Abstract
Systems biology holds the key for understanding biological systems on a system level. It eventually holds the key for the treatment and cure of complex diseases such as cancer, diabetes, obesity, mental disorders, and many others. The ‘-omics' technologies, such as genomics, transcriptomics, proteomics, and metabonomics, are among the major driving forces of systems biology. Featured as high-throughput, miniaturized, and capable of parallel analysis, protein microarrays have already become an important technology platform for systems biology. In this review, we will focus on the system level or global analysis of biological systems using protein microarrays. Four major types of protein microarrays will be discussed: proteome microarrays, antibody microarrays, reverse-phase protein arrays, and lectin microarrays. We will also discuss the challenges and future directions of protein microarray technologies and their applications for systems biology. We strongly believe that protein microarrays will soon become an indispensable and invaluable tool for systems biology.
Keywords: systems biology, proteome microarray, antibody microarray, reverse-phase protein array, lectin microarray
Introduction
Unlike traditional biology, in which one gene or protein is usually studied at a time, systems biology is trying to understand life on a system level. It is based on the solid foundation of modern biology. One of the very well recognized definitions of systems biology is from Leory Hood and co-workers [1]: systems biology does not investigate individual genes or proteins one at a time, as has been the highly successful mode of biology for the past 30 years. Rather, it investigates the behavior and relationships of all the elements in a particular biological system while it is functioning. The biological systems are very complicate. They are formed as a ‘network of networks'. Systems biology has the power of studying and understanding biological systems as a whole. It may eventually hold the key for the cure of complex diseases such as cancer, diabetes, obesity, mental disorders, and many others, which are usually caused by multiple changes of genes or proteins in more than one signaling pathway [2]. The ‘-omics' technologies, such as genomics, transcriptomics, proteomics, and metabonomics, are among the major driving forces of systems biology. Featured as high-throughput, miniaturized, and capable of parallel analysis, protein microarrays have already become an important technology platform for systems biology [3–6] (Fig. 1).
Figure 1.
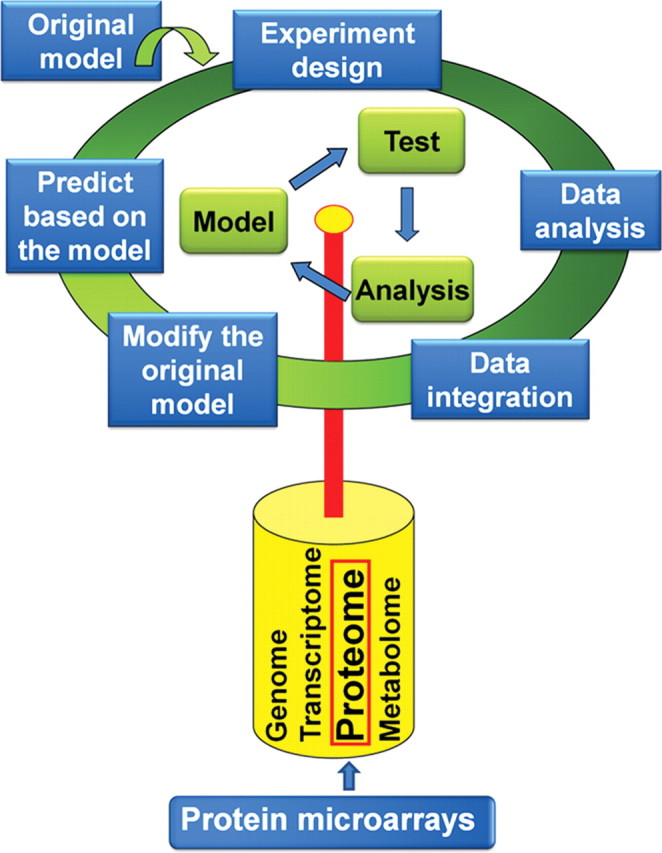
Protein microarray is a key technology for systems biology Theoretically, systems biology is a cycle of modeling–testing–data analysis–remodeling. Eventually, we can use the established model to predict the response of a given organism upon a given stimulation. High-throughput technologies, especially the ‘-omics' technologies, are the major driving force for systems biology. In the post-genome era, proteomics plays a fundamental role among those ‘-omics' technologies. Because of its high-throughput-analysis capability, miniaturized size, and minimal sample and reagent consumption, protein microarray technology has already become one of the most important technologies of proteomics.
Protein microarrays, also known as protein chips, are miniaturized, parallel assay systems that contain small amounts of purified proteins in a high-density format [7]. They allow the simultaneous determination of a variety of analytes from small amounts of samples in a single experiment. Early attempts to screen large numbers of protein/protein fragments for biochemical activities included spotting down bacterial strains and/or bacterial lysates of protein expression cDNA libraries on nylon membranes [8,9]. The major breakthrough came from a report by Zhu et al., in which protein microarrays consisting of 5800 unique yeast proteins on a modified microscopic slide were fabricated. Their utility has been shown by identifying calmodulin- and phospholipid-binding proteins [5]. After that, a variety of research groups have demonstrated numerous applications of protein chips in various biochemical assays, such as target identification of small molecules.
Protein microarrays are typically prepared by immobilizing proteins onto a microscope slide using a standard contact [5,10] or a noncontact microarrayer [11–13]. A variety of slide surfaces can be used. Popular types include aldehyde- and epoxy-derivatized glass surfaces, Fullmoon slides, and Schott NHS-derivatized slides for random attachment through amines [10,14], nitrocellulose [15,16], or gel-coated slides [17,18] for attachment through diffusion and adsorption/absorption, and nickel-coated slides for affinity attachment of HisX6-tagged proteins. After proteins are immobilized on the slides, they can be probed for a variety of functions/activities. The reaction signals are usually measured and recorded by fluorescent or radioisotope labeling.
Protein microarrays can be classified into two types according to their applications: analytical protein microarrays and functional protein microarrays. Analytical protein microarrays use well-characterized molecules that have known specific activities as immobilized probes, such as antibodies, peptide–MHC (major histocompatibility complex) complexes, or lectins. They have become one of the most powerful multiplexed detection platforms and can be used for monitoring protein expression, biomarker identification, cell surface marker/glycosylation profiling, clinical diagnosis, and environmental/food safety analysis. Traditionally, many different proteins, or even the total proteome of an organism, are spotted individually on a functional protein microarray. Often these immobilized proteins are not well characterized. Functional protein microarrays are mainly used to screen for various types of protein activities, including protein–protein, protein–lipid, protein–DNA, protein–drug, and protein–peptide interactions, to identify enzyme substrates, and to profile immune responses.
Till today, there are numerous formats of protein microarrays [19–25]. A variety of applications could be developed on these microarrays. We are not intending to cover the whole field of protein microarrays. Thus, we will focus on the basic concept and discuss the latest advances of protein microarray technology. Specifically, we will discuss four major types of protein microarrays: proteome microarrays, antibody microarrays, reverse-phase protein arrays (RPAs), and lectin microarrays. These microarrays are capable of global and high-throughput analysis for systems biology/systems biomedicine.
