Abstract
A growing body of evidence indicates that exosomes play a critical role in the cell–cell communication process. Exosomes are biological nanoparticles with an average diameter of 30–100 nm in size and are produced by almost all cell types in the human body; however, cancer cells contain higher concentrations of exosomes than healthy cells. They are released into all body fluids and contain double-stranded DNA (originated from nucleus and mitochondria), a variety of RNA species, and specific protein biomarkers that can be utilized as cancer biomarkers and therapeutic targets, and lipids. Therefore, the specific exosomes secreted by tumor cells could be used to predict the existence of the presence of a tumor in cancer patients. This review summarizes the role of exosomes in cancer development and their potential utility in the clinic.
Keywords: : biomarkers, body fluids, cancer, exosomes, extracellular vesicles, miRNA
Lay abstract
Intercellular communication between tumor cells and their neighboring cells and distant organs is key to cancer’s survival, progression, drug resistance and metastasis. Circulating exosomes carry a specific composition of proteins, lipids, RNA and DNA and can work as cargo to transfer this information from donor cells to target cells, leading to reprogramming of the recipient cells. Exosomes are small particles with big roles in cancer. The present review summarizes the significant aspects of exosomes in cancer development and how they might provide novel strategies for use in the clinic.
Cancer is the second highest cause of death and a major health problem worldwide, with approximately 18 million new cases and 9.6 million deaths annually; this is 16.5% of human deaths that occurred in 2017 according to WHO [1]. More than half of the world’s population will be afflicted by invasive cancer at some point during their lifetime [2]. Early-stage cancer diagnosis is key for positive prognosis, which can extend lifespan and reduce the number of disease-related death [3,4]. Cancer can have around 20 years of incubation period before it is detectable by ultrasound, x-ray-based computer tomography, endoscopy or other detection methods including tissue biopsy. However, the new and highly sensitive detection method, liquid biopsy-based exosome analysis, provides a promising platform for early diagnosis, therapeutic and prognostic process about a disease rather than the conventional tissue biopsy. Liquid biopsy, a new star of cancer detection, is growing in popularity because of their minimal invasiveness, ease of use, painless, lower sample volume, lower cost, more accuracy and high throughput for personalized cancer therapy [5,6,7]. It is important to know that most cancers can be more effectively treated if they are discovered in early stages.
Cell communication and transformation are essential in tumorigenesis: single tumor cells must interact with each other and with nontumor host cells to enhance tumor cell growth, survive, progress, angiogenesis and metastasis [8]. It is becoming increasingly clear that tumor cell-derived exosomes play a key role in this communication process through the transfer of various biomolecules including proteins, lipids, DNA and RNA, which can be transferred in active form from the donor to the recipient cells [9]. They are extremely stable and resistant against degradation enzymes such as RNases and can keep their contents intact for a longer time than other materials such as cells.
Almost all cancer cells harbor aberrant expressions of microRNAs (miRNAs), that a key component of the small noncoding RNA family. These cancer-specific upregulated or downregulated short noncoding miRNAs are extremely important for the cancer development by altering oncogenes, tumor suppressor genes and therefore cancer-related signalling pathways. Onco-miRNAs are aberrantly expressed in cancer cells and target the degradation of key tumor suppressor messenger RNA to promote tumor formation or growth. On the other hand, the elevated expressions of tumor suppressive miRNAs inhibit tumor growth by inactivating expressions of oncogenes and hence are in general downregulated in cancer cell growth [10].
Exosomes & microvesicles
Almost all our body’s cells release a various types of nanometer-sized membrane-derived lipid bilayer vesicles into the extracellular environment, which are collectively termed as extracellular vesicles (EVs) [1,12]. EVs have been detected in a various biological fluid, including saliva, amniotic fluid, breast milk, semen, nasal secretion, cerebrospinal fluid, lymph, tear, aqueous humor, urine and blood plasma or serum.
These membrane-derived EVs can be classified into three different classes depending on their cellular origin, sizes and dimensions, mode of release, their contents, and functions. These are named as exosomes (∼30–100 nm), microvesicles (∼100–1000 nm) and apoptotic bodies (∼500–3000 nm). Exosomes were first discovered by Pan and Johnstone in 1983 [13].
Differently from the other cellular vesicles such as apoptotic bodies (highly heterogeneous in size and composition) that formed at the final stages of apoptosis from the plasma membrane, exosomes are of endocytic origin and microvesicles (ectosomes, shedding vesicles) are formed a direct outward budding of the cell’s plasma membrane. Structurally, exosomes are the smallest EVs, display a cup-like shape when examined by transmission electron microscopy and are more homogeneous in shape than the other EVs (Figure 1). Exosomes are formed as intraluminal vesicles by a process that involves the endocytic pathway and are secreted upon fusion of late endosomes or multivesicular bodies (MVBs) with the plasma membrane and are released into the extracellular space (exocytosis) [14,15,16,17,18,19,20,21].
Figure 1. . The structure and content of exosome.
Exosomes contain various types of proteins, nucleic acids, lipids and metabolites.
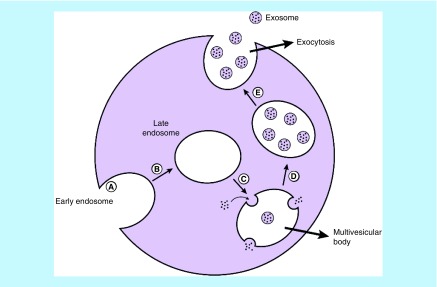
The formation of exosomes can be categorized into three different stages: formation of early endocytic vesicles from plasma membrane (early endosome), inward-budding of the endosomal vesicle membrane resulting in MVBs that consist of ILVs and fusion of these MVBs with either lysosome in which they are degraded, or the plasma membrane, which releases the vesicular contents, known as exosomes (late endosome) (Figure 2) [22]. During the exosome formation, cytoplasmic biomolecules including nucleic acids and proteins are trapped inside lumen. The contents of exosomes are sorted and loaded through either endosomal-sorting complexes required for transport system (ESCRT)-dependent (in cooperation with apoptosis-linked gene 2 interacting protein X (ALIX) and tumor-susceptibility gene 101 protein) or an ESCRT-independent (with tetraspanins proteins and lipids such as sphingosine-1-phosphate and ceramide dependent) mechanisms [23,24].
