Abstract
The Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3a locus encodes a 274 a.a. novel protein, and its expression has been confirmed in SARS patients. To study functional roles of 3a, we established a transgenic fly model for the SARS-CoV 3a gene. Misexpression of 3a in Drosophila caused a dominant rough eye phenotype. Using a specific monoclonal antibody, we demonstrated that the 3a protein displayed a punctate cytoplasmic localization in Drosophila as in SARS-CoV-infected cells. We provide genetic evidence to support that 3a is functionally related to clathrin-mediated endocytosis. We further found that 3a misexpression induces apoptosis, which could be modulated by cellular cytochrome c levels and caspase activity. From a forward genetic screen, 78 dominant 3a modifying loci were recovered and the identity of these modifiers revealed that the severity of the 3a-induced rough eye phenotype depends on multiple cellular processes including gene transcriptional regulation.
Keywords: Drosophila, Genetic screen, Transgenics, U274, X1
The Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) is a newly evolved coronavirus and is ascertained to be the etiological agent of the global atypical pneumonia pandemic in 2003. The SARS-CoV genome carries genes for the replicase enzymes, structural proteins, and several putative open reading frames (ORFs) with poorly defined functions [1], [2]. The 3a locus (also known as X1 [2], ORF3 [1], and U274 [3]) is unique to SARS-CoV and encodes a 274 a.a. novel protein. The 3a protein lacks sequence homology to any known proteins, but is predicted to carry three transmembrane domains and a C-terminal calcium-binding region [3], [4], [5], [6]. The detection of 3a protein [5] and 3a-specific IgG antibodies [6], [7], [8], [9] in SARS patients confirmed the cellular expression of 3a in SARS-CoV-infected cells and patients. The 3a protein localized preferentially on the Golgi apparatus in both transfected [5], [10] and SARS-CoV-infected cells [5]. In addition, 3a was also detected on the plasma membrane [3], [11]. Recent findings revealed that 3a interacted physically with other SARS-CoV structural proteins, including spike (S), membrane, and envelope [3], [6], and was also detected in newly packaged matured SARS-CoV virions [11], [12]. These findings would therefore imply that 3a functions as both a viral regulatory protein in infected cells and a structural protein in matured viral particles.
Many cellular proteins possess tyrosine-based YXXΦ motifs, which are sorting signals for internalization of proteins via endocytosis from the cell surface [13]. A functional YXXΦ signal has been identified in the porcine coronavirus but not SARS-CoV S protein [14]. On the other hand, 3a carries an analogous YXXΦ motif (160–163 a.a.) and it has also been reported to mediate protein internalization in cultured cells [3]. Because of the physical interaction between S and 3a [6], and the absence of an YXXΦ motif in the SARS-CoV S protein, it has therefore been hypothesized that one intracellular role of 3a is to modulate intracellular trafficking of S in SARS-CoV-infected cells [15].
Drosophila has been used to study the molecular functions of various viral gene products [16], [17], [18]. For example, the HIV-1 Tat protein was first found to interact with tubulin in Drosophila [16] and such interaction has subsequently been demonstrated in mammalian cells [19]. Therefore, Drosophila would be a feasible model to define intracellular regulatory actions of individual viral gene products [20]. Here, we established and characterized a 3a transgenic fly model to elucidate the cellular regulatory actions of SARS-CoV 3a locus in vivo. In brief, 3a misexpression caused a dominant rough eye phenotype, and we showed that genes which mediate clathrin-dependent endocytosis dominantly suppressed the 3a-induced rough eye phenotype. Furthermore, 3a misexpression caused apoptosis. From a forward genetic screen, we further found that 3a interacts genetically with genes that are involved in multiple cellular processes including gene transcription. In summary, our findings not only provide clues on the intracellular functions of 3a, but also demonstrate the feasibility of transgenic fly models in the studies of SARS-CoV 3a and other viral gene functions.
Materials and methods
Drosophila genetics. Fly strains were grown at 29 °C on standard cornmeal medium supplemented with dry yeast. The following fly lines were used in this study: gmr-GAL4; 34B-GAL4; UAS-DIAP1; UAS-emc (a kind gift of J. de Celis); UAS-GFP-lacZ; UAS-HSPA1L; UAS-P35; UAS-reaper; gmr-reaper; Csp R1 (a kind gift of K. Zinsmaier); CtBP 87De-10; dc3 EP2305; Eps15 EP2513; lwr 02858 (Bloomington Drosophila Stock Center); Nedd4 EY00500; Su(H)EY07695 (Bloomington Drosophila Stock Center); the Exelixis and DrosDel deficiency collections.
Generation of 3a transgenic lines. The 3a ORF was PCR amplified from viral cDNA to generate pUAST-3a and pUAST-EGFP-3a plasmids. The UAS-3a transgene produces a full-length non-tagged 3a protein, whereas the UAS-EGFP-3a transgene produces a full-length 3a protein with EGFP fused to its N-terminus. UAS-EGFP-3a E0 and UAS-3a F1 were selected for detailed analysis.
Genetic screen. We screened a total of 574 overlapping deletion lines to identify genomic regions that would modify the EGFP-3a-induced rough eye phenotype (Fig. 1 B) when present in only one copy. Five hundred and one of the deletion lines are from the Exelixis collection, and 73 are from the DrosDel collection. A total of 93 deletion lines representing 24 genomic regions showed reproducible modification of the EGFP-3a phenotype.