Proteome Microarray
A proteome microarray is defined as a protein microarray immobilized with the majority or all of the open reading frame (ORF)-coded proteins of an organism. It is highly suitable for the unbiased global discovery study. According to the fabrication strategy, the proteome microarray could be simply classified into two types: expression based and in vitro transcription and translation (IVTT) based (Fig. 2). A vast amount of effort and cost is required for constructing an expression-based proteome microarray. Since the proteins need to be affinity purified, this type of proteome microarray could be applied for all the possible protein-related studies. As compared with the expression-based proteome microarray, the IVTT-based proteome microarray bypasses the most time-consuming protein expression and purification step. Thus, the fabrication procedure of the IVTT-based strategy is simple. However, since the proteins have not been purified, application of this type of proteome microarray is almost limited to serum biomarker identification.
Figure 2.
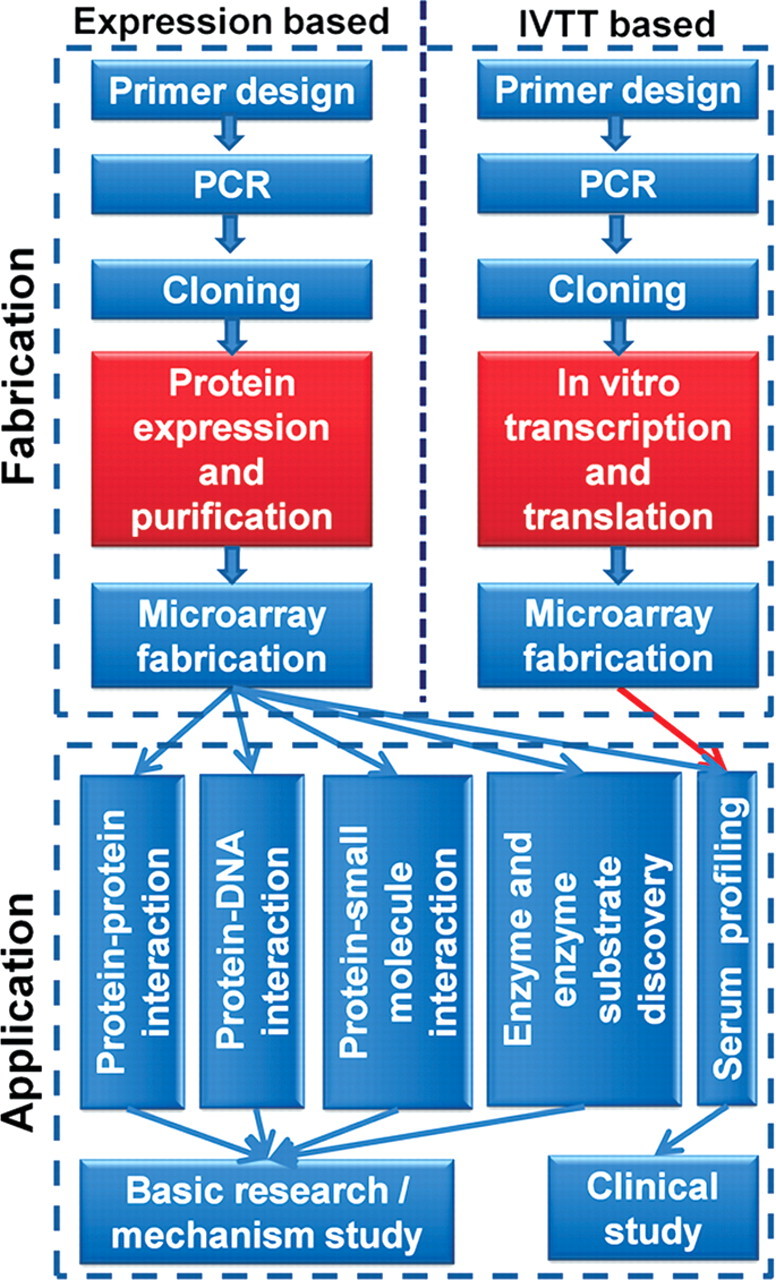
Proteome microarray fabrication and application There are two strategies for proteome microarray fabrication: the traditional protein-expression-based strategy and the IVTT-based strategy. The only difference between these two strategies during the process of protein microarray fabrication lies in the preparation of proteins for microarray printing. The expression-based strategy goes through the traditional cell culture-inducing-affinity purification procedure, while the IVTT-based strategy takes the cell-free IVTT system and without protein purification as a short-cut. As compared with the IVTT-based strategy, the expression-based strategy has many broad applications, especially for functional assays, although it takes much more time and labor. However, the application of protein microarrays fabricated by the IVTT-based strategy is only limited to serum profiling and biomarker identification.
Several expression-based proteome microarrays have been developed, such as the human protein microarray [26], the yeast proteome microarray [5,27], and the Escherichia coli proteome microarray [20]. These microarrays have recently been applied widely to discovery-based biology, including protein–protein, protein–lipid, protein–DNA, protein–drug, and protein–peptide interactions.
Protein–DNA interactions (PDIs) regulate a broad range of functions essential for cellular differentiation, function, and survival. As to profile sequence-specific PDIs globally, Hu et al. [28] have combined bioinformatics and a human transcriptional factor microarray to systematically characterize the human protein–DNA interactome. They identified 17,718 PDIs between 460 DNA motifs predicted to regulate transcription and 4191 human proteins of various functional classes. Among them, they recovered many known PDIs for transcription factors. Surprisingly, they also found over 300 unconventional DNA-binding proteins, i.e. RNA-binding proteins, mitochondrial proteins, and protein kinases. The most striking result was that a well-known kinase—ERK2—was found to have transcriptional factor activity.
Histone acetyltransferases (HATs) and histone deacetylases conduct many critical functions through nonhistone substrates in metazoans. In order to address the question of whether nonhistone HAT substrates exist in yeast, Lin et al. [29], by using yeast proteome microarrays, identified and validated many nonchromatin substrates of the essential nucleosome acetyltransferase of H4 complex. Among these, acetylation sites of phosphoenolpyruvate carboxykinase (Pck1p) were determined by tandem mass spectrometry. Acetylation at one site (Lys514) was crucial for its enzymatic activity and the ability of yeast cells to grow on nonfermentable carbon sources. Further data showed that Pck1p activity could be linked to yeast chronological life span.
Protein phosphorylation plays a central role in most cellular processes and biological functions. Trying to explore the high-throughput power of proteome microarrays, Ptacek et al. [30] accomplished a large-scale ‘Phosphorylome Project' using yeast proteome microarrays. Eighty-seven yeast kinases or kinase complexes were individually probed on the yeast proteome arrays. Totally, 1325 distinct protein substrates were identified, which represent 4129 phosphorylation events. These results provided the first global network that connected kinases to their potential substrates and offered a new opportunity to identify new signaling pathways or cross-talk between pathways.
