Abstract
In this study, peptides were selected to recognize staphylococcal enterotoxin B (SEB) which cause food intoxication and can be used as a biological war agent. By using commercial M13 phage library, single plaque isolation of 38 phages was done and binding affinities were investigated with phage-ELISA. The specificities of the selected phage clones showing high affinity to SEB were checked by using different protein molecules which can be found in food samples. Furthermore, the affinities of three selected phage clones were determined by using surface plasmon resonance (SPR) sensors. Sequence analysis was realized for three peptides showing high binding affinity to SEB and WWRPLTPESPPA, MNLHDYHRLFWY, and QHPQINQTLYRM amino acid sequences were obtained. The peptide sequence with highest affinity to SEB was synthesized with solid phase peptide synthesis technique and thermodynamic constants of the peptide–SEB interaction were determined by using isothermal titration calorimetry (ITC) and compared with those of antibody–SEB interaction. The binding constant of the peptide was determined as 4.2 ± 0.7 × 105 M−1 which indicates a strong binding close to that of antibody.
Keywords: Phage display, Peptide, ELISA, Surface plasmon resonance (SPR), Isothermal titration calorimetry (ITC)
Phage display techniques have aroused interest as a new tool in protein–ligand interaction studies since described by Smith [1]. This technology has been used to detect peptides which show high affinity to target molecules required for immunological and biological studies, drug discovery, pharmacology, plant science, and also inorganic materials [2]. Also through this technology, detection of ligands which are specific target proteins is accomplished without animal or human immunization system.
Recombinant protein and/or peptides structures which have recognize capability of target molecules are fused on Ff class (M13, f1, and fd) of filamentous phages. DNA fragments which code peptides are added pVIII, pIII or pVI gen regions of phages capsid proteins. Random DNA sequences that coded different peptides on phage genomes supply different peptides or proteins which have different antigen specificities [3], [4], [5]. The foreign DNA sequences can be originated from a natural source, or it can be advisedly designed and synthesized chemically [5].
Using a process of biopanning, selection of peptides from phage library is accomplished. Phage library is added to immobilized target molecules which in plates. After incubation non-binding phages washed away and selectively bound phage to target molecules are eluted and amplified in host bacteria. These amplified phages serve as input for another round of biopanning. Rounds of selection are generally repeated two or three times in same way and affinity of each phage clone from eluted phages to target molecules are ascertained by phage-ELISA and sequences of peptide are determined [3], [6].
Phage display technology has been used for selection and production of molecules that have antibody characteristics. Recombinant antibodies have been selected against various analytes like staphylococcal enterotoxin B [6], spores of Bacillus anthracis [7], Art v1 glycoprotein [8], rotavirus NSP4 enterotoxin [9], coronavirus [10], Clostridium botulinum neurotoxin serotype A [11], Salmonella typhimurium [12], and human Gonadotropin-releasing hormone promoter [13] by phage display technology. It also has been used to design high affinity super antigens for immunochemical application and immunotherapy [14] and to choose specific molecules to inorganic materials [2].
In this study, we have used a peptide-phage display library to identify peptides binding to SEB. SEB, produced by Staphylococcus aureus, is a pyrogenic toxin responsible for staphylococcal food poisoning in humans and has been an attractive choice of biological aerosol weapon due to its inherent stability and high intoxication effect [15]. Numerous immunological techniques have been applied to the detection of SEB such as enzyme-linked immunosorbent assays (ELISA) [16]. Because the traditional immunological methods are in most cases time-consuming and inconvenient, many novel and rapid immunosensors have been developed for the detection of SEB based on piezoelectric crystal sensors [17], [18], surface plasmon resonance (SPR) sensors [19], [20], [21], and fiber optic sensors [22].