Figure 2. . The formation and releasing of exosome.
(A) Exosome is derived from early endosome formed from plasma membrane. (B) Early endosome becomes late endosomes. (C) Then forms early multivesicular bodies. (D) Late multivesicular bodies. (E) Late multivesicular bodies can either get degraded by lysosomes or fuse with the membrane to release exosomes.
Both healthy and cancerous cells may release membrane-bound exosomes into the extracellular space and body fluids. However, cancer cells can produce about tenfold more exosomes when compared with normal healthy cells. The entry of exosome into recipient cells usually is made up the processes called endocytosis that can be measured by using methods such as confocal microscopy or flow cytometry. Endocytosis is an umbrella term for a range of molecular internalization pathways in the uptake of exosomes [25].
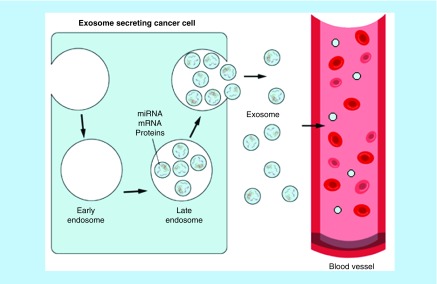
They are released from tumor cells (tumor-derived exosomes) into their surrounding extracellular space, and growing evidence indicates that these vesicles have multi functions including initiation of tumor progression, immune suppression, neovascularization, metastasis and drug resistance [26]. Exosomes are valuable sources for biomarker researches, as their contents are a wealth of information on the state of their cell of origin and function in biological processes and they are released in all biological fluids, including blood, tear, urine and saliva (Figure 3). Exosomes are also recognized as important mediators in cell to cell communication by transferring their contents (Figure 4). Indeed, many studies have now found evidence that exosomal contents, including double-stranded DNA, a variety of RNA species and specific protein biomarkers that are important as cancer predictive biomarkers for early cancer diagnosis and determination of prognosis. A total of 4.960 articles related to exosomes were identified in the study period from 1997 to 2017 by using Scopus and Web of Science [21].
Figure 3. . Exosomes biogenesis and sorting into the blood.
Exosome can be either fused with lysosomes for degradation or with plasma membrane thereby releasing exosomes to the extracellular space. Cell-released exosomes then can be taken up by neighboring recipient cells or travel through biological fluids such as blood, urine or saliva.
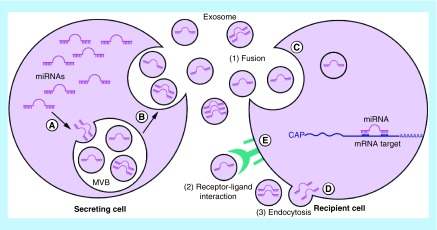
Figure 4. . Transfer of exosomal miRNA from donor cell to recipient cells.
(A) miRNAs are sorted to exosome during MVB formation. (B) Exosomes are released into the extracellular space. (C) Exosomal miRNAs can be delivered to recipient cells by endocytosis. (D) Fusion of the exosomes with the plasma membrane using soluble N-ethylmaleimide sensitive fusion into cells primarily uses receptor-mediated endocytosis. (E) Exosomes may also bind to a receptor and activate specific signaling pathways.
MVB: Multivesicular body.
Biological function of exosomes in cancer
Exosomes play a fundamental biological role in intercellular communication, induce physiological changes in recipient cells by transferring their cargo and have been implicated in many diseases such as cancer, cardiovascular diseases, autoimmune syndromes, neurodegenerative disorders and many others [2,27]. Tumor cell-released exosomes induce alterations in their recipient cells, thereby playing a crucial role in promotion of primary tumor development, stimulation of angiogenesis, activation of stromal fibroblasts, sculpting the cancer extracellular matrix adhesion, promotion of a premetastatic niche formation, suppression of the host immune response, resisting cell death and developing drug-resistance [17,28]. Tumor cell-released exosomes are also participating in the development of drug resistance to anticancer therapies and stimulate secretion of antiapoptotic proteins in tumor cells.
Exosomes contain various proteins such as ESCRT (ESCRT 0, I, II and III; which are required for transport), tetraspanins (transmembrane proteins induce vesicle formation), Rab GTPases (Rab7/Rab9/Rab11/Rab27/Rab35; which are essential for exosome release, tumor growth and metastasis), heat-shock proteins (HSP20/HSP60/HSP70/HSP90) and transforming growth factor β. Selecting, binding and uptaking of exosomes to the surface of recipient cells is mediated by different proteins such as tetraspanin family proteins (TSPAN: CD9, CD37, CD49, CD53, CD63, CD81 and CD82), immunoglobulins, proteoglycans, lectins and intercellular adhesion molecules (e.g., integrins with alpha subunits [ITGA], and with beta subunits [ITGB]) on the surface of both the exosome and the target cell. These attachments can then lead to direct delivery of exosomal cargo molecules into the recipient cell at new locations, and conceivably changing their biology (Table 1) [10,17,29,30,31].
Table 1. . Biological properties of some extracellular vesicles.
| Extracellular vesicles | Exosomes | Microvesicles (microparticles) | Apoptotic bodies |
| Size | 30–100 nm | 100–1000 nm | 500–3000 nm |
| Shape | Cup shaped | Irregular | Heterogeneous |
| Sedimentation | 100,000 × g or higher | 1200–100,000 × g | 1200–100,000 × g |
| Biomarkers | ALIX, TSG101, ESCRT-0, -I, -II and -III, tetraspanins-TSPAN/CD9/CD24/CD49/CD63/CD53/CD81/CD82, heat shock proteins- HSP20/HSP60/HSP70/HSP90, flotillins, GTPases- Rab11/Rab27/Rab31/Rab35. | Integrins-CD40/CD51/CD61, ligand, flotillin-2, metalloproteinase, selectins | Histones, annexin V positivity adhesion molecules, chemokines |
| Mode of release | Exocytosis, multivesicular endosomes | Exocytosis | Condensed apoptotic fragments |
ESCRT: Endosomal-sorting complexes required for transport system; TSG101: Tumor-susceptibility gene 101 protein.