Although the applications described above are mostly basic research, expression-based protein microarrays also have a profound impact on clinical research. When proteins on a functional protein microarray are viewed as potential antigens that may associate with particular diseases, it becomes a powerful tool in biomarker identification. In order to identify serum biomarkers for ovarian cancer, Hudson et al. [31] used protein microarrays and auto-antibodies from cancer patients to identify proteins that are aberrantly expressed in ovarian tissue. Sera from 30 cancer patients and 30 healthy individuals were used to probe on microarrays containing 5005 human proteins. Ninety-four antigens were identified, which exhibited enhanced reactivity from sera in cancer patients relative to control sera. The differential reactivity of four antigens was tested by immunoblotting and tissue microarrays. The combined signals from multiple antigens were proved to be a robust test to identify cancerous ovarian tissue.
A list of IVTT-based proteome microarrays for clinically significant pathogens has also been fabricated in recent years [32,33]. Compared with the expression-based strategy, the major difference of the IVTT-based strategy is that the proteins are expressed in an E. coli-based cell-free IVTT system, and the crude reactions containing expressed proteins were printed directly onto nitrocellulose-coated slides without purification. These protein microarrays are suitable for determining the antigen-specific humoral immune response triggered by the corresponding pathogens.
Mycobacterium tuberculosis is a high-mortality pathogen, killing almost two million people annually. The discovery of novel biomarkers for monitoring active tuberculosis (TB), and evaluating treatment outcome and vaccine development, is on the top priority list for TB study. Kunnath-Velayudhan et al. [34] fabricated an M. tuberculosis proteome microarray. The microarrays carried 4099 M. tuberculosis proteins, which corresponded to >99% of the ORFs in M. tuberculosis H37Rv DNA. Full-length M. tuberculosis proteins were detected in >95% of the printed spots, as assessed by monoclonal antibody reactivity to epitope fused to recombinant protein termini. More than 500 TB suspects’ sera collected at various sites worldwide with epidemiological and clinical parameters were probed on the microarray. The results showed that sera from worldwide TB suspects recognized ∼10% of the bacterial proteome. This result defines the M. tuberculosis immunoproteome, which is rich in membrane-associated and extracellular proteins.
Candida albicans is an opportunistic pathogen and little is known regarding the dynamics of its colonization and pathogenesis. Mochon et al. [35] developed a C. albicans protein microarray with 336 cell surface proteins to profile the immunoglobulin G response during commensal colonization and candidemia. Specific cell surface antigens were successfully identified for different phases (i.e. acute, early, and mid convalescence) of candidemia. A set of 13 cell surface antigens were identified, capable of distinguishing acute candidemia from healthy individuals and uninfected hospital patients with commensal colonization.
Malaria is a worldwide burden. The targets of protective antibodies and the basis of their inefficient acquisition are unknown. Addressing these knowledge gaps could accelerate malaria vaccine development. To this end, Crompton et al. [36] developed a protein microarray containing ∼23% of the Plasmodium falciparum (Pf) 5400-protein and used this microarray to probe plasma from 220 individuals between the ages of 2–10 and 18–25 years in Mali before and after the 6-month malaria season. Episodes of malaria were detected by passive surveillance over the 8-month study period. They found that most of the antibody reactivities to Pf proteins were short lived.
The above examples of IVTT-based proteome microarrays have been tested against a limit number of samples. These microarrays, if validated in a larger set of samples and in other epidemiological settings, could be proved to be a useful strategy for identifying novel diagnostic biomarkers and novel vaccine targets, and for a better understanding of the fundamental properties of the human immune response to pathogens.
Representative proteome microarrays are summarized in Table 1. Benefiting from the exploding sequencing power of the next-generation sequencing technologies, we are highly confident that more proteome microarrays for other important organisms will be developed in the near future, which will greatly facilitate the systems biology/biomedicine study of these organisms.
Table 1.
Published proteome microarrays
| Organism | Published year/PI | Proteins | Applications |
|---|---|---|---|
| Expression based | |||
| Yeast | 2001/M. Snyder [5] | 5800 | Interactions: protein–protein, protein–phospholipids [5]; protein–DNA [53]; phosphorylation network [30]; glycosylation [6,27]. |
| 2005/M. Snyder [27] | 5854 | ||
| Arabidopsis | 2005/B. Kersten [19] | 1690 | Phosphorylation [19]; protein–protein interaction [54,55]; PDI/transcriptional network [56]. |
| 2007/S.P. Dinesh-Kumar [54,55] | 1133 | ||
| 2008/X.W. Deng [56] | 802 | ||
| SARS -CoV | 2006/M. Snyder [57] | 100% of SARS-CoV and the majority of five other CoVs | Serum biomarker identification for SARS-CoV diagnostics [57]. |
| E. coli | 2008/H. Zhu [20] | 4256 | DNA repair protein identification [20]; inflammation bowel disease (IBD) biomarker screening [58]. |
| Human | 2005/ProtoArray® V1.0 | >1000 | Serum profiling [59]; kinase substrate and inhibitor screening [60]; ubiquitination and sumoylation [61]. |
| 2010/ProtoArray® V5.0 | >9000 | ||
| IVTT based | |||
| Vaccinia | 2005/P.L. Felgner [32,33] | 185 | Serum biomarker identification for diagnostics and vaccination. |
| E. coli | 2006/H. Yanagawa [62] | 4337 | Protein–protein interaction. |
| Francisella tularensis | 2007/P.L. Felgner [63] | 1741 | Serum biomarker identification for diagnostics and vaccination. |
| Coxiella burnetii | 2008/P.L. Felgner [64] | 1988 | Serum biomarker identification for diagnostics and vaccination. |
| P. falciparum | 2008/P.L. Felgner [65] | 250 | Serum biomarker identification for diagnostics and vaccination. |
| Burkholderia pseudomallei | 2009/P.L. Felgner [66] | 1205 | Serum biomarker identification for diagnostics and vaccination. |
| Chlamydia trachomatis | 2010/L.M. de la Maza [67] | 225 | Serum biomarker identification for diagnostics and vaccination. |
| C. albicans | 2010/H.P. Liu [35] | 336 | Serum biomarker identification for diagnostics and vaccination. |
| Brucella melitensis | 2010/P.L. Felgner [68] | 1406 | Serum biomarker identification for diagnostics and vaccination. |
| M. tuberculosis | 2010/M.L. Gennaro [34] | 4099 | Serum biomarker identification for diagnostics and vaccination. |
Antibody Microarray
Because of the specificity of the antibody–antigen binding and the commercial potential for clinical diagnostics, numerous antibody microarrays have been developed. And for most of them, only a handful of antibodies are arrayed on glass or membrane surfaces. The most exciting application of antibody microarrays for systems biology is proteome profiling and proteome comparison between paired samples, which is very similar to the gene expression microarray [37] (Fig. 3). Theoretically, if we can have one specific antibody for each human protein, we will be able to profile the human proteome globally in only a few hours. Once fabricated, this type of pan-antibody microarray will have unprecedented advantages over the currently dominant protein profiling technology, i.e. mass spectrometry. To achieve this goal, the biggest challenge we are facing is the high-throughput production of antibodies with high specificity and affinity. Because the traditional method for generating monoclonal antibodies is time-consuming and laborious, researchers have recently sought alternative approaches. For example, phage antibody-display, ribosome display, systematic evolution of ligands by exponential enrichment (SELEX), mRNA display, and affibody display have been developed to expedite the production of antibodies with high specificity [38–41]. All these methods involve the construction of a large library of viable regions with potential binding activity, which can then be selected by multiple rounds of affinity purification. The binding affinity of the resulting candidate clones can be further improved using maturation strategies. In a most recent study, Reddy et al. [42] bypassed the monoclonal antibody cell screening step by using high-throughput DNA sequencing and bioinformatics analysis to mine antibody variable region (V)-gene repertoires from bone marrow plasma cells of immunized mice. They found that generated antibodies using this strategy from six mice, each immunized with one of three antigens, were overwhelmingly antigen specific (21/27 or 78%). Another efficient strategy is to generate the so-called monospecific antibody [43], which is a compromise of polyclonal and monoclonal. The Human Proteome Atlas Project is majorly generating monospecific antibodies against human proteins, and its long-term goal is to have one validated antibody toward all nonredundant human proteins and provide a first draft of the human proteome by 2014. However, the ideal selection system that is not only fast, robust, sensitive, and of low cost but also automated and minimized has yet to be fully developed.
