Abstract
Gene therapy is progressively emerging as a promising and powerful therapeutic modality, and adeno-associated virus (AAV) is a major delivery vehicle for such therapies. Among the most significant challenges that limit AAV’s utility, however, is the immune response it elicits. Antibodies elicited by prior exposure to natural virus or vector can bind to an AAV vector, preventing it from entering the cell. Furthermore, even if AAV manages to infect a target cell, these cells can then be attenuated by lymphocytes. Improvements in our understanding of how the immune system responds to AAV have guided engineering of the capsid to reduce those responses, yielding capsid variants that are much stealthier and more effective. This review summarizes recent advances in understanding the immune response to AAV as well as highlights engineering methods that enhance AAV’s potential as a gene therapy vector.
Introduction
FDA approval of a vector based on adeno-associated virus (AAV) in a gene therapy to treat the blinding disorder Leber’s congenital amaurosis type 2 firmly established AAV as a powerful tool for therapeutic transgene delivery.[1] Moreover, several late stage clinical trials have further emphasized the versatility and efficacy of AAV in therapies for spinal muscular atrophy 1[2] and hemophilia A and B,[3] and numerous phase I/II clinical trials are in progress. One challenge that has faced the gene therapy field since its inception, however, is immune responses to vectors and their encoded products. AAV is no exception, and immune recognition of the virion capsid by neutralizing antibodies (NABs), cytotoxic T lymphocytes (CTLs) and even pattern recognition receptors (PRRs) are unresolved challenges to an efficacious treatment. Fortunately, a number of basic studies and vector engineering approaches are progressively offering knowledge and highlighting potential solutions to address these challenges.
AAV is an icosahedral virion particle from the family Parvoviridae that can only replicate in the presence of a secondary helper virus such as adenovirus and that compared to other viruses such as helper adenovirus and herpesvirus, is not highly proinflammatory. Its genome is flanked by inverted terminal repeats (ITRs) that serve as the packaging signal for AAV’s signal-stranded DNA genome, which consists of 3 open reading frames (ORFs): rep, which encodes 4 proteins responsible for helping with regulation, replication, and assembly; cap, which encodes 3 overlapping capsid proteins to form the 60-mer icosahedron capsid; and the AAP gene that is located as an alternate reading frame within cap and contributes to capsid assembly.[4] For therapeutic applications, the viral genes are placed on a separate helper construct and are replaced with a single-stranded gene cassette of up to 4.7kb of recombinant cargo. The flanking ITRs then function as the origin of replication and packaging signal such that the cassette is packaged into an infectious but replication incompetent AAV virion.
Immune recognition of AAV is an unresolved challenge to therapeutic efficacy. This recognition includes antibodies against the viral capsid, both due to prior natural exposure to AAV as well as subsequent antibody responses against an administered vector. Additional challenges include cytotoxic T lymphocyte (CTL) recognition of infected cells and innate immune responses. This review summarizes the current understanding of such immune responses and highlights recent advances in addressing them.