Exosomes can act as carriers to transport oncogenic proteins and nucleic acids from donor tumor cell to normal recipient target cells at distance from the originating cell. These horizontal (lateral) molecular transfers of exosomal factors can modulate cell signalling pathways in transformed and even untransformed cells. The formation of new blood vessel (angiogenesis) is an important part of preparing a site for future colonization by cancer cells (Figure 5). To achieve this, cancer cells secrete exosomes that act in diverse ways to induce neoangiogenesis at their premetastatic niche, and promoting cancerous cell migration [10]. It has been demonstrated that tumor cell-released exosomal miRNAs such as miR-9, miR-23a, miR-92a, miR-103, miR-105, miR-126, miR-132 miR-135b, miR-210, miR-221 and cytokines (e.g., interleukins: IL-6 and IL-8, TNF-α, transforming growth factor β, FGF2, and VEGF) are proangiogenic factors to promote neovascularization and metastasis [32,33,34,35,36,37,38]. For example, exosomal miR-9 secreted by tumor cells, activates the Janus kinase/signal transducers and activators of transcription JAK/SAT pathway by reducing cytocine signaling 5 (SOCS5) levels to promote tumor angiogenesis [39]. Additional work showed that miR-105 induces vascular leakiness and promoting metastasis [33].
Figure 5. . Primary tumor and angiogenesis.
(A) Cell proliferate to form primary tumor formation (without blood vessels). (B) Tumor mass increase and produce angiogenic factors that stimulate new blood vessel formation from the main blood vessel toward the tumor cells.
Metastasis is the main cause of mortality in cancer patients, accounting for more than 90% of all cancer-related deaths [2,40]. Metastasis is an enormously complex process by which cancer cells originating from a malignant primary tumor spread and colonize in distant organs within the body, establishing secondary tumors in different tissues. Recent studies have shown that tumor cell-derived exosomes play a prominent role in the pathology of tumor metastases by using tumor-signaling pathway such as caveolin-1, HIF-1a, miR-21, miR-105, miR-210, β-catenin and oncogenic kinases (e.g., mutated EGFR, RAS and MAP kinases) [2,41,42,43,44].
Exosome isolation techniques
For an effective use of exosomes as source of biomarker discovery in liquid biopsies, highly pure exosome samples are required [26]. Therefore, the choice of the suitable separation and isolation method is important. Unfortunately, due to their small size with diameters between 30 and 100 nm and low density, exosomes are extremely difficult to define, isolate and purify from other component in the blood plasma and requires major time and effort. There are two crucial points that must be controlled to achieve a high quality in exosome preparations: the appropriate collection and storage of the bodily fluid samples (such as source of fluids, preparation conditions and storage temperature) and the purity and yield of the isolated exosomes.
In blood, plasma is preferred for exosomal microRNAs (exo-miRNAs) analysis, as the preparation of plasma is less complex and slightly easier than that for serum, because serum contains high numbers of vesicles released by platelets in response to coagulation [45,46]. A recent exo-miRNAs analysis of 312 human plasma and serum samples collected from 13 healthy volunteers indicated that 78% of total RNAs in plasma and 53% of total RNAs in serum was exo-miRNAs [47]. Blood plasma is a yellow liquid consists of water with many substances dissolved in it such as: mineral salts and ions, low/high molecular weight components, gases and metabolites, and acts as the extracellular matrix of blood cells. It represents approximately 55% of the body’s total blood volume.
For exosome isolation, collected blood (0.5–1.0 ml) in potassium EDTA-coated tubes should be processed within 30 min after collection. Samples are cool centrifuged at 1500 × g for about 10 min to remove dead cells and then at 10,000 × g for about 10 min to remove the cellular debris and nonexosomal vesicles. Separated plasma sample aliquots should be used immediately, or stored at around -80°C until use.
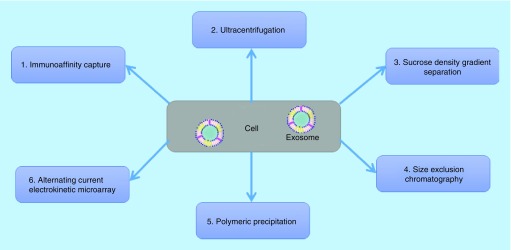
In recent years, various conventional protocols have been developed and applied to isolate and purify exosome from bodily fluids and cell culture media: ultracentrifugation-based technique at 100,000 × g [48], nano-membrane concentrator-based approach [49], immunoaffinity-based capture using monoclonal antibody-coupled nanobeads [50,51], sucrose density gradient separation using sucrose or Percoll [52], alternating current electrokinetic microarray chip technology (ACE) [3,53], nanowire-anchored microfluidic platforms [54,55,56] and utilizing a commercially available synthetic polymer-based precipitation reagents (Figure 6) [57,58]. Each of these methods has their own advantages and disadvantages for exosome isolation and purification from various biological samples. The ultracentrifugation-based technique is the classical and most commonly used (over 80%) isolation method. Overall, the preanalytical steps such as sample collection, storage, exosome concentration and processing time are important for the efficient and reliable method for the analysis of exosomes.
Figure 6. . Overview of the different exosome isolation and purification techniques.
Although advances have been used to isolate and analyze exosome miRNA, there remains a need for a rapid, sensitive and cost-effective gold standard method that generates an effective, pure isolation, detection, high yield extraction and accurate quantification of exo-miRNA from body fluids for research. This is because of the extremely low concentration of exo-miRNAs in body fluids (<0.01%) [59].
Physical characterization & molecular analysis techniques for exosome
Due to their small size (30–100 nm), accurate quantification and characterization of exosomes is technically challenging. Over the past several years, many techniques have been developed and applied to overcome these challenges [60]. Nanoparticle tracking analysis (NanoSight) is one of the best method used for exosome size and quantification.
The commonly employed physical characterization methods are microscopy based methods such as transmission electron microscopy, scanning electron microscopy, cryoelectron microscopy and atomic force microscopy [61,62]; dynamic light scattering [63]; nanoparticle tracking analysis [62,64]; tunable resistive pulse sensing [65]; and single EVs analysis [66].