Figure 3.
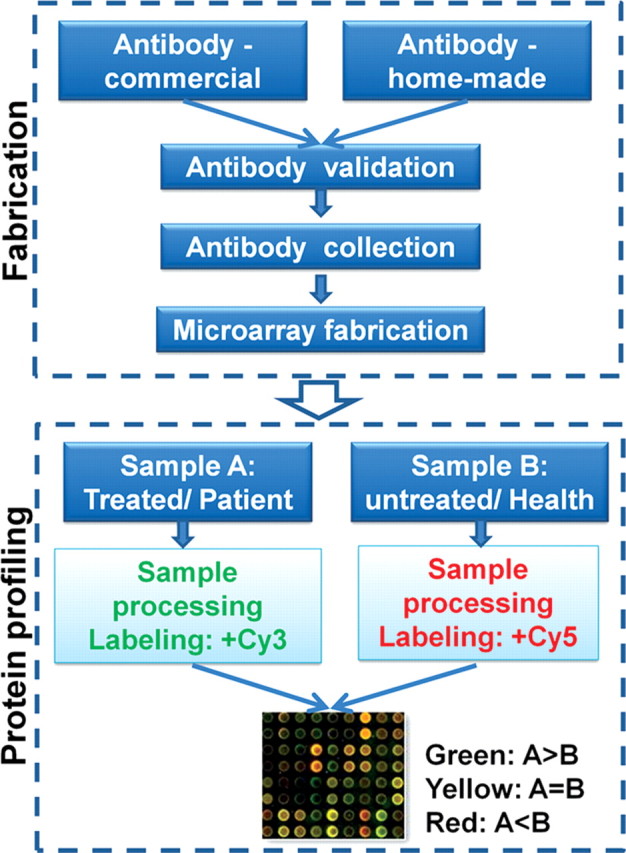
Antibody microarray for protein profiling and comparison The key for antibody microarrays is the quality of the antibodies. Antibodies could be either commercially obtained or home-made. They must pass the strict quality control step of both specificity and efficacy prior to microarray fabrication. The protein profiling part is similar to the gene expression microarray. In brief, the paired samples (treated/untreated or patient/health) were labeled with Cy3 and Cy5, respectively. Then the labeled samples were mixed at equal amounts and probed on the antibody microarray. The pseudo-color represents the ratio of a given protein between sample A and sample B.
Despite the challenge involved in obtaining specific antibodies, many studies using antibody microarrays have recently been reported, and also the list of commercial antibody microarrays is expanding rapidly. The representatives of published or commercial antibody microarrays are listed in Table 2. In a pioneer study by Haab et al. [44], the first high-density antibody microarray was used to test whether a linear relationship could be detected between an antibody and antigen pair in an array format. They investigated the ability of 115 well-characterized antibody–antigen pairs to react in high-density microarrays on modified glass slides: 30% of the pairs showed the expected linear relationships, indicating that a fraction of the antibodies was suitable for quantitative analysis. Sreekumar et al. [45] have created antibody arrays with 146 distinct antibodies against proteins involved in stress response, cell cycle progression, and apoptosis and used these arrays to monitor the alterations in protein quantity in LoVo colon carcinoma cells. The reference standards and samples were labeled separately using either Cy5 or Cy3 dyes, and the fluorescent signals of the bound proteins were detected. The authors were able to obtain differential expression profiles, with radiation-induced up-regulation of apoptotic regulators such as p53, DNA fragmentation factors, and tumor-necrosis-factor-related ligands.
Table 2.
Representative research groups and companies of antibody microarrays study
| PI/Company | Antibody/Number | Application |
|---|---|---|
| Academic | ||
| B.B. Haab [69,70] | Glycosylation and disease related | Serum protein profiling |
| Methodology establishment | ||
| J.D. Hoheisel [21] | Signaling pathway and disease related/810 | Methodology establishment |
| Disease proteomics | ||
| C. Wingren; C.A. Borrebaeck [71,72] | Signaling pathway related | Methodology establishment |
| Disease proteomics | ||
| L. Belov; R.I. Christopherson [73–75] | A variety of anti-CD antibodies | Live cell surface profiling, cell surface marker |
| Industrial | ||
| RayBiotech | Cytokine, angiogenesis, disease related/507 (reactivity: human) | Biotinylate sample for easy processing |
| Sigma | XPRESS Profiler725 Kit/725 (reactivity: human) | General |
| Full moon | Explorer Antibody Array/656 (reactivity: human) | General |
| Phospho Explorer Antibody Microarray/1318 (reactivity: human, mouse) | Phosphorylation event monitoring | |
| A variety of other antibodies | ||
| Kineuxs | Phosphorylation-related antibodies, Kinex™ 800 Antibody Microarray/810 (reactivity: human) | Phosphorylation event monitoring, phosphorylation network |
| SpringBio | A variety of important biological functions/722 (reactivity: human) | General |
| Clontech | Ab Microarray 500/507 (reactivity: human) | General |
The antibody microarray with a moderate or large set of antibodies is a very promising tool for protein profiling; however, there was no standard protocol. Thus the microarray data were hard to compare and interpret among labs. To address this problem, Alhamdani et al. [21] recently fabricated an antibody microarray with 810 antibodies, most of them related to signaling pathway and disease. Using this antibody microarray, they established a standardized protocol that could permit robust analyses of protein extracts from mammalian tissues and cells rather than body fluids. The factors optimized were buffer composition for surface blocking, blocking duration, protein handling and processing, labeling parameters such as type of dye, molar ratio of label versus protein, and dye removal, as well as incubation parameters such as duration, temperature, buffer, and sample agitation.