Immune Responses to AAV
Adaptive
Adaptive immunity includes both cellular and humoral components, and among the latter are neutralizing antibodies (NABs). NABs recognize surface residues of the capsid, and upon binding, both block viral infection of a target cell as well as mediate clearance of the recognized virion from the body. Due to exposure to natural AAVs, NABs against different AAV serotypes are typically present human serum as early as 1–3 years of age[5], [6] and can eliminate much if not all of the therapeutic efficacy following several route of administration, especially systemic, even at very low titers.[7] Even if a patient is seronegative to a given AAV variant, immune responses to administered vectors leads to seroconversion and renders re-administration much more difficult for applications that may require multiple administrations (e.g. when the target cell turns over).[8]
Adaptive cellular responses to AAV can lead to CTL-mediated attenuation of transduced cells. Specifically, cells infected by AAV proteolytically process capsid proteins and cross-present the resulting peptide epitopes on MHC-I complexes recognized by CTLs. The capsid is thought to primarily be processed by the proteasome, though the cell can still present capsid epitopes in mice deficient for the transporter associated with antigen processing (TAP−/−), indicating that endosomal degradative processing of capsid protein may also occur.[9] Following recognition of MHC-I presented capsid epitope, CTLs eliminate transduced cells and thereby attenuate transgene expression. Fortunately, transient steroidal immunosuppression, until viral capsid proteins are fully degraded, can attenuate this response and is increasingly being adopted in the clinic.[2], [10]
Innate
While not as heavily investigated as the adaptive immune system, innate immune responses can also be triggered by AAV virions that activate Pattern Recognition Receptors (PRRs), one of the immune system’s first lines of defense in recognizing foreign molecules (e.g. lipopolysaccharides, viral DNA, dsRNA). While AAV is not as inflammatory as other viruses such as adenovirus,[11] this response can exacerbate a subsequent adaptive immune response. The two innate pathways previously been shown to be active are the TLR9- and TLR2-MyD88, which both stimulate the production of interferons.[12], [13] While the primary focus has been on the acute post-infection response, recent work has shown that double-stranded RNA (dsRNA) can activate dsRNA sensor MDA5 and trigger an innate response 6–10 weeks after infection, even without a response at earlier time points.[14·] The 3’ ITR has been shown to function as a weak promoter that can generate RNA antisense to transgene mRNA, and Shao et al. postulate that it may take that long for sufficient antisense RNA to accumulate, bind to the sense mRNA, and trigger the response by activating RIG-1, which goes on to trigger a type-I interferon cascade.[14·] Overall, however, the innate immune response is primarily viewed as a trigger to activate the adaptive immune response, and transient immunosuppression has been found to mitigate how strongly an innate response can trigger the adaptive response.
AAV Vector Engineering
Rational Engineering
Several interventions have attempted to address the NAB issue. For example, empty capsids can function as decoys to saturate the circulating NABs and thus enable co-administered vector to infect the target cell population.[15] This elegant approach would require large scale production of empty capsids to add to genome-containing vectors at defined ratio, however, which represents additional regulatory and manufacturing complexity. Furthermore, Pei et al. have shown that empty capsids and even denatured AAV protein contaminants can be efficiently presented via MHC-I and trigger a T cell response,[9] such that the additional antigen load of a decoy capsid may increase the risk of an immune response.
In addition to using decoys, the vector capsid itself can be engineered to reduce antibody binding, aided by the considerable effort previously invested to characterize regions of the AAV that the immune system recognizes. In particular, antibody epitopes on the capsid are increasingly being mapped to aid such efforts.[16] Tse et al. found that NABs tend to target conserved residues among different subtypes near the axes of symmetry in AAV, particularly the three-fold axis. Mutation of these residues in AAV1 enabled a virion to evade specific subsets of antibodies as tested in both pre-immunized mice and NHPs, and combining multiple mutations reduced neutralization additively.[17··] Introduction of too many mutations, or the wrong combination of mutations, however, yielded a capsid that did not fold properly.[17··] One variant designed through a combination of these altered residues was found to avoid neutralization by mouse, NHP, and human antisera.[17··] Immunogenic epitopes on one of the least seropositive common variants, AAV rh.10, have been mutated to significant effect as well. By predicting immunogenic epitopes proximal to commonly ubiquitylated regions on the capsid surface and subjecting them to site-directed mutagenesis, a virion with a single S671A mutation was generated, underwent cell entry effectively, and translocated to the nucleus more rapidly than WT AAV rh.10. This variant also had higher transduction in vitro and in vivo and was 27- to 64- fold more resistant to NABs in mice passively immunized with human intravenous immunoglobulin (IVIG, pooled IgG from >1000 donors).[18]
As another strategy to disrupt epitopes, capsid residues or regions from different serotypes can be combined to yield a chimera with mixed properties of its parents. The first such variant was AAV2.5, which introduced five amino acid residues from AAV1 into AAV2, and thereby combined the receptor attachment behavior of AAV2, the muscle tropism of AAV1, and modified antigenic epitopes to reduce antibody neutralization.[19] Upon analyzing the structure of this capsid via cryo-EM, Burg et al. found that structural similarities between AAV2.5 and AAV1 in the VR-I and VR-IX regions along with mutations N706A, V809A, and T717N are what contribute to the 2–20-fold lower NAB titer AAV2.5 as compared to AAV2.[19], [20] Such sequence-guided strategies, potentially further refined by structural analysis, may enable the design of future chimeras with increased ability to evade NABs.[20]
