu Y., Hu Z., Thompson L.J., Yin L.Y., Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol . 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kullmann M., Schneikert J., Moll J., Heck S., Zeiner M., Gehring U., Cato A.C. RAP46 is a negative regulator of glucocorticoid receptor action and hormone-induced apoptosis. J. Biol. Chem. 1998;273:14620–14625. doi: 10.1074/jbc.273.23.14620. [DOI] [PubMed] [Google Scholar]
- 14.Szabo A., Langer T., Schroder H., Flanagan J., Bukau B., Hartl F.U. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc. Natl. Acad. Sci. USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosano H., Stensgard B., Charlesworth M.C., McMahon N., Toft D. The assembly of progesterone receptor-hsp90 complexes using purified proteins. J. Biol. Chem. 1998;273:32973–32979. doi: 10.1074/jbc.273.49.32973. [DOI] [PubMed] [Google Scholar]
- 16.Takayama S., Sato T., Krajewski S., Kochel K., Irie S., Millan J.A., Reed J.C. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell. 1995;80:279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 17.Song J., Takeda M., Morimoto R.I. Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat. Cell Biol. 2001;3:276–282. doi: 10.1038/35060068. [DOI] [PubMed] [Google Scholar]
- 18.Smithburn K.C., Hughes T.P., Burke A.W., Paul J.H. Am. J. Trop. Med. Hyg. 1940;20:471–492. [Google Scholar]
- 19.Sampson B.A., Ambrosi C., Carlot A., Reiber K., Veress J.F., Armbrustmacher V. The pathology of human West Nile Virus infection. Hum. Pathol. 2000;31:525–531. doi: 10.1053/hp.2000.8047. [DOI] [PubMed] [Google Scholar]
- 20.Chambers T.J., Hahn C.S., Galler R., Rice C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 21.Westaway E.G., Brinton M.A., Gaidamovich S., Horzinek M.C., Igarashi A., Kaariainen L., Lvov D.K., Porterfield J.S., Russel P.K., Trent D.W. Flaviviridae. Intervirology. 1985;24:183–192. doi: 10.1159/000149642. [DOI] [PubMed] [Google Scholar]
- 22.Chu J.J., Ng M.L. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 2004;78:10543–10555. doi: 10.1128/JVI.78.19.10543-10555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parquet M.C., Kumatori A., Hasebe F., Morita K., Igarashi A. West Nile virus-induced bax-dependent apoptosis. FEBS Lett. 2001;500:17–24. doi: 10.1016/s0014-5793(01)02573-x. [DOI] [PubMed] [Google Scholar]
- 24.Yang J.S., Ramanathan M.P., Muthumani K., Choo A.Y., Jin S.H., Yu Q.C., Hwang D.S., Choo D.K., Lee M.D., Dang K., Tang W., Kim J.J., Weiner D.B. Induction of inflammation by West Nile virus capsid through the caspase-9 apoptotic pathway. Emerg. Infect. Dis. 2002;8:1379–1384. doi: 10.3201/eid0812.020224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramanathan M.P., Chambers J.A., Pankhong P., Chattergoon M., Attatippaholkun W., Dang K., Shah N., Weiner D.B. Host cell killing by the West Nile Virus NS2B-NS3 proteolytic complex: NS3 alone is sufficient to recruit caspase-8-based apoptotic pathway. Virology. 2006;345:56–72. doi: 10.1016/j.virol.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 26.Oh W., Song J. Cooperative interaction of Hsp40 and TPR1 with Hsp70 reverses Hsp70-HspBp1 complex formation. Mol. Cells. 2003;16:84–91. [PubMed] [Google Scholar]
- 27.Hiscox J.A., Wurm T., Wilson L., Britton P., Cavanagh D., Brooks G. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75:506–512. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ning Bo., Shih Chiaho. Nucleolar localization of human hepatitis B virus capsid protein. J. Virol. 2004;78:13653–13668. doi: 10.1128/JVI.78.24.13653-13668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