In this study, SEB active peptide sequences were determined using phage display technique. Different from the common approach, not only the affinities of selected phage clones towards SEB molecule were investigated but also the interactions of clones with other molecules which can be found in real food samples were determined by phage-ELISA. The affinities of selected phage clones to SEB were also visualized using surface plasmon resonance system. After determining the amino acid sequences of selected peptides, the peptide with highest affinity was synthesized by solid phase peptide synthesis and the interaction of peptide with SEB molecule was studied by isothermal titration calorimetry (ITC). The binding constant (k) and thermodynamic binding parameters (ΔH, ΔG) were determined for the interactions of peptide–SEB and antibody–SEB.
Materials and methods
Culture and media conditions. M13 phage display (PhD) library and Escherichia coli ER2738, used as the host organism, were purchased from New England Biolabs Inc. (Ipswich, MA, USA). E. coli was grown in LB medium (Merck KgaA, Darmstadt, Germany) To determine the number of the phage titers, different dilutions of phage clones were mixed with top agar (10 g tryptone, 5 g yeast extract, 5 g NaCl, 1 g MgCl26H2O, and 7 g agarose in 1 l DI water) and were spreaded on to LB/IPTG/X-Gal agar. Monoclonal M13 antibody (GE Healthcare, UK) labeled with HRP (horseradish peroxidase) and polyclonal SEB antibody were obtained from Abcam plc. (Cambridge, UK). Stapylococcal enterotoxin B (SEB), bovine serum albumin (BSA), β-glycosidase, hemoglobin, glycerol, Tween 20, polyethylene glycol (PEG), glycine, 2,2′-azino-bis [3-ethylbenziazoline-6-sulfonicacid] (ABTS), and l-cystine were from Sigma–Aldrich Chemical Co. (St. Louis, MO, USA). Na2HPO4, KH2PO4, Tris, and NaCl were from J.T. Baker (Netherlands), used as PBS and TBS.
Phage display protocol. A commercial M13 phage display (PhD) library containing 1.5 × 1013 pfu ml−1 and has 2.7 × 109 transformants was used to screen against SEB. Target molecule SEB was prepared in ELISA plates for screening. The plates were prepared by incubating 150 μg ml−1 SEB. After an overnight incubation unbound and weakly bound SEB molecules were removed from the ELISA plate well surface by discarding the solution in the wells. Ten microliters of aliquots from PhD library was added on immobilized SEB molecules. After 1 h incubation unbound and weakly bound phages were removed by washing with TBST (0.1% Tween 20) buffer. Bound phages were eluted from the SEB surface by using glycine–HCl solution (pH 2.2). To neutralize the well content 1 M Tris–HCl (pH 9.1) was used. Eluted phage clones were amplified by infecting liquid E. coli culture. The amplified phage clones were separated by centrifugation at (10,000 rpm, 4 °C, for 10 min). Separated phage clones were purified and concentrated with PEG/NaCl. After incubation the phage clones were pelleted by centrifugation (10,000 rpm, 4 °C, for 15 min). The phage pellet was dissolved in 0.02% NaN3 in TBS buffer. Stock solutions of the phage clones were prepared in 50% glycerol.
Phage-ELISA protocol. Phage clones from the screening were quantitatively tested for their binding affinity using phage-ELISA. Phage clones were incubated in SEB coated ELISA plates for 1.5 h. Unbound and weakly bound phages were removed by washing with TBST (0.5% Tween 20) buffer. HRP-conjugated M13 antibody was brought in contact with phage–SEB complex and then the antibody was incubated for 1.5 h. Following the incubation none specifically bound antibody conjugates were removed by washing six times with TBST. Later 21 ml ABST stock solution was mixed with 36 μl H2O2 (30% v/v). Two hundred microliters of this substrate solution was distributed to each well and the enzymatic reaction took place for 60 min. The green color was measured at 405 nm by BIO-TEK EL 808 microplate reader (Biotek Instruments, Winooski, VT, USA).