Directed Evolution
Viral variants that evade the immune system can be generated by creating a library of variant capsids and applying a selective pressure, such as the addition of NABs, to enrich for infections virions with the capacity to evade antibody neutralization. Importantly, substantially improved variants can be generated in the absence of detailed mechanistic or structural knowledge, and subsequent mechanistic analysis of the resulting variants can yield biological insights. AAV capsid libraries have been generated using a range of approaches including error-prone PCR,[21], [22] random peptide display,[23],[24] and DNA loop shuffling.[25],[26] Recently, a computationally designed DNA shuffling strategy, SCHEMA, was used to create a new, structurally guided library. SCHEMA is an algorithm that was developed to create chimera libraries at crossover points predicted to minimize structural disruption.[27] This approach was implemented with AAV serotypes 2, 4, 5, 6, 8, and 9 at seven crossover positions and the resulting library was selected using a Cre-dependent selection strategy for increased transduction of neural stem cells (NSCs) in the subventricular zone of the adult brain. A resulting variant (SCH9) exhibited 24-fold higher GFP expression and 12-fold greater transduction volume than AAV9 in the murine brain[28··] and, presumably due to its chimeric nature, was 2–10 times more resistant to IVIG neutralization than the parent serotypes from which it was derived.[28··]
Chemical Capsid Modifications
Chemical complexation and modifications have also been introduced to the surface of AAV to avoid immune detection. For example, AAV has been shown to interact with extracellular vesicles and can reach difficult to target areas like the CNS.[29·] These exosome-associated AAVs are more resistant to NABs than naked AAV,[30]–[32] though scalable manufacturing of exosome-associated AAV will be a challenge. However, with the over-expression of a well-known exosome marker, tetraspanin CD9, exosome-associated AAV yield as well as efficiency increased by 26%.[33] Molecules such as polyethylene glycol (PEG) have also been tethered to the AAV surface to somewhat reduce antibody neutralization.[34] More recently, click chemistry was used with modified amino acids to attach DNA oligonucleotides, and subsequent incubation with lipofectamine enabled is complexation with the negatively charged DNA.[35·] The resulting coated capsid was more resistant to NABs than previously used PEG-conjugated capsids and maintained 80% activity even at antibody titers of 1:2.[35·] This complexed AAV was also able to retain complete activity after a 72hr incubation in pig sera as well as a subsequent 4-fold increase in editing rates from a delivered CRISPR/Cas9 construct as compared to the unconjugated particle.[35·]
Immune Response due to Delivery Routes
The route of delivery can also play a large role in immune recognition of AAV particles. AAV particles are often administered via intravascular (IV) injection – particularly for well-vascularized tissues such as the liver, heart, skeletal muscle, and even brain and spinal cord – though there are alternate routes being explored that are less “visible” to the immune system.[36], [37] Typically, there is a most clinically suitable route of administration for a given disease application, and the immune response can be a factor that is taken into consideration in such determinations. That is, NABs impact different routes of administration to different extents, and for example NAB titers greater than 1:10 have been shown to greatly reduce transduction from a systemic administration.[7] For the retina, a subretinal administration,[38] which is used in the first AAV FDA approval in the US,[1] is less exposed to antibody neutralization compared to intravitreal delivery,[39] though intravitreal delivery is less invasive and offers the potential for retinal wide transduction. The CNS has been targeted by both intrathecal (IT) or IV injections, which expose AAV particles to different levels of circulating NABs. IV delivery of AAV9 has led to therapeutic benefit in patients with spinal muscular atrophy (SMA1),[2] though the impact of neutralizing antibodies and the larger sized and accompanying doses of SMA2 patients may encourage IT administration. For IT, one study showed a 1:128 titer of neutralizing antibodies failed to inhibit delivery from the CSF in NHPs, which thus enabled broad transduction in the CNS.[36] Similar results were recently found in mice.[37]
Conclusion
With the first FDA approval of an AAV mediated gene therapy, a regulatory path to numerous subsequent likely approvals has been paved. However, the immune system still poses significant challenges for safety and especially efficacy. While CD8+ T cell responses to the capsid can be mitigated by transient immunosuppression, pre-existing NABs are a problem for initial vector administration, and immune responses to an administered vector can lead to rapid seroconversion problematic for vector re-administration. Fortunately, progressively increasing knowledge of AAV biology is enabling the rational design or chemical alteration of capsids to shield them from detection. Alternatively, directed evolution can rapidly generate variants that are resistant to neutralizing antibodies, and such selections can even be combined with evolution for targeted delivery to specific tissues and cells in vivo. AAV vectors can thus draw from a large and expanding tool box that is increasingly being translated towards patients and products.