Fig. 1.
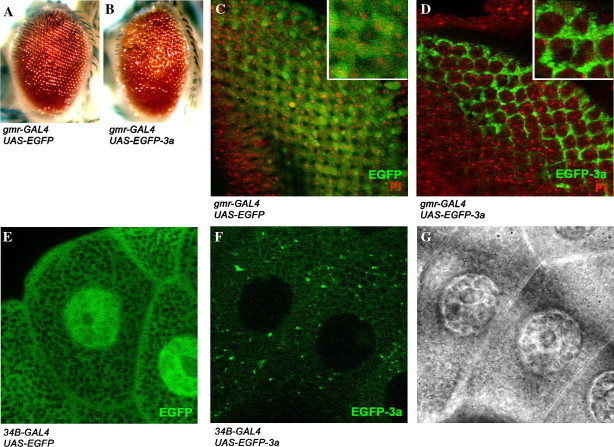
Misexpression of the SARS-CoV 3a gene in Drosophila. (A,B) Misexpression of the SARS-CoV 3a gene caused external eye disruption in adult Drosophila. Misexpression of the control EGFP transgene (A) showed normal external eye morphology, whereas misexpression of the SARS-CoV EGFP-3a transgene (B) caused a rough eye phenotype as characterized by loss of regularity of the external eye structure. (C–G) Subcellular localization of the EGFP-3a fusion protein in Drosophila. Misexpression of the EGFP control protein showed homogeneous intracellular green fluorescence signals in both third instar eye imaginal disc (C) and salivary gland (E) cells. A distinct punctate cytoplasmic expression pattern of the EGFP-3a fusion protein was observed in both eye imaginal disc (D) and salivary gland cells (F). (G) Location of salivary gland cell nuclei of (F). Propidium iodide (PI) was used to stain cell nuclei in eye imaginal disc cells (C,D).
Semi-quantitative RT-PCRs. Total RNA was prepared using Trizol reagent (Invitrogen). Primers used were dc3F: 5′ ATG GT TCT GGT GAT GCA 3′; dc3R: 5′ CTA CTT GTT TGA CTT GAG 3′; dc4F: 5′ ATG GGC GTT CCT GCT GGT 3′; dc4R: 5′ TTA CTT GT CGC CGA CTT 3′; emcF: 5′ TTC CGT TCA TGC CCA AGA ACA GGA 3′; emcR: 5′ GTT GGA CAG TTG CTG CTG TGA TTG 3′; GAPDHF: 5′ ACC GTC GAC GGT CCC TCT 3′; and GAPDHR: 5′ GTG TAG CCC AGG ATT CCC T 3′.
3a monoclonal antibodies. In brief, BALB/c mice were immunized intraperitoneally (i.p.) with 100 μg of the KLH-conjugated 3a peptide in complete Freund’s adjuvant (Sigma–Aldrich), followed by monthly i.p. boosts using the same amount of antigen in incomplete Freund’s adjuvant (Sigma–Aldrich). Preliminary screening of serum antibody activity and hybridoma fusion clones was determined by ELISA.
Western blot analysis. Primary antibodies used were anti-3a MAb X98 (1:40; this study), anti-cytochrome c clone 7H8.2C12 (1:500, Pharmingen), anti-Csp (1:50), and anti-β-tubulin E7 (1:2000; Developmental Studies Hybridoma Bank, Iowa City, IA, with funding from the NICHD).
Acridine orange staining and immunofluorescence. Acridine orange staining was performed as previously described [21]. Antibodies used for immunofluorescence include anti-3a MAbs X98 (1:20), anti-denatured cytochrome c clone 7H8.2C12 (1:200, Pharmingen), and anti-native cytochrome c clone 6H2.B2 (1:100; Pharmingen). Propidium iodide (PI) was used to label cell nuclei (Molecular Probes). Slides coated with both SARS-CoV-infected cells and non-infected cells were purchased from Euroimmun.
Results
To investigate 3a function in vivo, the 3a gene of the CUHK-W1 SARS-CoV isolate (AY278554) was used to generate pUAST-EGFP-3a and pUAST-3a constructs. When either EGFP- or non-tagged 3a transgene was misexpressed in the Drosophila eye, a rough eye phenotype was observed (Fig. 1B and supplementary Fig. 2B). When misexpressed in the third instar larval eye imaginal disc and salivary gland cells, both EGFP-3a and non-tagged 3a proteins showed similar punctate cytoplasmic localizations (Figs. 1D and F, and supplementary Fig. 2D) as in SARS-CoV-infected cells (supplementary Fig. 1C). Since both EGFP-3a and 3a transgenes showed comparable rough eye phenotype (Fig. 1B and supplementary Fig. 2B), EGFP-3a was mainly used in subsequent analyses.