The used molecular methods to analyze the concentration, quantitative and profile of exosomes are quantitative real time PCR [60], digital PCR (chip-based dPCR, droplet digital PCR, ddPCR) [60,67], western blotting, whole genome sequencing (next-generation sequencing) [68], exome-targeted sequencing (next-generation sequencing) [68], microarray profile [69] and ELISA [70].
Exosome-derived miRNAs as cancer biomarkers
The presence of the tumor at the earliest possible stage (0–1) should be detected by using a sensitive miRNA-based biomarker assay. In addition to tissue biopsy based current studies, investigation of circulating miRNA is a new expanding field in biomarker research because they possess all characteristics (miRNA profiling, diagnosis, prognosis, therapy response and predictive biomarkers), are detectable in liquid biopsy (biological fluids), and do not require both healthy and tumor biopsies from patients. Body fluid such as blood sample enables physicians and researchers to detect the development of cancer at an early stage.
Exosomes have been found to provide a protective and enriched stable source of miRNA in body fluids, preventing their biological molecules from degradation under nonphysiological conditions (multiple freeze-thaw cycles, long-term storage and extreme pH) [71,72]. It has been reported that exosomally derived miRNA remains stable at -20°C for up to 5 years and is resistant to freeze-thaw cycles [17,44,73,74,75]. It makes it a potential biomarker for cancer and other diseases. miRNAs have been implicated in the pathogenesis of many diseases including cancer and have also been shown to be taken up by either distal or nearby recipient cells as cargo in exosomes as a method of cell-to-cell communication to potentially influence the pathogenesis [60,76,77,78,79,80].
miRNAs are known as fundamental regulator of gene expression particularly in cancer, and play an important role in tumorigenesis, metastasis and resistance to various therapies. Over 80.000 articles related to miRNAs in title or keyword have been found by using PubMed [81]. It has been reported that a mammalian cell contains around 100,000 endogenous miRNA molecules per cell [82]. It has also been estimated that a single exosome can carry up to approximately 500 copies of miRNAs [46]. The amount of exosomes in normal human blood has been reported as around 109 exosomes/ml in cancer patients [83,84].
miRNAs are a major class of small, single-stranded, noncoding RNA molecules with a length between 20 and 22 nucleotides (NTs) in their mature form, which play important roles in virtually all biological pathways including cell growth, proliferation, differentiation, immunity response, apoptosis, metabolism and tumorigenesis. The latest release of miRBase (miRBase release v22: http://mirbase.org/) contains 2654 distinct mature miRNAs in humans [81,85], with each potentially having multiple mRNA targets.
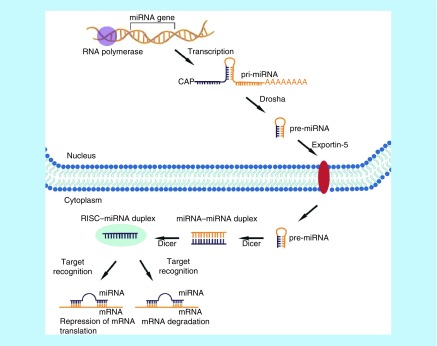
miRNA genes in humans and many other organisms are located in varying genomic contexts, which include intergenic (located in between protein coding genes) and intragenic (located in protein coding genes) as short noncoding RNA regions. Human miRNAs are transcribed from the corresponding miRNA genes containing their own promoters (transcribed independently) or intragenically from spliced portions of protein coding genes (transcribed dependently) [86]. This transcription is made with RNA polymerases (Pol) II and III, generating long noncoding primary miRNAs (1–3 kilobases). The primary miRNAs contain one or more miRNAs and are 5′ methyl capped (7MGpppG) and 3′ polyadenylated (AAAA.…) tail. These transcripts are further processed in the nucleus by the nuclear RNase III enzyme (DROSHA) and the double-stranded RNA-binding proteins, such as DiGeorge Syndrome Critical Region gene 8 (DGCR8), then, leading to premary miRNAs (pre-miRNA, around ≈ 70 NTs in length). After these pre-miRNAs are translocated from nucleus to the cytoplasm through the nuclear pore complex by a nuclear export factor, Exportin-5 (XPO5), they bind to the protein complex of RNase III, DICER and RNA-induced silencing complex, which includes argonaute proteins. DICER cleaves pre-miRNA into a double-stranded RNA of approximately 20–22 NTs in length as miRNA–miRNA dublex. In conjunction with RNA-induced silencing complex, a guide strand (one of the two miRNA strands in the dublex) helps to navigate the mature miRNAs (20–22 NTs) to the target mRNA with base pairing, consequently resulting in downregulation of target gene expression. Intracellular miRNAs are involved in the regulation of gene expression at the post-transcriptional level, acting as negative regulators of mRNA translation by binding to its complementary sequences (usually around 6–8 NTs) in either 5′ untranslated region or 3′ -untranslated region of their target mRNA molecules (Figure 7) [22,87,88,89,90]. The binding of miRNAs to their target mRNAs mainly leads to the mRNA degradation or inhibit expression of target proteins from mRNAs at the post-transcriptional level [78,91].
Figure 7. . The biogenesis of microRNA.
Noncoding miRNAs genes are transcribed in the nucleus into primary miRNAs, which are further processed into premary miRNA and then exported into the cytoplasm where they are finally converted into their matured forms. Mature miRNA then bind to its target mRNA with base pairing, acting as negative regulators of mRNA translation (either mRNA degradation or inhibition of protein expression from mRNA).
It is believed that miRNA controls about 60% of all protein-coding genes in human. Among the miRNA–mRNA regulatory relationships, many different miRNAs are often required to act cooperatively to target a single mRNA. On the other hand, a single miRNA can also affect the expression level of multiple mRNAs by targeting a transcription factor [92,93,94]. It is well known that miRNAs play crucial roles in the pathophysiology of many perhaps all human cancers. miRNAs can function either as tumor-suppressors or as oncogenes depending on the target mRNA and play important roles in tumor development, acquisition of drug resistance and metastasis. Many oncogenic miRNAs that have been reported to be aberrantly expressed in different cancer cells are responsible for sustaining a high cell proliferation rate, metastasis and stimulating oncogenic transcription factors [95,96].
Exosomes can be regarded as vehicles for transferring miRNAs to target recipient cells. Exosomes protect miRNAs from degradation, enabling them to be more stable than free miRNAs and to be efficiently integrated by specific recipient cells [97]. Therefore, within the cargo that exosomes carry, miRNAs can provide information about the identity of the cell type from which they are derived, the target and the cellular state, including therapy resistance.