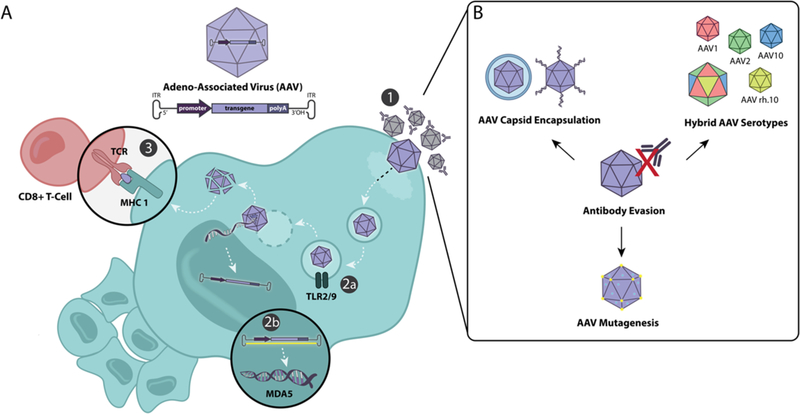
Figure 1:
A) An overview of the cell’s immune response to AAV infection. 1. Receptors on the AAV surface bind to the cell, and the virus is endocytosed. 2. An innate immune response is triggered by either a) viral proteins inside the AAV-containing vesicle activating Toll-like receptors (TLR) 2/9 or b) MDA5 sensors that recognize cytoplasmic double-stranded RNA. 3. An adaptive immune response is triggered when cytotoxic T Lymphocytes recognize viral peptides that have been cleaved and presented via MHC 1. B) One of the main hurdles of AAV infection is antibody neutralization and as such a number of strategies have been developed to prevent antibody binding.
Literature Annotations
Shao et al, Innate Immune Response – dsRNA has been shown to trigger a MDA5 response 6 weeks post infection, implying that transient immunosuppression may not address the entirety of the innate immune response as was previously assumed.
Tse L. V., et al, Structure-guided evolution – By identifying immunogenic hot spots on the capsid surface through 3D-modeling, structure guided evolution was used to evolve a number of libraries and isolate variants which were highly resistant to antibody neutralization
Ojala et al, Computationally guided evolution – Using an algorithm to find crossover points, a library was created from seven different AAV serotypes and variants with a number of improved characteristics, including antibody evasion were discovered.
Hudry et al, Exosome-associated AAV – One method of evading the neutralizing antibody recognition of AAV is enveloping it in an exosome. This study shows that exosome-associated AAV leads to higher gene transduction in the CNS compared to naked AAV
Katrekar et al, Chemically modified AAV – By chemically attaching oligonucleotides to the AAV capsid, PEG molecules were better able to coat the capsid, leading to significant improvements in neutralizing antibody evasion
References
- [1].O. of the Commissioner, “Press Announcements - FDA approves novel gene therapy to treat patients with a rare form of inherited vision loss.” [Online]. Available: https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm589467.htm. [Accessed: 26-Jul-2018].