Endocytosis and 3a function
A YXXΦ motif for rapid protein internalization has been identified in 3a [3], and similar sequences are also found in many cellular proteins, for example, the human transferrin receptor (TfR, [13]). It has been shown that internalization of TfR requires Eps15, an endocytic protein involved in clathrin-mediated endocytosis. When EGFP-3a was misexpressed in an Eps15 mutant background [22], a dominant suppression of the EGFP-3a-induced rough eye phenotype was observed (Fig. 2 C). Nedd4 is an E3 ubiquitin ligase and is able to monoubiquitinate Eps15 for its endocytic function [23]. We further found that a P-element insertion (Nedd4 EY00500) in the Nedd4 gene also dominantly suppressed the EGFP-3a-induced rough eye phenotype (Fig. 2D). Our data support previous findings that 3a is involved in protein trafficking [15], and our results highlight the involvement of clathrin-dependent endocytosis in 3a function.
Fig. 2.

Genes involved in endocytosis are related to the EGFP-3a-induced rough eye phenotype. Unlike the gmr-GAL4 control (A), EGFP-3a misexpression resulted in a rough eye phenotype (B). A mutant allele of the endocytic gene Eps15 (Eps15EP2513) dominantly suppressed the EGFP-3a-induced rough eye phenotype (C). Nedd4 monoubiquitinates Eps15 and such modification is essential for Eps15 function. A P-element insert line in the Nedd4 locus (Nedd4EY00500) showed dominant suppression of the EGFP-3a phenotype (D).
Misexpression of SARS 3a induces apoptosis in vivo
Apoptosis has been reported in SARS-CoV-infected cells [24], [25], [26], [27]. We therefore investigated whether the 3a-induced rough eye phenotype (Fig. 1B and supplementary Fig. 2B) was related to apoptotic cell death in flies. We performed acridine orange (AO) staining in third instar larval eye imaginal discs (Figs. 3 G–M) and observed increased numbers of AO-positive apoptotic cells in EGFP-3a-expressing eye discs (Fig. 3J). Consistently, eye discs misexpressed with the non-tagged 3a also showed increased numbers of AO-positive cells (supplementary Fig. 2F). When EGFP-3a was coexpressed with anti-apoptotic genes such as the caspase inhibitor P35, and the Drosophila inhibitor of apoptosis 1 (DIAP1), the rough eye phenotype was mostly suppressed (Figs. 3E and F). In addition, the number of AO-positive cells was also largely reduced when P35 or DIAP1 was coexpressed with EGFP-3a (Figs. 3K and L). Our data therefore confirmed that the SARS-CoV 3a gene is pro-apoptotic and its action can be counteracted by anti-apoptotic factors.
Fig. 3.

Apoptotic pathway is related to the EGFP-3a-induced rough eye phenotype. (A–F) Overexpression of anti-apoptotic genes suppresses the EGFP-3a-induced rough eye phenotype. gmr-GAL4 alone (A) and misexpression of EGFP-lacZ fusion protein by gmr-GAL4 (B) showed no external eye deformation. Overexpression of the pro-apoptotic gene reaper caused a rough eye phenotype and reduction in eye size (C). The EGFP-3a-induced rough eye phenotype (D) was suppressed when coexpressed with either P35 (E) or DIAP1 (F). (G–M) Coexpression of anti-apoptotic genes reduces the number of apoptotic cells in third instar larval eye discs. Comparable numbers of acridine orange-positive apoptotic cells were observed in gmr-GAL4 control eye discs (G) and discs misexpressed with a GFP-lacZ fusion protein (H). Expression of both reaper (I) and EGFP-3a (J) showed increased number of acridine orange-positive cells in eye discs. The EGFP-3a-induced apoptosis was suppressed by coexpression of baculoviral anti-apoptotic genes P35 (K), DIAP1 (L); and cytochrome c dc3 (M). Arrows indicate the location of the morphological furrows.
A genetic screen to define 3a functions in vivo
To further investigate the intracellular regulatory functions of 3a in vivo, we initiated a forward genetic screen to identify genes that would modify the EGFP-3a-induced rough eye phenotype (Fig. 1B). Based on overlapping chromosomal deletions, 24 genomic regions were isolated (see experimental procedures). By crossing all mutant lines available at the Bloomington Drosophila Stock Center that fall into these 24 modifying genomic regions to EGFP-3a flies, 78 individual genes (represented by 99 mutant lines) showed reproducible modification of the EGFP-3a-induced rough eye phenotype. Forty-six of these modifier genes (∼59%) have human orthologues and are also expressed in the human lung (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene). Based on their cellular functions, most of the EGFP-3a modifiers were classified into 5 categories including electron transport, gene transcription, protein posttranslational modification, calcium-binding, and nucleic acid-binding. Nineteen modifying loci are discussed here (Table 1 ).
Table 1.