Increasing evidence reveals that tumor cell-derived exosomes have become a central candidate for promoting tumor cell proliferation, invasion, angiogenesis, distant metastasis and remodeling of the tumor microenvironment through transmitting onco-miRNAs [98]. Angiogenesis is essential for malignant tumor growth and metastasis because new blood vessels offer extra oxygen and nutrients and also to remove waste products [99]. It has been demonstrated that cancer cell-released exosomal-miR-21, exo-miR-23; exo-miR-29; exo-miR-103 and exo-miR-210 promote proliferation, angiogenesis and tumor migration [93,94,98,100,101,102,103,104]. Increasing evidence reveals that exo-miR-21 may be a promising biomarker for many types cancer. Pakravan et al. reported that exo-miR-10 and exo-miR-100 promote suppression of angiogenesis and downregulation of VEGF in human breast cancer cell model [105,106].
Therefore, tumor-derived exosomes have pivotal roles in cancer progression; especially their miRNA cargoes contribute to manipulating transcriptome pool of target cells. It seems that discovery in the field of exosomal miRNAs biology could uncover the underlying mechanisms promoting the aggressive feature of tumors [98]. Nowadays, 2838 miRNAs have been described in exosomes released from various cell types (FunRich 3.0 released in 2016, www.exocarta.org). However, further studies are required to gain a better understanding of the role of exosomal miRNAs as biomarkers in carcinogenesis and cancer progression.
Exosomes in clinical applications
Many studies have demonstrated that exosomal miRNAs represent a very promising new therapeutic strategy for human cancer because of their important natural roles in many cellular processes combined with strong stability, tissue specific expression and secretion into all biological fluids [8,17,78,107]. Considering these findings, exo-miRNAs might play an important function during the transformation of normal cells into malignant cells [78]. Studies have identified circulating exo-miRNAs as potential diagnostic and prognostic biomarkers in therapeutic monitoring for cancers [108]. One of the therapeutic strategies of exosome is the inhibition of onco-miRNAs’ expression by delivery of antagonist tumor-suppressive miRNAs for the treatment of cancer. Exosome loaded with therapeutic anti-miRNA oligonucleotides complementary to the sequence of the targeted mature oncogenic miRNAs can be delivered either systemically or through local injection into the tumor. Another therapeutic strategy is the removal of exosomes from the body circulatory system or to prevent the fusion or uptake of exosomes by target cells to inhibit tumorigenesis. Exosome can be isolated from a patient’s fluids and after modification; it can be transferred back to the same patient for targeted cancer therapy [38,109,110].
Recent studies have demonstrated that tumor suppressor miRNA-loaded exosomes can be used against proangiogenetic mRNAs to inhibit tumor angiogenesis. Exosomes are also amenable to be used in genetic therapy, whereby desired therapeutic genetic materials can be delivered to target cells in certain diseases [10,22]. In addition, exosome-delivered some miRNAs can be considered ideal candidates in using specific gene knockdown to inhibit tumor growth.
Exosomes may represent not only the future biomarkers in medicine, but also a very valuable and effective ‘nanovector’ as transport vehicles for delivering targeted anticancer drugs with low immunogenity and toxicity than other drug-delivery vehicles in cancer therapy [7,111].
Since exosomes are small, nontoxic, nonimmunogenic and native to human as their membrane composition is similar to the cells in the body, and it can be used as drug delivery vehicle to the target cells. Drug-loaded exosome-based vehicle can cross the biological barriers, such as the blood–brain barrier, enabling targeted delivery of neuropharmacological agents into the brain. Exosomes isolated from bovine milk were loaded with anticancer agent withaferin A and used against breast and lung cancer in mice models. The study reported a significant higher efficacy of drug loaded into exosome when compared with the free drug [112]. Engineering designs permit the loading of exosomes with miRNAs, siRNAs, genes, small reactive biomolecules, peptides, antioxidants, and ligands, among other strategies to target delivery in cancer [113].
Currently, several clinical trials using exosome-based cancer therapy are ongoing in the worldwide. However, to use exosomes clinically, further research and proper validation are needed to resolve a number of contentious issues such as; purification and characterization in cancer treatment [17,114]. Together, the scope of using exosome is currently limited, likely, utilization of these biomolecules will soon be in place clinically.
Conclusion & future perspective
The discovery of exosomes as multicomponent signaling complexes mediating cell-to-cell communication between both neighboring (cell-to-cell) and distant cells (travel to distant) is an emerging area as a novel form of communication, as well as a delivery vehicle to carry their cargo. Due to their small size, natural products of the body cells, nontoxic characteristics and crossing the various biological barriers, they are an excellent delivery system for antitumor miRNAs and antitumor drugs in therapeutic tools [22,110,115]. Tumor-derived exosomal miRNA research is highly dynamic and promises novel approaches in cancer prevention, early detection, diagnosis and personalized therapy [17]. In general, there are distinct differences in tumor cell-derived exosomal miRNA expression patterns, compared with their healthy cells. Variations in expression profiles have also been shown to correlate with different tumor characteristics, such as tumor angiogenesis, invasion and metastasis.
It can be possible to inhibit exosome transport proteins such as ESCRT, ALIX and tetraspanins that decreased exosome secretion from cancer cell to neighbor normal cells and this may approach new targets for anticancer therapies to inhibit metastasis. Engineered exosomes could be used to deliver therapeutic agents including anticancer drugs and functional antitumor miRNAs to targeted cancer cells or tissues as personalized cancer therapy. Another possible therapeutic strategy, mature onco-miRNA formation can be inhibited by using inhibitor molecular against to Exportin-5 or DICER proteins as anticancer therapy. As another possible example is also to promote lysosomal degradation of exosomes in tumor cell.
As a result, exosomes are perfectly biocompatible, they reduced toxicity and immunogenity, display great stability in body fluids and can be loaded with specific molecules to targeted cells in cancer treatment. In conclusion, exosomes are small particles with big roles in cancer. Although rapid progress has been made in exo-miRNA detection methods, further efforts to get more sensitive, rapid and cost-effective methods are still needed to find more accurate characterization and functions of exosomal miRNAs from body fluids, thereby providing a strategy for the prevention, early diagnosis and treatment of cancer. This will provide tremendous opportunities for the clinical translation of engineered exosome delivery in targeted cancer therapy in the near future.