Cross-specificity experiments. In order to check the specificity of the SEB-binding peptides, BSA, β-glycosidase, whey, and hemoglobin were tested as target molecules for selected phage clones. For screening the affinity of clones, ELISA plate wells were coated with these target molecules (150 μg ml−1) and incubated for over night. Phage clones were added to target molecules and incubated for 1.5 h. Unbound clones were removed by washing six times with TBST (0.5% Tween 20) buffer. Phage-ELISA protocol described above was followed to quantify the binding affinity of SEB-binding peptides towards different proteins.
Surface plasmon resonance (SPR) sensor measurements. The affinity of selected phage clones to SEB was further analyzed by SPR sensor. Spreeta™ sensors (Texas Instruments, Dallas, TX) were used for SPR experiments. Spreeta™ sensors are capable of monitoring RI changes between 1.320 and 1.368 refractive index units (RI) and the resolution of the system is approximately 5 × 10−6 RI. The SPR system is comprised of a SPR sensor, a 3-channel flow cell and 12-bit DSP electronic control box. Goldman syringe pump (Biasis Ltd. Sti., Ankara, Turkey) controls the flow of reagents into the flow cell at a rate of 50 μl min−1. The changes in response unit (RU) (1 RU = 10−6 RI) with respect to time are plotted in the form of sensorgrams.
First, a baseline was established by injecting PBS (0.1 M, pH 7.4) to the sensor surface. Following the PBS injection, 650 μl of SEB (0.1 mg ml−1) in PBS was injected to the flow cell and its adsorption onto the gold surface was monitored. The unbound SEB was washed off by injecting PBS and non-specific sites on the sensor surface were blocked with 600 μl cystine (2 mM) in PBS.
The samples of 650 μl containing selected phage clones (1010 pfu ml−1) suspended in PBS were injected to the two sensing flow cells and PBS was injected to the control flow cell. The unbound phage clones were washed with PBS and the adsorption of the phage clones to the SEB immobilized sensor surface was monitored. Binding of phage clones to the sensor surface was marked by the corresponding RU changes. The net response was calculated by subtracting signals of sensing and control channels.
DNA sequencing. Phage particles purification, concentration and DNA isolation were carried out according to Sambrook et al. [23]. Sequencing analysis were done using by Beckmann Coulter CEQ™ 8000 cycle sequencer with −96 sequencing primer (New England BioLab, 5′-GCCCTCATAGTTAGCGTAACG-3′) and 5′-CAGGGATAGCAAGCCCAATA-3′.
Isothermal titration calorimetry (ITC) measurements. After DNA sequencing, the peptide of phage clone number 24 was synthesized by solid phase peptide synthesis (GenScript Corp., Piscataway, NJ, USA). ITC was used to determine the thermodynamic properties of peptide–SEB interaction. ITC experiments were performed at 25 °C using a TAM III microcalorimeter (TA Instruments, New Castle, Delaware, USA). Before analysis, the ITC calibration was performed [24] by diluting aqueous sucrose solution (Sigma Co., St. Louis, MO). The ΔH of dilution determined was within the acceptable limits, confirming that the instrument was functioning properly.
Peptide and SEB were suspended in PBS (0.1 M, pH 7.4) and the reference ampoule was filled with PBS solution. Peptide solution (0.35 mM) was placed in 250 μl syringe and 800 μl of SEB solution (0.035 mM) was placed in the sample ampoule. The peptide solution was injected into the stirred sample ampoule containing SEB solution in 25 injections (7 μl/injection) by a computer-controlled pump. Heat released with each injection was measured by the calorimeter in dynamic correction mode. Blank experiments were performed by titrating peptide solution into PBS and PBS into SEB solution. In order to determine the thermodynamic constants of antibody–SEB interaction, SEB solution (0.0175 mM) was placed in syringe and 800 μl of anti-SEB solution (0.0014 mM) was placed in the sample ampoule. The SEB solution was injected into the sample ampoule in 25 injections (7 μl/injection). Binding constants (k), the change in enthalpy (ΔH), the change in free energy (ΔG) and the change in entropy (ΔS) were determined by using the software package of TAM Assistant supplied by TA Instruments. Binding stoichiometry was estimated as 1:1 for both two interactions.