- [2].Mendell JR et al. , “Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy,” 10.1056/NEJMoa1706198, 01-Nov-2017 [Online]. Available: https://www.nejm.org/doi/10.1056/NEJMoa1706198?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub%3Dwww.ncbi.nlm.nih.gov. [Accessed: 06-Sep-2018]. [DOI] [PubMed]
- [3].George LA, “Hemophilia gene therapy comes of age,” Blood Adv, vol. 1, no. 26, pp. 2591–2599, December 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Knipe DM and Howley PM, Fields’ Virology. Lippincott Williams & Wilkins, 2007. [Google Scholar]
- [5].Calcedo R et al. , “Adeno-Associated Virus Antibody Profiles in Newborns, Children, and Adolescents,” Clin. Vaccine Immunol, vol. 18, no. 9, pp. 1586–1588, September 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fu H et al. , “Differential Prevalence of Antibodies Against Adeno-Associated Virus in Healthy Children and Patients with Mucopolysaccharidosis III: Perspective for AAV-Mediated Gene Therapy,” Hum. Gene Ther. Clin. Dev, vol. 28, no. 4, pp. 187–196, October 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang L et al. , “Impact of Pre-Existing Immunity on Gene Transfer to Nonhuman Primate Liver with Adeno-Associated Virus 8 Vectors,” Hum. Gene Ther, vol. 22, no. 11, pp. 1389–1401, April 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tenenbaum L, Lehtonen E, and Monahan P, “Evaluation of Risks Related to the Use of Adeno-Associated Virus-Based Vectors,” Curr. Gene Ther, vol. 3, no. 6, pp. 545–565, December 2003. [DOI] [PubMed] [Google Scholar]
- [9].Pei X et al. , “Efficient Capsid Antigen Presentation From Adeno-Associated Virus Empty Virions In Vivo,” Front. Immunol, vol. 9, April 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nathwani AC et al. , “Adenovirus-Associated Virus Vector–Mediated Gene Transfer in Hemophilia B,” N. Engl. J. Med, vol. 365, no. 25, pp. 2357–2365, December 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zaiss A-K, Liu Q, Bowen GP, Wong NCW, Bartlett JS, and Muruve DA, “Differential Activation of Innate Immune Responses by Adenovirus and Adeno-Associated Virus Vectors,” J. Virol, vol. 76, no. 9, pp. 4580–4590, May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhu J, Huang X, and Yang Y, “The TLR9-MyD88 pathway is critical for adaptive immune responses to adeno-associated virus gene therapy vectors in mice,” J. Clin. Invest, vol. 119, no. 8, pp. 2388–2398, August 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hösel M et al. , “Toll-like receptor 2–mediated innate immune response in human nonparenchymal liver cells toward adeno-associated viral vectors,” Hepatology, vol. 55, no. 1, pp. 287–297. [DOI] [PubMed] [Google Scholar]
- [14·].Shao W et al. , “Double-stranded RNA innate immune response activation from long-term adeno-associated virus vector transduction,” JCI Insight, vol. 3, no. 12, June 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mingozzi F et al. , “Overcoming Preexisting Humoral Immunity to AAV Using Capsid Decoys,” Sci. Transl. Med, vol. 5, no. 194, pp. 194ra92–194ra92, July 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tseng Y-S et al. , “Generation and characterization of anti-Adeno-associated virus serotype 8 (AAV8) and anti-AAV9 monoclonal antibodies,” J. Virol. Methods, vol. 236, pp. 105–110, October 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17··].Tse LV et al. , “Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion,” Proc. Natl. Acad. Sci. U. S. A, vol. 114, no. 24, pp. E4812–E4821, June 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Selot R, Arumugam S, Mary B, Cheemadan S, and Jayandharan GR, “Optimized AAV rh.10 Vectors That Partially Evade Neutralizing Antibodies during Hepatic Gene Transfer,” Front. Pharmacol, vol. 8, July 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bowles DE et al. , “Phase 1 Gene Therapy for Duchenne Muscular Dystrophy Using a Translational Optimized AAV Vector,” Mol. Ther, vol. 20, no. 2, pp. 443–455, February 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Burg M et al. , “Atomic structure of a rationally engineered gene delivery vector, AAV2.5,” J. Struct. Biol, May 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huttner NA et al. , “Genetic modifications of the adeno-associated virus type 2 capsid reduce the affinity and the neutralizing effects of human serum antibodies,” Gene Ther, vol. 10, no. 26, pp. 2139–2147, December 2003. [DOI] [PubMed] [Google Scholar]