Reverse-phase Protein Array
In contrast to protein microarrays that immobilize individual proteins, in 2001 Paweletz et al. [46] developed the so-called ‘reverse-phase' protein array that relies on immobilized lysate, which represents the state of individual tissue cell populations undergoing disease transitions. Technically, tiny amounts of (a) cellular lysates, from intact cells or laser capture microdissected cells, and (b) body fluids such as serum, cerebrospinal fluid, urine, vitreous, saliva, etc. are immobilized as individual spots on a microarray. This microarray is then incubated with a single specific antibody to detect expression of the target protein across many samples. A single microarray can accommodate hundreds to thousands of samples that are printed in a series of replicates. Detection is usually performed using a primary and a secondary labeled antibody. Multiplexing is achieved by probing multiple arrays spotted with the same lysate with different antibodies simultaneously. In addition, since RPAs are utilizing cell lysate, they can provide direct quantifiable information on protein post-translational modifications. Thus, the greatest advantage of RPAs is that they allow for high-throughput, multiplexed, and sensitive detection of proteins from small amounts of input material. RPAs have increased sensitivity and are capable of detecting proteins in the picogram or even the attogram range [47]. The high sensitivity of RPAs allows for the detection of low-abundance proteins or biomarkers such as phosphorylated signaling proteins from very small amounts of starting material such as biopsy samples. Using laser capture microdissection, lysates can be analyzed from as few as 10 cells [48]. Another advantage of RPAs is the uniformity of results, as all samples on the chip are probed with the same set of primary and secondary antibody. This allows for the quantification of differences in protein levels across all samples. The biggest limitation of RPA is that its success is highly dependent on the quality of the antibody set used. All the antibodies for RPA have to be extensively validated, to ensure its reactivity and specificity.
RPA has a wide range of possible applications from basic biology, translational and clinical research, such as quantitative analysis of protein expression in cancer cells, body fluids or tissues for biomarker profiling, cell signaling analysis and clinical prognosis, diagnosis, or therapeutic prediction. It has also been used for monitoring protein dynamics in response to various stimuli or doses of drugs at multiple time points. Some other applications of RPA include exploring and mapping protein signaling pathways (Fig. 4) and understanding a candidate drug's molecular mechanism. It has also been suggested as a potential early screening test in cancer patients to facilitate or guide therapeutic decision making. For example, Paweletz et al. [46] used RPA to analyze the pro-survival checkpoint proteins in patient-matched samples at the microscopic transition stage from histologically normal prostate epithelium to prostate intraepithelial neoplasia (PIN) and then to invasive prostate cancer. Cancer progression was associated with an increased phosphorylation of serine/threonine protein kinase (Akt), suppression of apoptosis pathways, and decreased phosphorylation of extracellular-signal-regulated kinases. At the transition from histologically normal epithelium to PIN, a significant increase in phosphorylated Akt and a concomitant suppression of downstream apoptosis pathways were observed, preceding the transition into invasive carcinoma. Using the same strategy, two promising diagnostic markers (villin and moesin) for distinguishing colon from ovarian adenocarcinomas [49] were also revealed. Similarly, the same group constructed another reverse protein microarray [50] on which the lysates represented 60 human cancer cell lines (NCI-60). Fifty-two mouse monoclonal antibodies were probed individually to test the expression of 52 proteins on the microarrays. Using this reverse-protein microarray, the authors found that cell-structure-related proteins almost invariably showed a high correlation between mRNA and protein levels across the NCI-60 cell lines, while non-cell-structure-related proteins showed a poor correlation.
Figure 4.
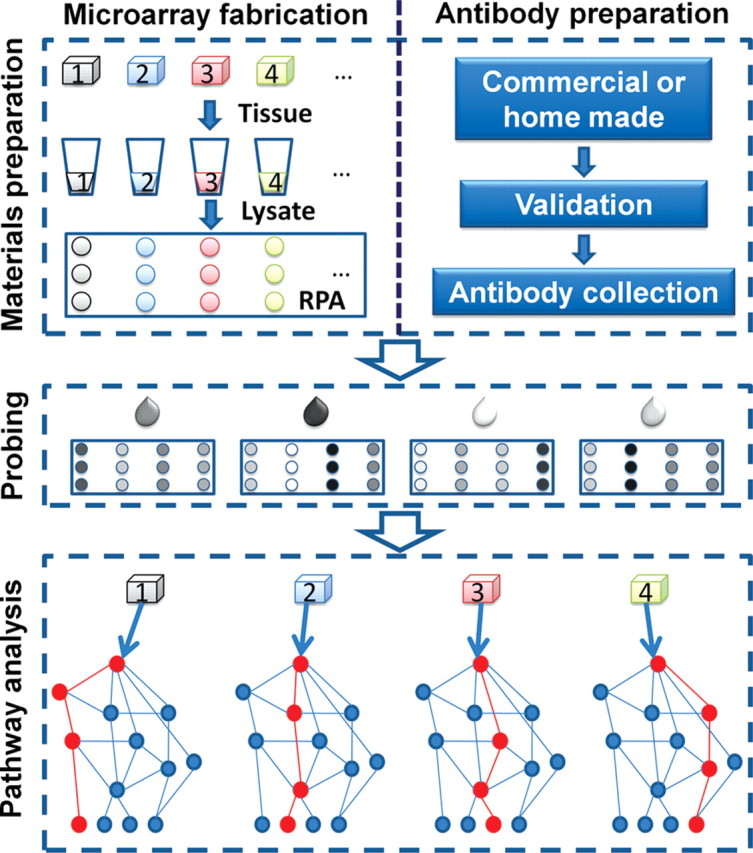
RPA for signaling pathway analysis High-quality tissue samples (usually from patients) and high-quality antibodies are the prerequisite for the success of RPA. The tissue will be lysed and printed directly onto a membrane-coated substrate slide for fabricating RPA. The high-quality antibodies of one or a few related specific signaling pathways will be probed individually onto the RPA. With the help of bioinformatics, significant differences of specific pathways among different tissues will be easily identified.
Lectin Microarray
One of the latest and most interesting developments of protein microarrays is the cell-surface-specific protein microarray for monitoring the cell surface characteristics globally. These microarrays have the potential to elucidate the behavior of both normal and abnormal cells by allowing researchers to characterize cells on a global scale from a complex mixture at the molecular level.
Kuschel et al. [51] have generated an extracellular matrix (ECM) protein microarray by using a piezoelectric microarray printer to print ECM protein solutions onto nitrocellulose-coated glass slides. Five thousand or fewer cells could be applied to an array of 4 × 4 mm2, consisting of 64 microspots. Using this ECM microarray, differences in the adhesive properties of three cell lines to 14 different ECM proteins were identified.