19 loci that dominantly modify the EGFP-3a-induced rough eye phenotype
| Category | Gene | Allele | Modification (Strength)a | Molecular functionb | Human homologueb | Human lung expressionc |
|---|---|---|---|---|---|---|
| Electron transport | Cyt-c-d (dc3) | Cyt-c-dEP2305 | Su(+) | Electron transporter activity; oxidoreductase activity; electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity | Cytochrome c, somatic (CYCS) | Yes |
| Calcium-binding | CalpB | EP(3)875 | Su(++) | Calmodulin binding; calpain activity; calcium ion binding | Calpain 9 (CAN9) | Yes |
| Csp | EP(3)659 | Su(+) | ATPase activity, coupled | DnaJ (Hsp40) homolog, subfamily C, member5 (DNAJC5) | Yes | |
| EP(3)3141 | Su(+) | |||||
| Past1 | Past1EY01852 | E(−) | Calmodulin binding; calcium ion binding | EH-domain containing 1 (EHD1) | Yes | |
| Protein post-translational modification | lwr | lwr02858 | Su(+) | SUMO conjugating enzyme activity; ligase activity; protein binding; ubiquitin conjugating enzyme activity | Ubc9 | Yes |
| Nedd4 | Nedd4EY00500 | Su(+) | Ubiquitin–protein ligase activity | Neural precursor cell expressed, developmentally down-regulated 4-like (NEDD4L) | Yes | |
| Gene transcription | CG12313 | EP(3)3367 | Su(++) | — | Carboxyl-terminal domain, RNA polymerase II, polypeptide A small phosphatase 1 (CTDSP1) | Yes |
| CG6994 | EP(3)691 | Su(++) | Transcription factor activity | — | — | |
| emc | emc[D] | NM | Transcription corepressor activity | Inhibitor of DNA binding | Yes | |
| emc03620 | Su(++) | |||||
| CtBP | Protein C-terminus binding; transcription corepressor activity | C-terminal-binding protein 2 (CTBP2) | Yes | |||
| CtBPEP3352 | Su (+) | |||||
| Nucleic acid binding | CG16940 | CG16940KG02284 | E(−) | Nucleic acid binding; exoribonuclease activity | — | — |
| CG9705 | CG9705KG07795 | E(−) | RNA binding; nucleic acid binding | Calcium regulated heat stable protein 1, 24 kDa (CARHSP1) | Yes | |
| Aats-ile | Aats-ile00827 | Su(+) | RNA binding; isoleucine-tRNA ligase activity | Isoleucine-tRNA synthetase (IARS) | Yes | |
| danr | P{GT1}BG01545 | E(−) | DNA binding | — | — | |
| dan | EP(3)798 | Su(++) | DNA binding | — | — | |
| Unknown functions | CG16971 | CG16971EP3581 | Su(++) | — | — | — |
| l(3)L3809 | l(3)L3809L3809 | Su(++) | — | — | — | |
| CG10252 | PsEY02072 | Su(++) | — | — | — | |
| CG10365 | CG10365KG00107 | E(−) | — | Hypothetical protein MGC4504 | Yes |
Strengths of modification: NM, no modification; Su(++), strong suppressor; Su(+), weak suppressor; E(−), weak enhancer.
Description of “Molecular Function” was adapted from FlyBase reports (http://www.flybase.net/).
Based on information obtained from the NCBI UniGene database.
Cytochrome c dominantly modifies 3a-induced rough eye phenotype
One of the 3a modifiers identified from our screen is cytochrome c (Fig. 4 C). In Drosophila, there are 2 cytochrome c genes (dc3 and dc4) and they are both located at the 36A10-B3 region (Fig. 4E). A deletion line Df(2L)Exel6039, which uncovers this region, dominantly enhanced the EGFP-3a-induced rough eye phenotype (Fig. 4C). We further showed that cytochrome c protein levels were significantly reduced in this deletion line (Fig. 4F). Conversely, overexpression of the endogenous dc3 gene (Figs. 4G and H), by means of an EP insert line dc3 EP2305, dominantly suppressed the EGFP-3a phenotype (Fig. 4D). Our data therefore suggest that the severity of the EGFP-3a-induced rough eye phenotype correlates with cellular cytochrome c levels. In support of this, a reduction in the number of AO-positive cells was observed when EGFP-3a was coexpressed with dc3 EP2305 (Fig. 3M). Further, we showed that EGFP-3a overexpression did not affect mRNA expression levels of both dc3 and dc4 genes (Fig. 4G).
Fig. 4.

Cellular cytochrome c levels correlate with EGFP-3a-induced rough eye phenotype in Drosophila. Misexpression of EGFP-3a, but not EGFP (A), caused a rough eye phenotype (B). A chromosomal deletion Df(2L)Exel6039 of the cytochrome c gene region enhanced the EGFP-3a phenotype (C). In contrast, a P-element insert line, dc3EP2305, in the dc3 locus suppressed (D) the EGFP-3a-induced rough eye phenotype. (E) Genomic structure of cytochrome c genes in Drosophila shows that a P-element insert line, EP2305, is located in the 5′ untranslated region of the dc3 locus. (F) Western blot analysis showed a significant reduction of cytochrome c protein levels in the deletion line, Df(2L)Exel6039. (G) RT-PCR analysis showed overexpression of the dc3 gene in the P-element insert line, dc3EP2305. (H) Immunofluorescence of cytochrome c protein in salivary gland cells. Overexpressed cytochrome c protein, via dc3EP2305, was only detected by antibodies that recognize the denatured, but not native, cytochrome c.