Result and discussion
Phage-ELISA
Toxins, which cause intoxications and accepted as bioterrorism agents, were usually detected by microbiological techniques. Because those conventional techniques are comparatively time-consuming, lots of novel methods were developed to detect toxins in a much more rapid and easier way [4]. In this study, phage-display technique was used to select the peptides which can be used as the recognition agents in the detection of staphylococcal enterotoxin B (SEB). Phage clones from the third round of biopanning were isolated and affinities of peptides to SEB were determined by ELISA. The results (Fig. 1 ) show that 16 of 38 phage clones were positive ones. Phage clone number 24, 9, and 6 showed higher SEB-binding affinity and were chosen for further analysis.
Fig. 1.
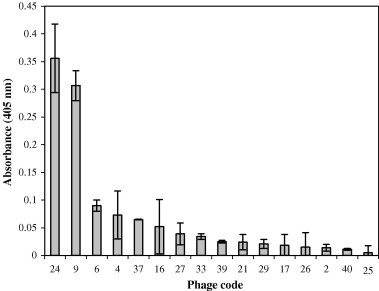
The affinities of phage clones to SEB molecule realized by phage-ELISA.
Cross reactivity with other protein molecules
The specificity of the selected phage clones to SEB was tested by ELISA using different proteins found in food matrices frequently. The affinities of phage clones 24, 9, and 6 to SEB, albumin, whey, hemoglobin, and β-glycosidase were determined (Fig. 2 ). Phage clone 24 showed minimal cross reactivity with other proteins when compared with SEB. For phage clones 9 and 6, significant cross reactivity was observed with hemoglobin and β-glycosidase. None of the phage clones bound significantly to the bovine serum albumin (BSA) and whey, although BSA is a highly hydrophobic protein.
Fig. 2.
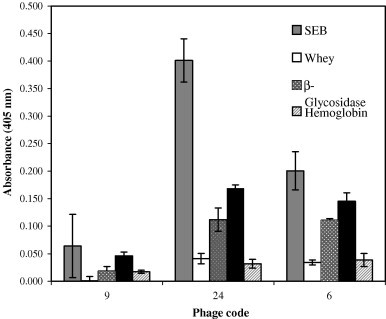
Cross-specificity of selected phage clones with other protein molecules.
SPR measurements
In order to investigate the affinities of selected phage clones to SEB, phage clones were allowed to bind to the SEB immobilized on the sensor surface. Fig. 3 A shows the immobilization of SEB molecule via physical adsorption and blocking the surface by cystine. Response unit (RU) was increased during the adsorption of SEB and a small decrease in signal was observed after washing with PBS due to the removal of loosely bound molecules. Cystine solution was used for blocking the non-specific sites on the sensor surface and a remarkable RU change was observed due to the binding of cystine molecules to the sensor surface.
Fig. 3.
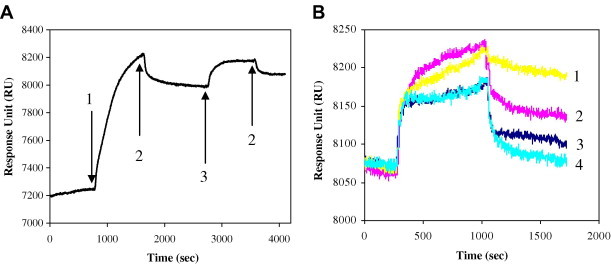
(A) SPR sensorgram showing the immobilization of SEB. Injection of (1) SEB (0.1 mg ml−1); (2) PBS and (3) cystine (2 mM). (B) Overlay plot of sensorgrams showing the binding of phage clones. (1) Phage clone 3; (2) phage clone 9; (3) phage clone 6; and (4) non-binding phage clone (25).