- [22].Maheshri N, Koerber JT, Kaspar BK, and Schaffer DV, “Directed evolution of adeno-associated virus yields enhanced gene delivery vectors,” Nat. Biotechnol, vol. 24, no. 2, pp. 198–204, February 2006. [DOI] [PubMed] [Google Scholar]
- [23].Varadi K et al. , “Novel random peptide libraries displayed on AAV serotype 9 for selection of endothelial cell-directed gene transfer vectors,” Gene Ther, vol. 19, no. 8, pp. 800–809, August 2012. [DOI] [PubMed] [Google Scholar]
- [24].Chan KY et al. , “Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems,” Nat. Neurosci, vol. 20, no. 8, pp. 1172–1179, August 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kienle E et al. , “Engineering and Evolution of Synthetic Adeno-Associated Virus (AAV) Gene Therapy Vectors via DNA Family Shuffling,” J. Vis. Exp. JoVE, no. 62, April 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li C et al. , “Development of Patient-specific AAV Vectors After Neutralizing Antibody Selection for Enhanced Muscle Gene Transfer,” Mol. Ther, vol. 24, no. 1, pp. 53–65, January 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Voigt CA, Martinez C, Wang Z-G, Mayo SL, and Arnold FH, “Protein building blocks preserved by recombination,” Nat. Struct. Mol. Biol, vol. 9, no. 7, pp. 553–558, July 2002. [DOI] [PubMed] [Google Scholar]
- [28··].Ojala DS, Sun S, Santiago-Ortiz JL, Shapiro MG, Romero PA, and Schaffer DV, “In Vivo Selection of a Computationally Designed SCHEMA AAV Library Yields a Novel Variant for Infection of Adult Neural Stem Cells in the SVZ,” Mol. Ther, vol. 26, no. 1, pp. 304–319, January 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29·].Hudry E et al. , “Exosome-associated AAV vector as a robust and convenient neuroscience tool,” Gene Ther, vol. 23, no. 4, pp. 380–392, April 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Meliani A et al. , “Enhanced liver gene transfer and evasion of preexisting humoral immunity with exosome-enveloped AAV vectors,” Blood Adv, vol. 1, no. 23, pp. 2019–2031, October 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Maguire CA et al. , “Microvesicle-associated AAV Vector as a Novel Gene Delivery System,” Mol. Ther, vol. 20, no. 5, pp. 960–971, May 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].György B et al. , “Rescue of Hearing by Gene Delivery to Inner-Ear Hair Cells Using Exosome-Associated AAV,” Mol. Ther, vol. 25, no. 2, pp. 379–391, February 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schiller LT, Lemus-Diaz N, Rinaldi Ferreira R, Böker KO, and Gruber J, “Enhanced Production of Exosome-Associated AAV by Overexpression of the Tetraspanin CD9,” Mol. Ther. Methods Clin. Dev, vol. 9, pp. 278–287, March 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lee GK, Maheshri N, Kaspar B, and Schaffer DV, “PEG conjugation moderately protects adeno-associated viral vectors against antibody neutralization,” Biotechnol. Bioeng, vol. 92, no. 1, pp. 24–34, October 2005. [DOI] [PubMed] [Google Scholar]
- [35·].Katrekar D, Moreno AM, Chen G, Worlikar A, and Mali P, “Oligonucleotide conjugated multi-functional adeno-associated viruses,” Sci. Rep, vol. 8, February 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gray SJ, Kalburgi SN, McCown TJ, and Samulski RJ, “Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates,” Gene Ther, vol. 20, no. 4, pp. 450–459, April 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bey K et al. , “Efficient CNS targeting in adult mice by intrathecal infusion of single-stranded AAV9-GFP for gene therapy of neurological disorders,” Gene Ther, vol. 24, no. 5, pp. 325–332, May 2017. [DOI] [PubMed] [Google Scholar]
- [38].Bennett J et al. , “Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: a follow-on phase 1 trial,” The Lancet, vol. 388, no. 10045, pp. 661–672, August 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kotterman MA, Yin L, Strazzeri JM, Flannery JG, Merigan WH, and Schaffer DV, “Antibody neutralization poses a barrier to intravitreal adeno-associated viral vector gene delivery to non-human primates,” Gene Ther, vol. 22, no. 2, pp. 116–126, February 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]