Abstract
C1 domains are lipid-binding modules that regulate membrane activation of kinases, nucleotide exchange factors, and other C1-containing proteins to trigger signal transduction. Despite annotation of typical C1 domains as diacylglycerol (DAG) and phorbol ester sensors, the function of atypical counterparts remains ill-defined. Here, we assign a key role for atypical C1 domains in mediating DAG fatty acyl specificity of diacylglycerol kinases (DGKs) in live cells. Activity-based proteomics mapped C1 probe binding as a principal differentiator of type 1 DGK active sites that combined with global metabolomics revealed a role for C1s in lipid substrate recognition. Protein engineering by C1 domain swapping demonstrated that exchange of typical and atypical C1s is functionally tolerated and can directly program DAG fatty acyl specificity of type 1 DGKs. Collectively, we describe a protein engineering strategy for studying metabolic specificity of lipid kinases to assign a role for atypical C1 domains in cell metabolism.
INTRODUCTION
The chemical makeup of lipids can have profound effects on cell biology and physiology. Fatty acid (FA) length and unsaturation are important determinants of membrane fluidity, permeability and signaling activity of metabolic products. Cells must tightly regulate the ratio of saturated (devoid of cis double bonds) and unsaturated (mono- or poly-) FAs in order to maintain an optimal balance of fluidity to permit trafficking of proteins and lipids without compromising structural integrity and barrier function of membranes. Metabolic enzymes that regulate the FA composition and dynamics of membranes are directly involved in fine tuning membrane curvature, budding, and fusion that broadly impact transport, metabolism, and signal transduction1, 2, 3. The ability of metabolic enzymes to select lipid substrates based on FA composition at membranes remains a poorly understood process in need of new methodologies to uncover the structure-function relationships of lipids in vivo.
A case example is the diacylglycerol kinase (DGK) family of lipid kinases that catalyze ATP-dependent phosphorylation of diacylglycerol4 (DAG) to produce phosphatidic acid5 (PA, Fig. 1a). To date, 10 mammalian DGKs have been identified and divided into 5 subtypes (Supplementary Fig. 1) based on primary protein sequence that differentiates isoforms on the basis of regulatory domains remote from the conserved lipid kinase domain (Pfam PF007816). Isoform-selective targeting of DGKs remains difficult as inhibitors often block the conserved catalytic domain, leading to off-target activity7. The non-catalytic domains of DGKs modulate Ca2+ activation8, subcellular localization9, 10, oligomerization11, and protein-protein interactions12 and presumably control when and where these enzymes can access DAG substrates for phosphorylation. Understanding DGK specificity is important because altered function of individual isoforms has been implicated in impaired tumor immune responses13, 14, 15, 16, 17, 18, 19, Parkinson’s disease20, 21, bipolar disorder22, 23, 24, 25, 26, asthma27, and metabolic disease28, 29, 30, 31, 32, suggesting that distinct DAG and/or PA species are mediating specific biology. Paradoxically, DGKε, which lacks defined regulatory domains compared with other DGKs is the only isoform with reported DAG fatty acyl chain specificity33, 34, 35.