Phage clones (phage 24, phage 9, and phage 6), selected by phage-ELISA technique, were injected to the sensor surface on which SEB was immobilized and the change in the RU induced by the binding of phage clones is shown in Fig. 3B. A phage clone (phage 25), which showed low affinity to the SEB in phage-ELISA, was also injected to the sensor surface in order to investigate the selectivity of the SPR sensor. The concentrations of all phage clones used in SPR measurements were kept constant at 1010 pfu ml−1.
Following the injection of phage clones, RU signal increased due to the refractive index difference between PBS and phage solution. During the period of adsorption, RU of phage clones 24 and 9 increased while the signal of phage clones 6 and 25 did not change significantly, although the concentrations of phage solutions were same. This shows the high affinity of phage clones 24 and 9 to the SEB molecule, because when phages bind to the SEB molecule, they get closer to the surface of the sensor which increases the RU. The RU difference before the injection of phage solution and after washing with PBS indicates the amount of phage clones bound to the sensor surface. The change in RU induced by phage clone 24 was highest, followed by phage clones 9 and 6 which were similar to the results of phage-ELISA. The change in response for non-binding phage clone (25) was too small which shows the specificity of the SPR sensor.
Sequence analysis
The nucleotide sequences of the random regions of SEB-binding phage clones (phage clone number 24, 9, and 6) and weakly or non-binding phage clones (phage clone number 12 and 25) were determined. The nucleotide sequences and corresponding amino acid sequences of those phage clones were reported in Table 1 . The random regions of SEB-binding phage clones do not share any consensus sequence. This indicates that SEB-binding phage clones show affinity to different epitopes of the SEB molecule. While phage clone 9 is rich in glutamine, phage clone 6 is rich in proline.
Table 1.
DNA and amino acid sequences of peptides which show high affinity to SEB molecule
| Phage code | DNA sequences | 12-mer peptides |
|---|---|---|
| 24 | ATGAATCTTCATGATTATCATAGGCTGTTTTGGTAT | MNLHDYHRLFWY |
| 9 | CAGCATCCTTAGATTAATTAGACGCTGTATCGTATG | QHPQINQTLYRM |
| 6 | TGGTGGCGTCCTCTGACTCCTGAGTCGCCGCCTGCG | WWRPLTPESPPA |
Isothermal titration calorimetry (ITC) measurements
Detection of thermodynamic parameters for peptide–SEB interaction by ITC provides valuable information about the binding of peptide to SEB. Raw ITC profiles obtained by the titration of peptide to the SEB solution and the corrected injection heats are shown in Fig. 4 . The interaction between peptide–SEB was exothermic with a binding constant (k) of 4.2 ± 0.7 × 105 M−1. An enthalpy change (ΔH) of –32.0 ± 2.0 kJ mol−1 and free energy change (ΔG) of −32.12 kJ mol-1 was obtained from the ITC data. The interaction between antibody–SEB was also exothermic with a higher binding constant (k = 1.6 ± 0.8 × 107 M−1). The ΔH and ΔG values for antibody–SEB interaction were −369 ± 29 kJ mol−1 and −41.06 kJ mol−1, respectively. The interactions were accompanied by a large change in enthalpy and a large negative value in ΔG represents a high affinity [25]. Although the binding constant of the peptide is lower than that of the antibody, it still indicates a strong binding to SEB molecule.
Fig. 4.
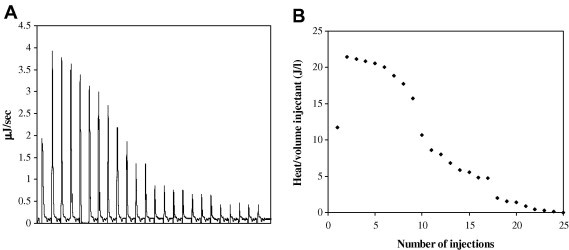
ITC profiles for peptide–SEB interaction. (A) Heat flow against time resulting from sequential injections and (B) normalized heat data against injections.