Executive summary.
Exosomes are small particles with big roles in cancer. They are generated by both normal and tumor cells and are found in all body fluids. The specific exosomes secreted by tumor cells that contain biomarkers can be used to predict the existence of the presence of a tumor in cancer patients.
The liquid biopsy, a new star of cancer detection, is growing in popularity because of its minimal invasiveness, ease of use, painlessness, lower sample volume, lower cost, higher accuracy and high-throughput.
Tumor cell-derived exosomal miRNA research is highly dynamic and promises novel approaches in cancer prevention, early detection, diagnosis and personalized therapy.
Currently, several clinical trials using exosome-based cancer therapy are ongoing. However, to use exosomes clinically, further research and proper validation are needed to resolve a number of contentious issues such as purification and characterization in cancer treatment.
Further studies are required to gain a better understanding of the role of exosomes in carcinogenesis and cancer progression. This will provide tremendous opportunities for the clinical translation of engineered exosome delivery in targeted cancer therapy in the near future.
Acknowledgments
The author wish to thank IP Salihoglu for the drawing of some figures.
Footnotes
Financial & competing interests disclosure
The author has no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Creative Commons Attribution 4.0 License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68(6), 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Markopoulos GS, Roupakia E, Tokamani M. et al. A step-by-step microRNA guide to cancer development and metastasis. Cell. Oncol. 40(4), 303–339 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Liu X, Wu Y. et al. MicroRNAs in biofluids are novel tools for bladder cancer screening. Oncotarget 8(19), 32370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, Li C, Zhou T. et al. Role of exosomal proteins in cancer diagnosis. Mol. Cancer 16(1), 145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tadimety A, Closson A, Li C, Yi S, Shen T, Zhang JX. Advances in liquid biopsy on-chip for cancer management: technologies, biomarkers, and clinical analysis. Crit. Rev. Cl. Lab. Sci. 55(3), 140–162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B. Clinical analysis of OncomiR – therapeutic targeting of tumorigenesis and tumor disease. Int. J. Genomics Proteomics Metabolomics Bioinformatics 2(2), 28–35 (2017). [Google Scholar]
- 7.Bu H, He D, He X, Wang K. Exosomes: isolation, analysis, and applications in cancer detection and therapy. Chembiochem 20(4), 451–461 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 5(1), 31292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 21(1), 9 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Dakubo GD. Advanced Technologies for Body Fluid Biomarker Analyses. : Cancer Biomarkers in Body Fluids: Principles. Dakubo GD. (). Springer Nature, Basel, Switzerland, 55–74 (2016). [Google Scholar]
- 12.He C, Zheng S, Luo Y, Wang B. Exosome theranostics: biology and translational medicine. Theranostics 8(1), 237–255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 33(3), 967–978 (1983). [DOI] [PubMed] [Google Scholar]
- 14.Ludwig AK, Giebel B. Exosomes: small vesicles participating in intercellular communication. Int. J. Biochem. Cell Biol. 44(1), 11–15 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200(4), 373–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giebel B, Helmbrecht C. Methods to analyze EVs. Methods Mol. Biol. 1–20 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Tomasetti M, Lee W, Santarelli L, Neuzil J. Exosome-derived microRNAs in cancer metabolism: possible implications in cancer diagnostics and therapy. Exp. Mol. Med. 49(1), e285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 29, 116–125 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Dreyer F, Baur A. Biogenesis and functions of exosomes and extracellular vesicles. Methods Mol. Biol. 1448, 201–216 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Yeung BZ, Cui M. et al. Exosome is a mechanism of intercellular drug transfer: application of quantitative pharmacology. J. Control. Release 268, 147–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B, Xing D, Zhu Y, Dong S, Zhao B. The state of exosomes research: a global visualized analysis. BioMed Res. Int. 1–10 (2019) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm. Sin. B. 6(4), 287–296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 5, 442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Souza-Schorey C, Schorey JS. Regulation and mechanisms of extracellular vesicle biogenesis and secretion. Essays Biochem. 62(2), 125–133 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Mulcahy LA, Pink RC, Carter DRF. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 3(1), 24641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fontana S, Giallombardo M, Alessandro R. Technical aspects for the evaluation of exosomes and their content. : Liquid Biopsy in Cancer Patients. Russo A, Giordano A, Rolfo C. (). Humana Press, Cham: 61–70 (2017). [Google Scholar]
- 27.Antonyak MA, Cerione RA. Microvesicles as mediators of intercellular communication in cancer. Methods Mol. Biol. 1165, 147–173 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J. Clin. Invest. 126, 1163–1172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parolini I, Federici C, Raggi C. et al. Microenvironmental pH is a keyfactor for exosome traffic in tumor cells. J. Biol. Chem. 284, 34211–34222 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Hong Y, Cho E, Kim GB, Kim IS. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. J. Extracell. Vesicles 7(1), 1440131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19(4), 213 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Umezu T, Tadokoro H, Azuma K, Yoshizawa S, Ohyashiki K, Ohyashiki JH. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood 124(25), 3748–3757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W, Fong MY, Min Y. et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25(4), 501–515 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinelli C. Exosomes: new biomarkers for targeted cancer therapy. : Molecular Oncology: Underlying Mechanisms and Translational Advancements. Farooqi AA, İsmail M. (). Springer International Publishing, 129–157 (2017). [Google Scholar]