Glycosylation of bacterial cell surfaces is a critical factor in symbiosis, pathogenesis, cell–cell interactions, and immune evasion. Hsu et al. [52] have fabricated a lectin microarray with 21 lectins in profiling surface lipopolysaccharides (LPS) in bacterial cells. When labeled E. coli cells were incubated on the lectin microarrays, the lectins were able to capture bacterial cells onto the chip surfaces via interactions with LPS on the surface of bacteria. Two closely related E. coli strains, JM109 and HB101, could be distinguished by their differences in cell surface glycosylation. In addition, dynamic changes in the surface glycosylation of the neonatal meningitis-associated E. coli strain RS218 were observed.
Almost all the known membrane proteins of mammalian cells are glycosylated. These surface glycans are highly related to cell–cell communication, host–pathogen interaction, development, cancer metastasis [25], etc. To high-throughput profile the live surface glycan, Tao et al. [25] have constructed a lectin microarray with 94 lectins for defining mammalian cell surface glycan signatures. Using the lectin microarray the authors established a binary analysis of cell binding and hierarchical organization of 23 mammalian cell lines. The array was also used to document changes in cell surface glycosylation during cell development and differentiation of primary murine immune system cells. To establish the biological and clinical importance of glycan signatures, the lectin microarray was applied in two systems. Breast cancer cells and their corresponding cancer stem-like cells were compared on the lectin microarray. Interestingly, three lectins, Lycopersicon esculentum (tomato) lectin (LEL), Aleuria aurantia lectin (AAL), and Wheat germ lectin (WGA), were identified as only binding to parental cancer cells. Using LEL-conjugated magnetic beads, the cancer stem cells could be enriched dramatically after two rounds of depletion of the parental cells. Finally, the stem-cell characteristics were validated through tumor-growing experiments on nonobese diabetic/severe combined immunodeficient mice.
Perspectives
Protein microarray technology has been shown to be a useful tool for systems biology studies. Femtomolar sensitivity has been achieved for analytical protein microarrays, and the number of applications of functional protein microarrays has grown dramatically. Novel applications utilizing protein microarrays and new protein microarray technologies are continually emerging. However, there are still several issues that need to be resolved before protein chip technology can be widely applied. First, the traditional cloning–expressing–purification–printing approach is still the gold standard for making protein microarrays, especially for proteome microarrays. Because of the sophisticated expertise required and the high cost of production, it is almost impossible for most laboratories to make their own microarrays, but the price for commercial protein chips is unacceptably high. A variety of promising strategies have already been tested to bypass the traditional procedure. However, none of them are close to being used for the large-scale fabrication of protein microarrays. Thus, to make protein microarray technology more applicable, a simpler and more powerful strategy is needed. Second, there is no widely accepted experimental standardization protocol for protein microarrays. Fortunately, this issue is now being investigated by the Human Proteome Organization, which is developing guidelines for experimental design and data annotation. Third, at present most protein microarray results are only semi-quantitative. In order to reach the goal of accurate quantification, which is very critical for most systems biology studies, new technology is eagerly anticipated. Fourth, clinical biobanking (clinical sample collecting, management, and sharing) lags far behind technology development. One of the ultimate goals of systems biology is to understand the biological systems and to facilitate the cure of complex diseases. As a key technology for systems biology, protein microarrays have been largely applied for clinical related research. To ensure the success of this kind of study, a high-quality sample set is a prerequisite. There are already some organizations/consortia specific for biobanking, such as (BiobankUK, Stockport, Greater Manchester, UK) and (Victorian Cancer Biobank, Carlton Victoria, Australia). However, for China, although we have the largest population and the richest clinical sample resource, we still do not have any internationally well-recognized organization for clinical biobanking. To lead or at least secure a prestigious position in the worldwide competition of systems biology in future, and thus contribute more to clinical research in the long run, there is an urgent need for us to make extensive efforts on clinical biobanking.
Although still in its developing stage, protein microarrays will no doubt prove to be one of the most powerful tools for systems biology. Improvements in our ability to generate large sets of high-quality proteins and antibodies or their mimetics will play a key role in quantitative analysis and promote the extension of this technology to many model organisms.
Funding
This work was supported by grants from the State Key Development Program for Basic Research of China (2010CB529205), the Program for New Century Excellent Talents in University (NCET-09-551), the Shanghai ‘Phosphor' Science Foundation (10QA1403800), the National Natural Science Foundation of China (31000388), SRF for ROCS, SEM, and the fund from the State Key Laboratory of Oncogenes and Related Genes (91-10-12).
References
- 1.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 2.Auffray C, Chen Z, Hood L. Systems medicine: the future of medical genomics and healthcare. Genome Med. 2009;1:2. doi: 10.1186/gm2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tao SC, Chen CS, Zhu H. Applications of protein microarray technology. Comb Chem High Throughput Screen. 2007;10:706–718. doi: 10.2174/138620707782507386. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H, Snyder M. Protein chip technology. Curr Opin Chem Biol. 2003;7:55–63. doi: 10.1016/S1367-5931(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 5.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 6.Kung LA, Tao SC, Qian J, Smith MG, Snyder M, Zhu H. Global analysis of the glycoproteome in Saccharomyces cerevisiae reveals new roles for protein glycosylation in eukaryotes. Mol Syst Biol. 2009;5:308. doi: 10.1038/msb.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CS, Zhu H. Protein microarrays. Biotechniques. 2006;40:423. doi: 10.2144/06404TE01. 425, 427. [DOI] [PubMed] [Google Scholar]
- 8.Bussow K, Cahill D, Nietfeld W, Bancroft D, Scherzinger E, Lehrach H, Walter G. A method for global protein expression and antibody screening on high-density filters of an arrayed cDNA library. Nucleic Acids Res. 1998;26:5007–5008. doi: 10.1093/nar/26.21.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lueking A, Horn M, Eickhoff H, Bussow K, Lehrach H, Walter G. Protein microarrays for gene expression and antibody screening. Anal Biochem. 1999;270:103–111. doi: 10.1006/abio.1999.4063. [DOI] [PubMed] [Google Scholar]