Conclusion
By using phage display technique, three peptides with high affinities to SEB molecule were selected. As the result of cross-specificity test, although there were significant amount of phages clones bound non-specifically to the other protein molecules, the selected phage clones showed higher affinity to SEB than the others. The non-specific binding of phage clones may be due to the hydrophobic regions in the phage coat protein. The SPR sensorgrams show that, phage 24 and phage 9 bind to SEB with a strong affinity. After DNA sequencing, the peptide of the phage clone with highest affinity was synthesized and the binding constant of peptide to SEB was determined by ITC. The results indicate that the selected peptides can be used as recognizing agents in biosensors. In addition to their high affinity, the small size they have, will be an important advantage in enhancing the sensitivity of biosensors like SPR. In further studies, the selected peptides will be combined with SPR sensors allowing the detection of SEB with a sensitivity comparing favorably to that of immunosensors.
Acknowledgments
This work was supported by a grant project 2006 K 120 640 06 05 of Republic of Turkey Prime Ministry State Planning Organization (DPT). Esra Acar Soykut, one of the authors of article, was financially supported by The Scientific and Technological Research Council of Turkey (TUBITAK-BIDEB).
References
- 1.Smith G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 2.Sarikaya M., Tamerler C., Jen A.K.Y., Schulten K., Baneyx F. Molecular biomimetics: nanotechnology through biology. Nat. Mater. 2003;2:577–585. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- 3.Smith G.P., Petrenko V.A. Phage display. Chem. Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 4.Petrenko V.A., Vodyanoy V.J. Phage display for detection of biological threat agents. J. Microbiol. Methods. 2003;53:253–262. doi: 10.1016/s0167-7012(03)00029-0. [DOI] [PubMed] [Google Scholar]
- 5.Petrenko V.A., Sorokulova I.B. Detection of biological threats. A challenge for directed molecular evolution. J. Microbiol. Methods. 2004;58:147–168. doi: 10.1016/j.mimet.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Goldman E.R., Pazirandeh M.P., Mauro J.M., King K.D., Frey J.C., Anderson G.P. Phage-displayed peptides as biosensor reagents. J. Mol. Recognit. 2000;13:382–387. doi: 10.1002/1099-1352(200011/12)13:6<382::AID-JMR511>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Williams D.D., Benedek O., Turnbough C.L. Species-specific peptide ligands for the detection of Bacillus anthracis spores. Appl. Environ. Microbiol. 2003;69:6288–6293. doi: 10.1128/AEM.69.10.6288-6293.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruber P., Dedic A., Gadermaier G., Wagner S., Breiteneder H., Ferreira F. Production of recombinant single-chain Fv antibody fragments specific for the hydroxyproline-rich domain of natural Art v 1 by phage display. J. Allergy Clin. Immunol. 2004;113:S300. [Google Scholar]
- 9.Rodriguez-Diaz J., Monedero V., Perez-Martinez G., Buesa J. Single-chain variable fragment (scFv) antibodies against rotavirus NSP4 enterotoxin generated by phage display. J. Virol. Methods. 2004;121:231–238. doi: 10.1016/j.jviromet.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z., Yi G., Qi Y., Liu Y., Yan J., Qian J., Du E., Ling W. Identification of single-chain antibody fragments specific against SARS-associated coronavirus from phage-displayed antibody library. Biochem. Biophys. Res. Commun. 2005;329:437–444. doi: 10.1016/j.bbrc.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma H., Zhou B., Kim Y.S., Janda K.D. A cyclic peptide-polymer probe for the detection of Clostridium botulinum neurotoxin serotype A. Toxicon. 2006;47:901–908. doi: 10.1016/j.toxicon.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Steingroewer J., Bleya T., Bergemannb C., Boschke E. Biomagnetic separation of Salmonella typhimurium with high affine and specific ligand peptides isolated by phage display technique. J. Magn. Magn. Mater. 2007;311:295–299. [Google Scholar]