- 35.Mentkowski KI, Snitzer JD, Rusnak S, Lang JK. Therapeutic potential of engineered extracellular vesicles. AAPS J. 20(3), 50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma T, Chen Y, Chen Y. et al. MicroRNA-132, delivered by mesenchymal stem cell-derived exosomes, promote angiogenesis in myocardial infarction. Stem Cells Int. 1–18 (2018) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fortunato O, Gasparini P, Boeri M, Sozzi G. Exo-miRNAs as a new tool for liquid biopsy in lung cancer. Cancers 11(6), 888 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-mediated metastasis: communication from a distance. Dev. Cell 49(3), 347–360 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Zhuang G, Wu X, Jiang Z. et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 31(17), 3513–3523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clancy J, D'Souza-Schorey C. Extracellular vesicles in cancer: purpose and promise. Cancer J. 24(2), 65–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melo SA, Luecke LB, Kahlert C. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523(7559), 177–182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma A, Khatun Z, Shiras A. Tumor exosomes: cellular postmen of cancer diagnosis and personalized therapy [J]. Nanomedicine (Lond). 11(4), 421–437 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Syn N, Wang L, Sethi G, Thiery JP, Goh BC. Exosome-mediated metastasis: from epithelial–mesenchymal transition to escape from immunosurveillance. Trends Pharmacol. Sci. 37(7), 606–617 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Wang N, Xie L. Diagnostic and therapeutic applications of tumor-associated exosomes. Precision Rad. Oncol. 1(1), 34–39 (2017). [Google Scholar]
- 45.O'Neill S, O'Driscoll L. Profiling circulating miRNAs from the plasma of individuals with metabolic syndrome. Methods Mol. Biol. 13, 141–149 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Sauter ER. Exosomes in blood and cancer. Transl. Cancer Res. 6(8), S1316–S1320 (2017). [Google Scholar]
- 47.Max KE, Bertram K, Akat KM. et al. Human plasma and serum extracellular small RNA reference profiles and their clinical utility. Proc. Natl Acad. Sci. USA 115(23), E5334–E5343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pisitkun T, Shen R, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc. Natl Acad. Sci., USA 101(36), 13368–13373 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheruvanky A, Zhou H, Pisitkun T. et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol. Renal Physiol. 292(5), F1657–1661 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rupp AK, Rupp C, Keller S. et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol. Oncol. 122(2), 437–446 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Chen BY, Sung CWH, Chen C, Cheng CM, Lin DPC, Hsu MY. Advances in exosomes technology. Clin. Chim. Acta 493, 14–19 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Tauro BJ, Greening DW, Mathias RA. et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 56(2), 293–304 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Ibsen SD, Wright J, Lewis JM. et al. Rapid isolation and detection of exosomes and associated biomarkers from plasma. ACS Nano 11(7), 6641–6651 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Wunsch BH, Smith JT, Gifford SM. et al. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nat. Nanotechnol. 11(11), 936–940 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Yasui T, Yanagida T, Ito S. et al. Unveiling massive numbers of cancer-related urinary-microRNA candidates via nanowires. Sci. Adv. 3(12), e1701133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, Lyden D. Asymmetric-flow field-flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat. Prot. 14(4), 1027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol. Biol. 728, 235–246 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Merchant ML, Rood IM, Deegens JK, Klein JB. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat. Rev. Nephrol. 13(12), 731–749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barutta F, Tricarico M, Corbelli A. et al. Urinary exosomal microRNAs in incipient diabetic nephropathy. PLoS ONE 8(11), e73798 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bellingham SA, Shambrook M, Hill AF. Quantitative analysis of exosomal miRNA via qPCR and digital PCR. Exosomes & Microvesicles: Methods & Protocols Hill AF. (). Humana Press, NY, USA, 1545, 55–70 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Cizmar P, Yuana Y. Detection and characterization of extracellular vesicles by transmission and cryo-transmission electron microscopy. : Extracellular Vesicles. Humana Press, NY, NY, 221–232 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Dong H, Chen H, Jiang J, Zhang H, Cai C, Shen Q. Highly sensitive electrochemical detection of tumor exosomes based on aptamer recognition-induced multi-DNA release and cyclic enzymatic amplification. Anal. Chem. 90(7), 4507–4513 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Sitar S, Kejžar A, Pahovnik D. et al. Size characterization and quantification of exosomes by asymmetrical-flow field-flow fractionation. Anal. Chem. 87(18), 9225–9233 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Van der Pol E, Coumans FA, Grootemaat AE, Gardiner C, Sargent IL, Harrison P. et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flowcytometry, nanoparticle tracking analysis, and resistive pulse sensing. J. Thromb. Haemos. 12(7), 1182–1192 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Maas SL, Broekman ML, de Vrij J. Tunable resistive pulse sensing for the characterization of extracellular vesicles. : Exosomes and Microvesicles. Hill AF. (). Humana Press, NY, USA, 1545, 21–33 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Lee K, Fraser K, Ghaddar B. et al. Multiplexed profiling of single extracellular vesicles. ACS Nano 12(1), 494–503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang C, Ding Q, Plant P. et al. Droplet digital PCR improves urinary exosomal miRNA detection compared to real-time PCR. Clin. Biochem. 67, 54–59 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Liu X, Yuan W, Yang L, Li J, Cai J. miRNA profiling of exosomes from spontaneous hypertensive rats using next-generation sequencing. J. Cardiovas. Transl. Res. 12(1), 75–83 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Lim JH, Song MK, Cho Y, Kim W, Han SO, Ryu JC. Comparative analysis of microRNA and mRNA expression profiles in cells and exosomes under toluene exposure. Toxicol. in Vitro 41, 92–101 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Salimu J, Webber J, Gurney M, Al-Taei S, Clayton A, Tabi Z. Dominant immunosuppression of dendritic cell function by prostate-cancer-derived exosomes. J. Extracell. Vesicles 6(1), 1368823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fabris L, Calin GA. Circulating free xeno-microRNAs – the new kids on the block. Mol. Oncol. 10, 503–508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Momen-Heravi F, Getting SJ, Moschos SA. Extracellular vesicles and their nucleic acids for biomarker discovery. Pharmacol. Therapeut. 192, 170–187 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ. et al. The microRNA spectrum in 12 body fluids. Clin. Chem. 56, 1733–1741 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 5(1), 31292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. : Seminars in Cell & Developmental Biology. Academic Press, 40, 41–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng L, Hill AF. Small RNA library construction for exosomal RNA from biological samples for the Ion Torrent PGM™ and Ion S5™ System. Methods Mol. Biology 71–90 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y. et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 28, 1721–1726 (2010). [DOI] [PubMed] [Google Scholar]