Transcriptional dysregulation in 3a flies
The C-terminal-binding-protein (CtBP) is a transcriptional corepressor protein. From our screen, we found that a chromosome deletion line (87D8-10) and a mutant allele of CtBP [28] both dominantly suppressed the EGFP-3a phenotype (Figs. 5 C and D). It has been reported that CtBP undergoes SUMOylation and such protein posttranslational modification is mediated through an E2 ubiquitin conjugating enzyme, Ubc9 [29]. Consistent with this view, we found that a mutant allele of the fly orthologue of Ubc9, lesswright (lwr), showed dominant suppression of the EGFP-3a phenotype (Fig. 5E). It has recently been reported that CtBP cooperates with another transcriptional repressor, Suppressor of Hairless Su(H), to determine cell fates in Drosophila [30]. We further demonstrated that a mutant allele of Su(H) dominantly suppressed the EGFP-3a phenotype (Fig. 5F). Our data thus consolidate the genetic interaction of 3a with cellular transcription machineries.
Fig. 5.

EGFP-3a interacts with gene transcription regulatory machineries. Misexpression of the SARS-CoV 3a gene disrupted the normal external eye structure (A; gmr-GAL4) and caused a rough eye phenotype in Drosophila (B). The EGFP-3a-induced rough eye phenotype was dominantly suppressed by a chromosome deletion Df(3R)Exel8157 (C) which uncovers the 87D8-10 region, and by a null allele of C-terminal-binding protein (CtBP87De-10) (D). The EGFP-3a-induced rough eye phenotype was suppressed by the Ubc9 mutant lesswright (lwr02858) which is an E2 enzyme that SUMOylates CtBP (E). A P-element allele of Suppressor of hairless (Su(H)EY07695), inserted in the 5′ untranslated region of Su(H), dominantly suppressed EGFP-3a-induced rough eye phenotype (F).
It has been shown that CtBP represses gene transcription of other transcription regulators, including the helix–loop–helix transcription regulator Id1 [31]. The extra macrochaetae (emc) gene is the Drosophila orthologue of Id1 [32]. A chromosomal deletion (Df(3L)Exel6086) which uncovers the emc gene (the 61C9 region) dominantly suppressed the EGFP-3a-induced rough eye phenotype (Fig. 6 B). Interestingly, we showed that emc mRNA expression level was reduced in EGFP-3a flies (Fig. 6D) and overexpression of emc suppressed the EGFP-3a phenotype (Fig. 6C). Our data therefore suggest that 3a alters cellular gene transcription in vivo by interfering with transcriptional regulators including emc.
Fig. 6.
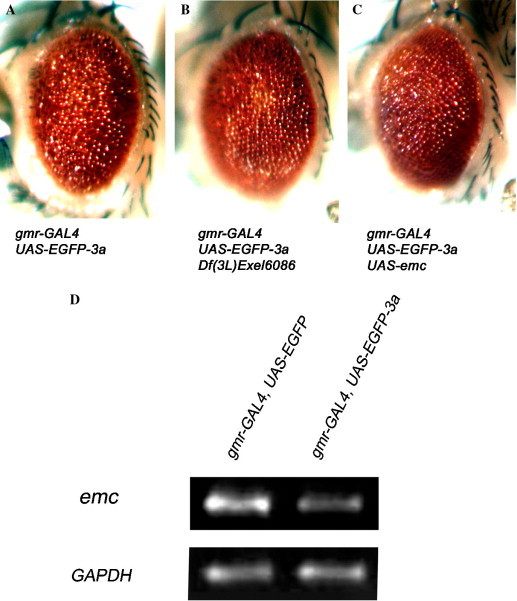
extra macrochaetae dominantly modifies EGFP-3a-induced rough eye phenotype. Misexpression of EGFP-3a caused a rough eye phenotype (A). A chromosomal deletion Df(3L)Exel6086 which uncovers the extra macrochaetae (emc) genomic region 61C9 (B), and an UAS-emc line (C) showed dominant suppression of the EGFP-3a phenotype. (D) RT-PCR analysis showed misexpression of EGFP-3a caused down-regulation of emc gene transcription.
Discussion
Expression of the SARS-CoV 3a protein has been detected both intracellularly in infected cells [5], [10] and in matured viral particles [11], [12]. This indicates that 3a is a viral protein which would bear both structural [11], [12] and intracellular regulatory [3], [15] functions. In this study, we established a 3a transgenic fly model and investigated the intracellular regulatory roles of 3a in vivo. Using monoclonal antibodies against specific 3a protein sequences, we observed comparable punctate cytoplasmic expression pattern of 3a in both Drosophila cells (Fig. 1 and supplementary Fig. 2) and SARS-CoV-infected Vero E6 cells (supplementary Fig. 1). Similar to mammalian cells, the 3a protein was also demonstrated to undergo parallel biochemical modifications in insect cells and also became assembled into viral-like particles [12]. Altogether, these data clearly illustrate the feasibility of the use of invertebrate insect model systems to investigate functional roles of 3a.