- 13.Xiao Y., Zhou Y., Wang J., Yu M., Wang G., Jin J., Xiao J. Selection and identification of human Gonadotropin-releasing hormone promoter binding peptides by phage display-CEMSA. J. Mol. Recognit. 2007;20:51–57. doi: 10.1002/jmr.811. [DOI] [PubMed] [Google Scholar]
- 14.Enever C., Tomlinson I.M., Lund J., Levens M., Holliger P. Engineering high affinity superantigens by phage display. J. Mol. Biol. 2005;347:107–120. doi: 10.1016/j.jmb.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 15.Ler S.G., Lee F.K., Gopalakrishnakone P. Trends in detection of warfare agents: detection methods for ricin, staphylococcal enterotoxin B and T-2 toxin. J. Chromatogr. A. 2006;1133:1–12. doi: 10.1016/j.chroma.2006.08.078. [DOI] [PubMed] [Google Scholar]
- 16.Bhatti A.R., Siddiqui Y.M., Micusan V.V. Highly sensitive fluorogenic enzyme-linked immunosorbent assay: detection of staphylococcal enterotoxin B. J. Microbiol. Methods. 1994;19:179–187. [Google Scholar]
- 17.Harteveld J.L.N., Nieuwenhuizen M.S., Wils E.R.J. Detection of staphylococcal enterotoxin B employing a piezoelectric crystal immunosensor. Biosens. Bioelectron. 1997;7:661–667. doi: 10.1016/s0956-5663(97)00024-9. [DOI] [PubMed] [Google Scholar]
- 18.Lin H.C., Tsai W.C. Piezoelectric crystal immunosensor for the detection of staphylococcal enterotoxin B. Biosens. Bioelectron. 2003;18:1479–1483. doi: 10.1016/s0956-5663(03)00128-3. [DOI] [PubMed] [Google Scholar]
- 19.Naimushin A.N., Soelberg S.D., Nguyen D.K., Dunlap L., Bartholomew D., Elkind J., Melendez J., Furlong C.E. Detection of Staphylococcus aureus enterotoxin B at femtomolar levels with a miniature integrated two-channel surface plasmon resonance (SPR) sensor. Biosens. Bioelectron. 2002;17:573–584. doi: 10.1016/s0956-5663(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 20.Slavík R., Homola J., Brynda E. A miniature fiber optic surface plasmon resonance sensor for fast detection of staphylococcal enterotoxin B. Biosens. Bioelectron. 2002;17:591–595. doi: 10.1016/s0956-5663(02)00013-1. [DOI] [PubMed] [Google Scholar]
- 21.Homola J., Dostálek J., Chen S., Rasooly A., Jiang S., Yee S.S. Spectral surface plasmon resonance biosensor for detection of staphylococcal enterotoxin B in milk. Int. J. Food Microbiol. 2002;7:61–69. doi: 10.1016/s0168-1605(02)00010-7. [DOI] [PubMed] [Google Scholar]
- 22.King K.D., Anderson G.P., Bullock K.E., Regina M.J., Saaski E.W., Ligler F.S. Detecting staphylococcal enterotoxin B using an automated fiber optic biosensor. Biosens. Bioelectron. 1999;14:163–170. doi: 10.1016/s0956-5663(98)00108-0. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J., Fritsch E.F., Maniatis T. second ed. Cold Spring Habour Laboratory Press; New York: 1989. Molecular Cloning—A Laboratory Manual. [Google Scholar]
- 24.Wadsö I., Goldberg R.N. Standards in isothermal microcalorimetry (IUPAC technical report) Pure Appl. Chem. 2001;73:1625–1639. [Google Scholar]
- 25.V.V. Andrushchenko, M.H. Aarabi, L.T. Nguyen, E.J. Prenner, H.J. Vogel, Thermodynamics of the interactions of tryptophan-rich cathelicidin antimicrobial peptides with model and natural membranes, Biochim. Biophys. Acta, in press. [DOI] [PubMed]