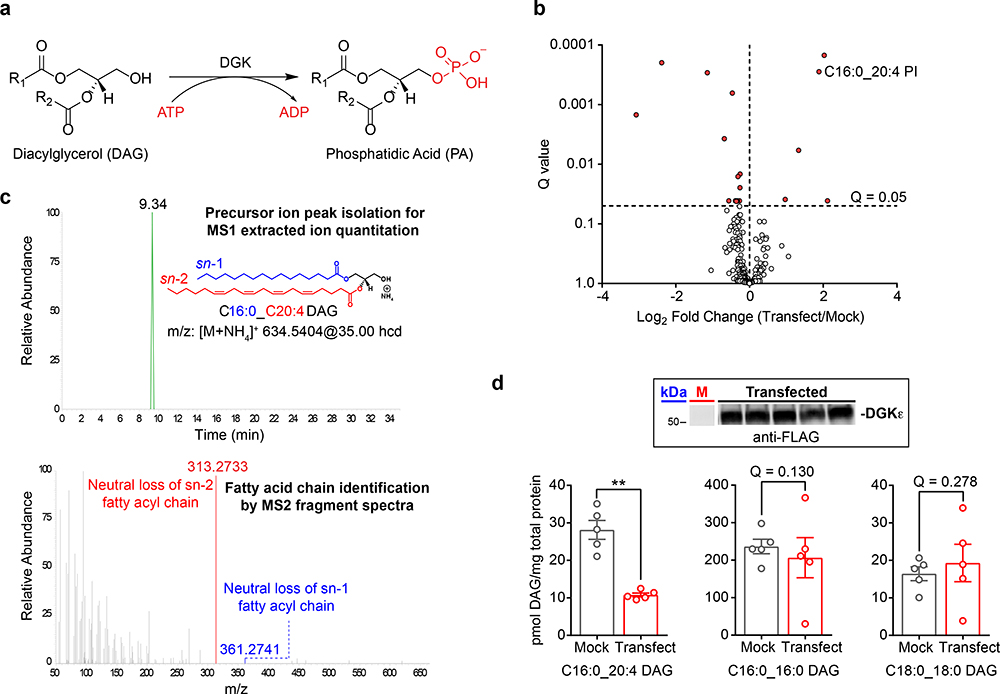
Figure 1. LC-MS/MS metabolomics for deciphering DGK isoform fatty acyl specificity.
a) DGKs catalyze ATP-dependent phosphorylation of diacylglycerol to generate phosphatidic acid. b) Results from an untargeted lipidomics analysis of human DGKε overexpressed in HEK293T cells (n=5 biological samples). Significantly altered lipids were identified by a Q-value < 0.05 following a Benjamini-Hochberg correction of a two-sided binomial test. c) MS1 chromatogram (top) and MS2 fragmentation spectrum (bottom) obtained for identification and quantitative analysis of the DGKε substrate C16:0_C20:4 DAG. d) Targeted lipidomics analysis showed recombinant human DGKε substrate specificity for C16:0_C20:4 compared with C16:0_C16:0 and C18:0_C18:0 DAGs in live cells. Significance was determined using a Benjamini-Hochberg correction following a two-sided binomial test (**Q < 0.01). Data shown represents mean ± s.e.m.; (n=5 biological samples). Inset: western blot shows reproducible transient overexpression of recombinant DGKε in HEK293T cells (anti-FLAG antibody); “M” represents mock lane. Full images of blots are shown in Supplementary Fig. 24. All data shown are representative of two experiments (n=2 biologically independent experiments). hcd, higher energy collisional dissociation; PI, phosphatidylinositol.
DGKε shows preference for sn-2 arachidonic acid-esterified DAGs33 (C20:4). The preference of DGKε for stearic acid (C18:0) at the sn-1 position led to discovery of its role in phosphorylating 1-stearoyl-2-arachidonoylglycerol (SAG) in the phosphatidylinositol (PI) cycle36. DGKε is unique among DGK isoforms as the smallest member composed of a hydrophobic domain, tandem cysteine-rich (C1) domains, and the conserved lipid kinase domain. The small size of DGKε shows that acyl chain specificity can be achieved with a minimum domain topology. The C1 domains are promising candidates to function as lipid recognition modules based on their role in translocating target proteins to membranes during cellular activation37, 38 (e.g. protein kinase C or PKC). Unlike “typical” PKC C1s, the C1 domains of DGKs lack conserved amino acids needed for membrane association (presumably via DAG recognition)37, 38. With the exception of DGKβ and DGKγ, all DGK isoforms contain “atypical” C1s and the function of these domains with regards to metabolic function remains ill-defined in living systems.
Here, we annotate a new role for C1 domains in mediating DAG fatty acyl chain specificity of DGKs. We utilized chemical proteomics and metabolomics to reveal differences in molecular recognition of DGK active sites that correlate and potentially explain the distinct DAG substrate assignments in live cell gain-of-function studies. We performed C1 domain swapping experiments in type 1 DGKs to demonstrate that DAG fatty acyl chain specificity of DGKs is directly encoded by the identity of respective C1 domains independent of subcellular localization. The implication of our findings is establishment of C1 domains as a key component of DGK active sites that directs lipid substrate specificity and suggests that atypical and typical C1 domains are functionally interchangeable in terms of lipid kinase metabolic activity.