3a and endocytosis
The YXXΦ tetra-peptide motif is essential for the rapid internalization of proteins from the plasma membrane [13]. The 3a protein possesses such conserved YXXΦ motif (160–163 a.a.) and has been shown to be able to mediate specific internalization of 3a-specific IgG molecules to the cell interior from the culture medium [3]. Therefore, 3a has therefore been suggested to play some roles in protein trafficking in viral-infected cells [3], [15]. Here, we provided genetic evidence to support a trafficking role of 3a and further linked it to clathrin-mediated endocytosis (Fig. 2). Eps15 has been shown to play an important role in the endocytosis of TfR, a cellular YXXΦ-containing protein [33]. Nedd4 is an E3 ubiquitin ligase and has also been implicated in endocytosis and virus budding [34]. Nedd4 is able to monoubiquitinate Eps15 for its endocytic function [23]. We showed that mutant alleles of both Eps15 and Nedd4 dominantly suppressed the EGFP-3a phenotype (Fig. 2). Our data therefore support that 3a is functionally related to clathrin-mediated endocytosis. Given the physical interaction between 3a and other viral proteins (the S protein in particular, [6]), our results underscore the role of 3a in the intracellular trafficking of SARS-CoV proteins during the viral life cycle. In humans, both Eps15 and Nedd4 are expressed in the human lung (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene). This further highlights the functional relevance of these proteins to SARS-CoV pathogenesis.
3a is pro-apoptotic
We have recently shown that 3a induces apoptotic cell death in Vero E6 cells [35]. Here, we demonstrated the pro-apoptotic properties of 3a in vivo in our transgenic model (Fig. 3). We further showed that 3a-induced apoptosis can be suppressed by coexpression of anti-apoptotic genes including the inhibitor of apoptosis protein (IAP) DIAP1 (Figs. 3F and L), and the caspase inhibitor P35 (Figs. 3E and K). Interestingly, upregulation of gene expression of an IAP family protein, BIRC3, was also observed in SARS-CoV-infected cells [36]. Consistent with our findings that overexpression of the caspase inhibitor P35 suppressed the EGFP-3a-induced rough eye phenotype, caspase activation was also observed when 3a was expressed in Vero E6 cells [35].
Cytochrome c plays key roles in apoptosis via the mitochondrial pathway [37]. Since cytochrome c is a potent 3a modifier (Fig. 3), our data would suggest that 3a induces apoptosis through the mitochondria. Suppression of the EGFP-3a phenotype was observed when cytochrome c was overexpressed (Fig. 4D). Interestingly, we could only detect the overexpressed cytochrome c protein in the dc3 EP2305 line with antibodies that recognize the denatured, but not native, cytochrome c proteins (Fig. 4H). This indicates that the overexpressed cytochrome c protein was not in its native conformation. It has previously been reported that overexpression of the non-native apocytochrome c (cytochrome c protein without the heme group) can block apoptosis in mammalian cells [38]. We therefore reasoned that the dc3 EP2305 dominant suppression of the EGFP-3a phenotype is the result of the inhibition of apoptosis. Consistent with this view, we also observed reduced numbers of AO-positive apoptotic cells when EGFP-3a was coexpressed with dc3 EP2305 (Fig. 3M).
3a alters gene transcription
We found that loss-of-function of CtBP, a transcriptional repressor, dominantly suppressed the EGFP-3a-induced rough eye phenotype (Figs. 5C and D). Similar to CtBP, we showed that two other transcriptional regulators Su(H) and emc also dominantly suppressed the EGFP-3a phenotype (Fig. 5, Fig. 6) Further, CtBP has been shown to repress transcription of an emc orthologue, Id1, in mammalian cells [31]. It is therefore expected that the reduction of CtBP activity would lead to de-repression of emc. Consistent with this view, we observed suppression of the EGFP-3a phenotype when emc was overexpressed (Fig. 6C). We also observed that several EP lines (including EP3620 and EP0415) that have been mapped upstream of the emc ORF were capable of suppressing the EGFP-3a-induced rough eye phenotype (data not shown).
We further showed that the mRNA expression level of emc was reduced in EGFP-3a flies (Fig. 6D). This further supports that 3a alters gene transcription. Since emc is a transcriptional regulator, down-regulation of its expression would in turn affect the expression of downstream emc-responsive genes. It has also recently been shown that 3a altered gene expression in cultured cells [39]. Interestingly, transcriptional alteration of some of the 3a modifiers identified in our study (e.g., DIAP1 and emc) has also been detected in SARS-CoV-infected cells [36], [40]. However, we found that 3a does not affect gene expression level of all 3a modifiers. For example, we did not detect any alteration of dc3 mRNA expression in EGFP-3a flies (Fig. 4G). This would suggest that 3a is a multi-functional protein which affects multiple cellular processes.
In summary, we established and characterized a Drosophila transgenic model for the SARS-CoV 3a gene. To investigate the intracellular roles of 3a, we performed a genetic screen to isolate dominant 3a modifier genes. Our data indicate that 3a interacts genetically with multiple cellular machineries (Table 1). We further highlighted that 3a is functionally related to clathrin-mediated endocytosis (Fig. 2) and its misexpression causes apoptosis (Fig. 3, Fig. 4). Besides, 3a also interacts genetically with various transcriptional regulators (Fig. 5) and it alters cellular transcription activity (Fig. 6). This is the first example illustrating the use of transgenic fly models to investigate the functional roles of 3a and other SARS-CoV viral gene products in vivo.