- 10.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 11.Delehanty JB, Ligler FS. Method for printing functional protein microarrays. Biotechniques. 2003;34:380–385. doi: 10.2144/03342mt02. [DOI] [PubMed] [Google Scholar]
- 12.Delehanty JB. Printing functional protein microarrays using piezoelectric capillaries. Methods Mol Biol. 2004;264:135–143. doi: 10.1385/1-59259-759-9:135. [DOI] [PubMed] [Google Scholar]
- 13.Jones VW, Kenseth JR, Porter MD, Mosher CL, Henderson E. Microminiaturized immunoassays using atomic force microscopy and compositionally patterned antigen arrays. Anal Chem. 1998;70:1233–1241. doi: 10.1021/ac971125y. [DOI] [PubMed] [Google Scholar]
- 14.Kusnezow W, Jacob A, Walijew A, Diehl F, Hoheisel JD. Antibody microarrays: an evaluation of production parameters. Proteomics. 2003;3:254–264. doi: 10.1002/pmic.200390038. [DOI] [PubMed] [Google Scholar]
- 15.Kramer A, Feilner T, Possling A, Radchuk V, Weschke W, Burkle L, Kersten B. Identification of barley CK2alpha targets by using the protein microarray technology. Phytochemistry. 2004;65:1777–1784. doi: 10.1016/j.phytochem.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Stillman BA, Tonkinson JL. FAST slides: a novel surface for microarrays. Biotechniques. 2000;29:630–635. doi: 10.2144/00293pf01. [DOI] [PubMed] [Google Scholar]
- 17.Angenendt P, Glokler J, Murphy D, Lehrach H, Cahill DJ. Toward optimized antibody microarrays: a comparison of current microarray support materials. Anal Biochem. 2002;309:253–260. doi: 10.1016/S0003-2697(02)00257-9. [DOI] [PubMed] [Google Scholar]
- 18.Charles PT, Goldman ER, Rangasammy JG, Schauer CL, Chen MS, Taitt CR. Fabrication and characterization of 3D hydrogel microarrays to measure antigenicity and antibody functionality for biosensor applications. Biosens Bioelectron. 2004;20:753–764. doi: 10.1016/j.bios.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 19.Feilner T, Hultschig C, Lee J, Meyer S, Immink RG, Koenig A, Possling A, et al. High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol Cell Proteomics. 2005;4:1558–1568. doi: 10.1074/mcp.M500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Chen CS, Korobkova E, Chen H, Zhu J, Jian X, Tao SC, He C, et al. A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli. Nat Methods. 2008;5:69–74. doi: 10.1038/nmeth1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alhamdani MS, Schroder C, Hoheisel JD. Analysis conditions for proteomic profiling of mammalian tissue and cell extracts with antibody microarrays. Proteomics. 2010;10:3203–3207. doi: 10.1002/pmic.201000170. [DOI] [PubMed] [Google Scholar]
- 22.Gulmann C, Sheehan KM, Conroy RM, Wulfkuhle JD, Espina V, Mullarkey MJ, Kay EW, et al. Quantitative cell signalling analysis reveals down-regulation of MAPK pathway activation in colorectal cancer. J Pathol. 2009;218:514–519. doi: 10.1002/path.2561. [DOI] [PubMed] [Google Scholar]
- 23.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 24.Hsu KL, Mahal LK. A lectin microarray approach for the rapid analysis of bacterial glycans. Nat Protoc. 2006;1:543–549. doi: 10.1038/nprot.2006.76. [DOI] [PubMed] [Google Scholar]
- 25.Tao SC, Li Y, Zhou J, Qian J, Schnaar RL, Zhang Y, Goldstein IJ, et al. Lectin microarrays identify cell-specific and functionally significant cell surface glycan markers. Glycobiology. 2008;18:761–769. doi: 10.1093/glycob/cwn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lueking A, Possling A, Huber O, Beveridge A, Horn M, Eickhoff H, Schuchardt J, et al. A nonredundant human protein chip for antibody screening and serum profiling. Mol Cell Proteomics. 2003;2:1342–1349. doi: 10.1074/mcp.T300001-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin YY, Lu JY, Zhang J, Walter W, Dang W, Wan J, Tao SC, et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 31.Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc Natl Acad Sci USA. 2007;104:17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies DH, McCausland MM, Valdez C, Huynh D, Hernandez JE, Mu Y, Hirst S, et al. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J Virol. 2005;79:11724–11733. doi: 10.1128/JVI.79.18.11724-11733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci USA. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunnath-Velayudhan S, Salamon H, Wang HY, Davidow AL, Molina DM, Huynh VT, Cirillo DM, et al. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc Natl Acad Sci USA. 2010;107:14703–14708. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mochon AB, Ye J, Kayala MA, Wingard JR, Clancy CJ, Nguyen MH, Felgner P, et al. Serological profiling of a Candida albicans protein microarray reveals permanent host-pathogen interplay and stage-specific responses during candidemia. PLoS Pathog. 2010;6:e1000827. doi: 10.1371/journal.ppat.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crompton PD, Kayala MA, Traore B, Kayentao K, Ongoiba A, Weiss GE, Molina DM, et al. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci USA. 2010;107:6958–6963. doi: 10.1073/pnas.1001323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoll D, Templin MF, Schrenk M, Traub PC, Vohringer CF, Joos TO. Protein microarray technology. Front Biosci. 2002;7:c13–c32. doi: 10.2741/stoll. [DOI] [PubMed] [Google Scholar]
- 39.Templin MF, Stoll D, Schrenk M, Traub PC, Vohringer CF, Joos TO. Protein microarray technology. Trends Biotechnol. 2002;20:160–166. doi: 10.1016/S0167-7799(01)01910-2. [DOI] [PubMed] [Google Scholar]
- 40.Cahill DJ. Protein and antibody arrays and their medical applications. J Immunol Methods. 2001;250:81–91. doi: 10.1016/S0022-1759(01)00325-8. [DOI] [PubMed] [Google Scholar]
- 41.Haab BB. Advances in protein microarray technology for protein expression and interaction profiling. Curr Opin Drug Discov Devel. 2001;4:116–123. [PubMed] [Google Scholar]
- 42.Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. 2010;28:965–969. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson P, Paavilainen L, Larsson K, Odling J, Sundberg M, Andersson AC, Kampf C, et al. Towards a human proteome atlas: high-throughput generation of mono-specific antibodies for tissue profiling. Proteomics. 2005;5:4327–4337. doi: 10.1002/pmic.200500072. [DOI] [PubMed] [Google Scholar]
- 44.Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2:RESEARCH0004. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sreekumar A, Nyati MK, Varambally S, Barrette TR, Ghosh D, Lawrence TS, Chinnaiyan AM. Profiling of cancer cells using protein microarrays: discovery of novel radiation-regulated proteins. Cancer Res. 2001;61:7585–7593. [PubMed] [Google Scholar]