- 78.Takahashi RU, Prieto-Vila M, Hironaka A, Ochiya T. The role of extracellular vesicle microRNAs in cancer biology. Clin. Chem. Lab. Med. (CCLM) 55(5), 648–656 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev. Biol. 302(1), 1–12 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110(1), 13–21 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 47(D1), D155–D162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 104, 18097–18102 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jia S, Zhang R, Li Z, Li J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget 8(33), 55632 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marrugo-Ramírez J, Mir M, Samitier J. Blood-based cancer biomarkers in liquid biopsy: a promising non-invasive alternative to tissue biopsy. Int. J. Mol. Sci. 19(10), 2877 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alles J, Fehlmann T, Fischer U. et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 47(7), 3353–3364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr. Biol. 24(16), R762–R776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ji W, Sun B, Su C. Targeting MicroRNAs in cancer gene therapy. Genes 8(1), 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maltby S, Plank M, Ptaschinski C, Mattes J, Foster PS. MicroRNA function in mast cell biology: protocols to characterize and modulate microRNA expression. Methods Mol. Biol. 1220, 287–304 (2015). [DOI] [PubMed] [Google Scholar]
- 89.Yarlagadda S, Thota A, Bansal R, Kwon J, Korc M, Kota J. Methods for microRNA profiling in cancer. : Epigenetics and Gene Expression in Cancer, Inflammatory and Immune Diseases. Stefanska B, MacEwan DJ. (). Humana Press, NY, USA, 97–113 (2017). [Google Scholar]
- 90.MacEwan DJ, Shah NM, Antoine DJ. MicroRNAs in Therapy and Toxicity. : Epigenetics and Gene Expression in Cancer, Inflammatory and Immune Diseases. Stefanska B, MacEwan DJ. (Eds). Humana Press, NY, USA, 155–167 (2017). [Google Scholar]
- 91.Dilsiz N, Balik A, Mutaf F, Yesil C, Borazan E. Differential expression of miRNAs in colorectal cancer. J. Gastrointest. Cancer Stromal Tumors 1:1, 1–13 (2016). [Google Scholar]
- 92.Wang Z, Liu Y. Predicting functional MicroRNA–mRNA interactions. Methods Mol. Biol. 117–126 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Hsu YL, Hung JY, Chang WA. et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene 36(34), 4929 (2017). [DOI] [PubMed] [Google Scholar]
- 94.Wang XF, Zhang XW, Hua RX. et al. Mel-18 negatively regulates stem cell-like properties through downregulation of miR-21 in gastric cancer. Oncotarget 7(39), 63352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu J, Sun H, Wang X. et al. Increased exosomal microRNA-21 and microRNA-146a levels in the cervicovaginal lavage specimens of patients with cervical cancer. Int. J. Mol. Sci. 15, 758–773 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cui S, Cheng Z, Qin W, Jiang L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer 116, 46–54 (2018). [DOI] [PubMed] [Google Scholar]
- 97.Vautrot V, Chanteloup G, Elmallah M. et al. Exosomal miRNA: small molecules, big impact in colorectal cancer. J. Oncol. (2019) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rahbarghazi R, Jabbari N, Sani NA. et al. Tumor-derived extracellular vesicles: reliable tools for cancer diagnosis and clinical applications. Cell Commun. Signal. 17(1), 73 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martial S. Involvement of ion channels and transporters in carcinoma angiogenesis and metastasis. Am. J. Physiol. Cell Physiol. 310, C710–C727 (2016). [DOI] [PubMed] [Google Scholar]
- 100.Liu Y, Luo F, Wang B. et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 370(1), 125–135 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Fang JH, Zhang ZJ, Shang LR. et al. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology 68(4), 1459–1475 (2018). [DOI] [PubMed] [Google Scholar]
- 102.Lin XJ, Fang JH, Yang XJ. et al. Hepatocellular carcinoma cell-secreted Exosomal MicroRNA-210 promotes angiogenesis in vitro and in vivo. Mol. Ther. Nucleic Acids 11, 243–252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sruthi TV, Edatt L, Raji GR. et al. Horizontal transfer of miR-23a from hypoxic tumor cell colonies can induce angiogenesis. J. Cell. Physiol. 233(4), 3498–3514 (2018). [DOI] [PubMed] [Google Scholar]
- 104.Bao L, You B, Shi S. et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene 37, 2873–2889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pakravan K, Babashah S, Sadeghizadeh M. et al. MicroRNA-100 shuttled by mesenchymal stem cell-derived exosomes suppresses in vitro angiogenesis through modulating the mTOR/HIF-1a/VEGF signaling axis in breast cancer cells. Cell. Oncol. 40(5), 457–470 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Singh R, Pochampally R, Watabe K, Lu Z, Mo YY. Exosomemediated transfer of miR-10b promotes cell invasion in breast cancer. Mol. Cancer 13(1), 256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takahashi Y, Nishikawa M, Shinotsuka H, Matsui Y, Ohara S, Imai T. et al. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J. Biotechnol. 165, 77–84 (2013). [DOI] [PubMed] [Google Scholar]
- 108.Daly M, O'Driscoll L. MicroRNA profiling of exosomes. : MicroRNA Profiling: Methods and Protocols. Rani S. (). Humana Press, NY, USA: 1509, 37–46 (2017). [DOI] [PubMed] [Google Scholar]
- 109.Wahlgren J, Karlson TDL, Brisslert M. et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 40(17), e130–e130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Samanta S, Rajasingh S, Drosos N, Zhou Z, Dawn B, Rajasingh J. Exosomes: new molecular targets of diseases. Acta Pharm. Sin. 39, 501–513 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cappello F, Logozzi M, Campanella C. et al. Exosome levels in human body fluids: a tumor marker by themselves? Eur. J. Pharm Sci. 96, 93–98 (2017). [DOI] [PubMed] [Google Scholar]
- 112.Munagala R, Aqil F, Jeyabalan J, Gupta RC. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 371, 48–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tai YL, Chen KC, Hsieh JT, Shen TL. Exosomes in cancer development and clinical applications. Cancer Sci. 109(8), 2364–2374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim JH, Kim E, Lee MY. Exosomes as diagnostic biomarkers in cancer. Mol. Cell. Toxicol. 14(2), 113–122 (2018). [Google Scholar]
- 115.Datta A, Kim H, McGee L. et al. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: A drug repurposing strategy for advanced cancer. Sci. Rep. 8(1), 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]