Acknowledgments
We thank Erich Buchner, Jose de Celis, Shigeo Hayashi, Konrad Zinsmaier, and the Bloomington Drosophila Stock Center for antibodies and fly stocks; Susan Zusman for the generation of transgenic lines. We thank Patrick Law, Ben Chan, Parry Lee, and Philip Yeung for technical support; Cahir O’Kane, Tony Ip, and members of the LDR for critical comments on the manuscript. This work was supported, in part, by grants from the Health, Welfare and Food Bureau of Hong Kong (Research Fund for the Control of Infectious Diseases; 02040302); the Research Grants Council of Hong Kong (Special Competitive Earmarked Grant for SARS Research; CUHK4536/03M); and the Faculty of Medicine, CUHK (SARS Research Supplementary Grant: 6901535).
Footnotes
Abbreviations: a.a., amino acid; AO, acridine orange; Csp, cysteine string protein; dc3, cytochrome-c-distal; emc, extra macrochaetae; SARS-CoV, severe acute respiratory syndrome-Coronavirus; TfR, transferrin receptor.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbrc.2005.09.098.
Appendix A. Supplementary data
Supplementary Figures.
References
- 1.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 2.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 3.Tan Y.J., Teng E., Shen S., Tan T.H., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh A.D., Gupta D., Jameel S. Bioinformatic analysis of the SARS virus X1 protein shows it to be a calcium-binding protein. Curr. Sci. 2004;86:842–844. [Google Scholar]
- 5.Yu C.J., Chen Y.C., Hsiao C.H., Kuo T.C., Chang S.C., Lu C.Y., Wei W.C., Lee C.H., Huang L.M., Chang M.F., Ho H.N., Lee F.J. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 2004;565:111–116. doi: 10.1016/j.febslet.2004.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng R., Yang R.F., Shi M.D., Jiang M.R., Xie Y.H., Ruan H.Q., Jiang X.S., Shi L., Zhou H., Zhang L., Wu X.D., Lin Y., Ji Y.Y., Xiong L., Jin Y., Dai E.H., Wang X.Y., Si B.Y., Wang J., Wang H.X., Wang C.E., Gan Y.H., Li Y.C., Cao J.T., Zuo J.P., Shan S.F., Xie E., Chen S.H., Jiang Z.Q., Zhang X., Wang Y., Pei G., Sun B., Wu J.R. Characterization of the3a protein of SARS-associated coronavirus in infected vero E6 cells and SARS patients. J. Mol. Biol. 2004;341:271–279. doi: 10.1016/j.jmb.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan M., Chen H.Y., Foo S.Y., Tan Y.J., Goh P.Y., Wee S.H. Recombinant protein-based enzyme-linked immunosorbent assay and immunochromatographic tests for detection of immunoglobulin G antibodies to severe acute respiratory syndrome (SARS) coronavirus in SARS patients. Clin. Diagn. Lab. Immunol. 2004;11:287–291. doi: 10.1128/CDLI.11.2.287-291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu M., Shi Y., Guo Z., Chen Z., He R., Chen R., Zhou D., Dai E., Wang X., Si B., Song Y., Li J., Yang L., Wang J., Wang H., Pang X., Zhai J., Du Z., Liu Y., Zhang Y., Li L., Wang J., Sun B., Yang R. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect. 2005 doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan Y.J., Goh P.Y., Fielding B.C., Shen S., Chou C.F., Fu J.L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E., Lim S.G., Hong W. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan X., Li J., Shan Y., Yang Z., Zhao Z., Chen B., Yao Z., Dong B., Wang S., Chen J., Cong Y. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005;109:191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome Coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen S., Lin P.S., Chao Y.C., Zhang A., Yang X., Lim S.G., Hong W., Tan Y.J. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 2005;330:286–292. doi: 10.1016/j.bbrc.2005.02.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trowbridge I.S., Collawn J.F., Hopkins C.R. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu. Rev. Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- 14.Schwegmann-Wessels C., Al-Falah M., Escors D., Wang Z., Zimmer G., Deng H., Enjuanes L., Naim H.Y., Herrler G. A novel sorting signal for intracellular localization is present in the S protein of a porcine coronavirus but absent from severe acute respiratory syndrome-associated coronavirus. J. Biol. Chem. 2004;279:43661–43666. doi: 10.1074/jbc.M407233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan Y.J. The severe acute respiratory syndrome (SARS)-coronavirus 3a protein may function as a modulator of the trafficking properties of the spike protein. Virol. J. 2005;2:5. doi: 10.1186/1743-422X-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battaglia P.A., Zito S., Macchini A., Gigliani F. A Drosophila model of HIV-Tat-related pathogenicity. J. Cell Sci. 2001;114:2787–2794. doi: 10.1242/jcs.114.15.2787. [DOI] [PubMed] [Google Scholar]
- 17.Leulier F., Marchal C., Miletich I., Limbourg-Bouchon B., Benarous R., Lemaitre B. Directed expression of the HIV-1 accessory protein Vpu in Drosophila fat-body cells inhibits Toll-dependent immune responses. EMBO Rep. 2003;4:976–981. doi: 10.1038/sj.embor.embor936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battaglia P.A., Ponti D., Naim V., Venanzi S., Psaila R., Gigliani F. The HIV-Tat protein induces chromosome number aberrations by affecting mitosis. Cell Motil. Cytoskeleton. 2005;61:129–136. doi: 10.1002/cm.20070. [DOI] [PubMed] [Google Scholar]