- 46.Paweletz CP, Charboneau L, Bichsel VE, Simone NL, Chen T, Gillespie JW, Emmert-Buck MR, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 47.Ramaswamy A, Lin E, Chen I, Mitra R, Morrisett J, Coombes K, Ju Z, et al. Application of protein lysate microarrays to molecular marker verification and quantification. Proteome Sci. 2005;3:9. doi: 10.1186/1477-5956-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheehan KM, Calvert VS, Kay EW, Lu Y, Fishman D, Espina V, Aquino J, et al. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics. 2005;4:346–355. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Nishizuka S, Chen ST, Gwadry FG, Alexander J, Major SM, Scherf U, Reinhold WC, et al. Diagnostic markers that distinguish colon and ovarian adenocarcinomas: identification by genomic, proteomic, and tissue array profiling. Cancer Res. 2003;63:5243–5250. [PubMed] [Google Scholar]
- 50.Nishizuka S, Charboneau L, Young L, Major S, Reinhold WC, Waltham M, Kouros-Mehr H, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci USA. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuschel C, Steuer H, Maurer AN, Kanzok B, Stoop R, Angres B. Cell adhesion profiling using extracellular matrix protein microarrays. Biotechniques. 2006;40:523–531. doi: 10.2144/000112134. [DOI] [PubMed] [Google Scholar]
- 52.Hsu KL, Pilobello KT, Mahal LK. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat Chem Biol. 2006;2:153–157. doi: 10.1038/nchembio767. [DOI] [PubMed] [Google Scholar]
- 53.Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- 54.Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M, et al. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci USA. 2007;104:4730–4735. doi: 10.1073/pnas.0611615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Popescu SC, Snyder M, Dinesh-Kumar S. Arabidopsis protein microarrays for the high-throughput identification of protein-protein interactions. Plant Signal Behav. 2007;2:416–420. doi: 10.4161/psb.2.5.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gong W, He K, Covington M, Dinesh-Kumar SP, Snyder M, Harmer SL, Zhu YX, et al. The development of protein microarrays and their applications in DNA–protein and protein–protein interaction analyses of Arabidopsis transcription factors. Mol Plant. 2008;1:27–41. doi: 10.1093/mp/ssm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu H, Hu S, Jona G, Zhu X, Kreiswirth N, Willey BM, Mazzulli T, et al. Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc Natl Acad Sci USA. 2006;103:4011–4016. doi: 10.1073/pnas.0510921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen CS, Sullivan S, Anderson T, Tan AC, Alex PJ, Brant SR, Cuffari C, et al. Identification of novel serological biomarkers for inflammatory bowel disease using Escherichia coli proteome chip. Mol Cell Proteomics. 2009;8:1765–1776. doi: 10.1074/mcp.M800593-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schweitzer B, Meng L, Mattoon D, Rai AJ. Immune response biomarker profiling application on ProtoArray protein microarrays. Methods Mol Biol. 2010;641:243–252. doi: 10.1007/978-1-60761-711-2_14. [DOI] [PubMed] [Google Scholar]
- 60.Schnack C, Hengerer B, Gillardon F. Identification of novel substrates for Cdk5 and new targets for Cdk5 inhibitors using high-density protein microarrays. Proteomics. 2008;8:1980–1986. doi: 10.1002/pmic.200701063. [DOI] [PubMed] [Google Scholar]
- 61.Del Rincon SV, Rogers J, Widschwendter M, Sun D, Sieburg HB, Spruck C. Development and validation of a method for profiling post-translational modification activities using protein microarrays. PLoS One. 2010;5:e11332. doi: 10.1371/journal.pone.0011332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oishi Y, Yunomura S, Kawahashi Y, Doi N, Takashima H, Baba T, Mori H, et al. Escherichia coli proteome chips for detecting protein-protein interactions. Proteomics. 2006;6:6433–6436. doi: 10.1002/pmic.200600341. [DOI] [PubMed] [Google Scholar]
- 63.Eyles JE, Unal B, Hartley MG, Newstead SL, Flick-Smith H, Prior JL, Oyston PC, et al. Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics. 2007;7:2172–2183. doi: 10.1002/pmic.200600985. [DOI] [PubMed] [Google Scholar]
- 64.Beare PA, Chen C, Bouman T, Pablo J, Unal B, Cockrell DC, Brown WC, et al. Candidate antigens for Q fever serodiagnosis revealed by immunoscreening of a Coxiella burnetii protein microarray. Clin Vaccine Immunol. 2008;15:1771–1779. doi: 10.1128/CVI.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, Randall A, et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008;8:4680–4694. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Felgner PL, Kayala MA, Vigil A, Burk C, Nakajima-Sasaki R, Pablo J, Molina DM, et al. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc Natl Acad Sci USA. 2009;106:13499–13504. doi: 10.1073/pnas.0812080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Molina DM, Pal S, Kayala MA, Teng A, Kim PJ, Baldi P, Felgner PL, et al. Identification of immunodominant antigens of Chlamydia trachomatis using proteome microarrays. Vaccine. 2010;28:3014–3024. doi: 10.1016/j.vaccine.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang L, Leng D, Burk C, Nakajima-Sasaki R, Kayala MA, Atluri VL, Pablo J, et al. Large scale immune profiling of infected humans and goats reveals differential recognition of Brucella melitensis antigens. PLoS Negl Trop Dis. 2010;4:e673. doi: 10.1371/journal.pntd.0000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haab BB, Geierstanger BH, Michailidis G, Vitzthum F, Forrester S, Okon R, Saviranta P, et al. Immunoassay and antibody microarray analysis of the HUPO Plasma Proteome Project reference specimens: systematic variation between sample types and calibration of mass spectrometry data. Proteomics. 2005;5:3278–3291. doi: 10.1002/pmic.200401276. [DOI] [PubMed] [Google Scholar]
- 70.Hamelinck D, Zhou H, Li L, Verweij C, Dillon D, Feng Z, Costa J, et al. Optimized normalization for antibody microarrays and application to serum-protein profiling. Mol Cell Proteomics. 2005;4:773–784. doi: 10.1074/mcp.M400180-MCP200. [DOI] [PubMed] [Google Scholar]
- 71.Wingren C, Borrebaeck CA. Antibody microarray analysis of directly labelled complex proteomes. Curr Opin Biotechnol. 2008;19:55–61. doi: 10.1016/j.copbio.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 72.Wingren C, Ingvarsson J, Dexlin L, Szul D, Borrebaeck CA. Design of recombinant antibody microarrays for complex proteome analysis: choice of sample labeling-tag and solid support. Proteomics. 2007;7:3055–3065. doi: 10.1002/pmic.200700025. [DOI] [PubMed] [Google Scholar]
- 73.Belov L, de la Vega O, dos Remedios CG, Mulligan SP, Christopherson RI. Immunophenotyping of leukemias using a cluster of differentiation antibody microarray. Cancer Res. 2001;61:4483–4489. [PubMed] [Google Scholar]
- 74.Belov L, Huang P, Chrisp JS, Mulligan SP, Christopherson RI. Screening microarrays of novel monoclonal antibodies for binding to T-, B- and myeloid leukaemia cells. J Immunol Methods. 2005;305:10–19. doi: 10.1016/j.jim.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 75.Barber N, Gez S, Belov L, Mulligan SP, Woolfson A, Christopherson RI. Profiling CD antigens on leukaemias with an antibody microarray. FEBS Lett. 2009;583:1785–1791. doi: 10.1016/j.febslet.2009.03.018. [DOI] [PubMed] [Google Scholar]