- 19.Chen D., Wang M., Zhou S., Zhou Q. HIV-1 Tat targets microtubules to induce apoptosis, a process promoted by the pro-apoptotic Bcl-2 relative Bim. EMBO J. 2002;21:6801–6810. doi: 10.1093/emboj/cdf683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spresser C.R., Carlson K.A. Drosophila melanogaster as a complementary system for studying HIV-1-related genes and proteins. J. Neurosci. Res. 2005;80:451–455. doi: 10.1002/jnr.20460. [DOI] [PubMed] [Google Scholar]
- 21.Hay B.A., Wassarman D.A., Rubin G.M. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 22.Tang H., Rompani S.B., Atkins J.B., Zhou Y., Osterwalder T., Zhong W. Numb proteins specify asymmetric cell fates via an endocytosis- and proteasome-independent pathway. Mol. Cell. Biol. 2005;25:2899–2909. doi: 10.1128/MCB.25.8.2899-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polo S., Sigismund S., Faretta M., Guidi M., Capua M.R., Bossi G., Chen H., De Camilli P., Di Fiore P.P. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 24.Mizutani T., Fukushi S., Murakami M., Hirano T., Saijo M., Kurane I., Morikawa S. Tyrosine dephosphorylation of STAT3 in SARS coronavirus-infected Vero E6 cells. FEBS Lett. 2004;577:187–192. doi: 10.1016/j.febslet.2004.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. Importance of Akt signaling pathway for apoptosis in SARS-CoV-infected Vero E6 cells. Virology. 2004;327:169–174. doi: 10.1016/j.virol.2004.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem. Biophys. Res. Commun. 2004;319:1228–1234. doi: 10.1016/j.bbrc.2004.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan H., Xiao G., Zhang J., Hu Y., Yuan F., Cole D.K., Zheng C., Gao G.F. SARS coronavirus induces apoptosis in Vero E6 cells. J. Med. Virol. 2004;73:323–331. doi: 10.1002/jmv.20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barolo S., Stone T., Bang A.G., Posakony J.W. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 2002;16:1964–1976. doi: 10.1101/gad.987402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kagey M.H., Melhuish T.A., Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 30.Castro B., Barolo S., Bailey A.M., Posakony J.W. Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless. Development. 2005;132:3333–3344. doi: 10.1242/dev.01920. [DOI] [PubMed] [Google Scholar]
- 31.Lin X., Liang Y.Y., Sun B., Liang M., Shi Y., Brunicardi F.C., Shi Y., Feng X.H. Smad6 recruits transcription corepressor CtBP to repress bone morphogenetic protein-induced transcription. Mol. Cell. Biol. 2003;23:9081–9093. doi: 10.1128/MCB.23.24.9081-9093.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrell J., Modolell J. The Drosophila extra macrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix–loop–helix protein. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- 33.Benmerah A., Poupon V., Cerf-Bensussan N., Dautry-Varsat A. Mapping of Eps15 domains involved in its targeting to clathrin-coated pits. J. Biol. Chem. 2000;275:3288–3295. doi: 10.1074/jbc.275.5.3288. [DOI] [PubMed] [Google Scholar]
- 34.Hicke L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell. Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 35.Law P.T.W., Wong C.H., Au T.C.C., Chuck C.P., Kong S.K., Chan P.K.S., To K.F., Lo A.W.I., Chan J.Y.W., Suen Y.K., Chan H.Y.E., Fung K.P., Waye M.M.Y., Sung J.J.Y., Lo Y.M., Tsui S.K.W. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- 36.Tang B.S., Chan K.H., Cheng V.C., Woo P.C., Lau S.K., Lam C.C., Chan T.L., Wu A.K., Hung I.F., Leung S.Y., Yuen K.Y. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome Coronavirus and human Coronavirus 229E. J. Virol. 2005;79:6180–6193. doi: 10.1128/JVI.79.10.6180-6193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polster B.M., Fiskum G. Mitochondrial mechanisms of neural cell apoptosis. J. Neurochem. 2004;90:1281–1289. doi: 10.1111/j.1471-4159.2004.02572.x. [DOI] [PubMed] [Google Scholar]
- 38.Martin A.G., Fearnhead H.O. Apocytochrome c blocks caspase-9 activation and Bax-induced apoptosis. J. Biol. Chem. 2002;277:50834–50841. doi: 10.1074/jbc.M209369200. [DOI] [PubMed] [Google Scholar]
- 39.Tan Y.J., Tham P.Y., Chan D.Z., Chou C.F., Shen S., Fielding B.C., Tan T.H., Lim S.G., Hong W. The severe acute respiratory syndrome coronavirus 3a protein up-regulates expression of fibrinogen in lung epithelial cells. J. Virol. 2005;79:10083–10087. doi: 10.1128/JVI.79.15.10083-10087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cinatl J., Jr., Hoever G., Morgenstern B., Preiser W., Vogel J.U., Hofmann W.K., Bauer G., Michaelis M., Rabenau H.F., Doerr H.W. Infection of cultured intestinal epithelial cells with severe acute respiratory syndrome coronavirus. Cell. Mol. Life Sci. 2004;61:2100–2112. doi: 10.1007/s00018-004-4222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures.