RESULTS
Metabolomic evaluation of DGK substrate specificity
We employed a tandem liquid chromatography-mass spectrometry (LC-MS/MS) platform39 for comparing diacylglycerol (DAG) fatty acyl specificity across all 10 mammalian DGKs (Supplementary Fig. 2). We reasoned that a gain of function approach would permit systematic evaluation of live cell changes in the lipidome in response to DGK activity that would help normalize differences in endogenous expression of isoforms. Individual human DGK isoforms were transiently overexpressed in HEK293T cells and recombinant protein expression confirmed by western blot. In general, we observed maximal overexpression at 48 hours for several DGK isoforms and selected this time point for our lipidomics study (Supplementary Fig. 3). We also found that overexpression of recombinant DGKs does not affect expression of an endogenous DGK (Supplementary Fig. 4). Using our approach, we could detect on average ~600 distinct lipids per sample, based on fatty acyl composition, across 25 lipid classes in both positive and negative ion modes by global untargeted analyses (Supplementary Table 1). Lipid identifications were performed as shown in Supplementary Fig. 5 and described in Methods.
We proceeded with proof of concept studies on DGKε, which is the only reported isoform with DAG fatty acyl specificity33, 34, 35. Our untargeted analyses identified several lipid species with levels that were significantly altered in recombinant DGKε- versus mock-transfected HEK293T cells (Q < 0.05, Fig. 1b and Supplementary Table 2). Notably, recombinant overexpression of DGKε resulted in elevations in cellular levels of a C16:0_C20:4 phosphatidylinositol (PI), which supports previous findings of DGKε involvement in the PI cycle34, 35. We used a targeted parallel reaction monitoring (PRM39) approach to quantitate 34 DAG and 30 PA species with FA identities that were assigned by neutral loss fragmentation of fatty acids from the [M+NH4]+ DAG adduct ions (Fig. 1c and Supplementary Fig. 6). As expected based on previous reports40, we discovered that recombinant DGKε overexpression resulted in reductions in cellular levels of the arachidonic acid-containing DAG species (C16:0_C20:4, Fig. 1d). In contrast, saturated DAGs including C16:0_C16:0 and C18:0_C18:0 species were largely unaffected showing DGKε-mediated changes were fatty acyl chain specific (Fig. 1d).
Collectively, our metabolomics strategy to assign endogenous DAG substrates for DGKs, which are identified by cellular depletion, confirms specificity of DGKε for arachidonoyl-containing DAGs in live cells.
Assigning DAG acyl specificity to the DGK superfamily
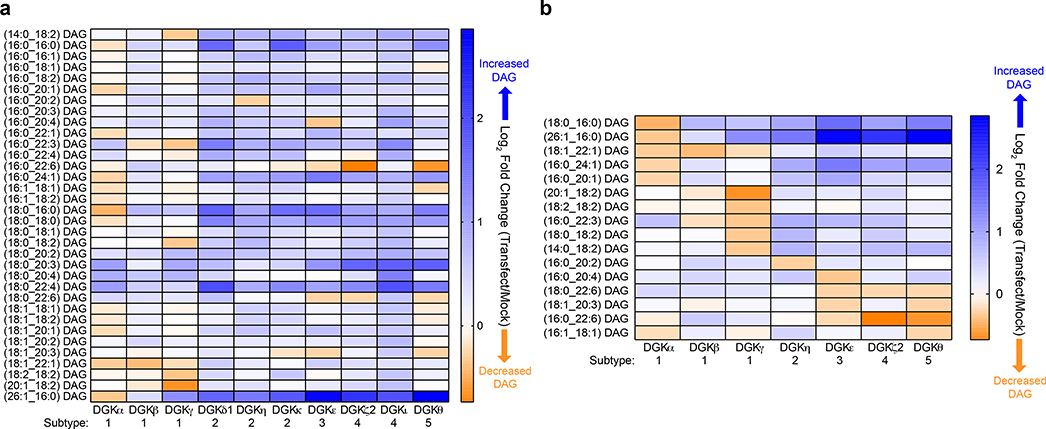
We next applied our metabolomics platform to assign DAG substrates to additional DGK isoforms overexpressed in live HEK293T cells (Fig. 2a, Supplementary Fig. 7). To compare substrate fatty acyl specificity across the DGK family, we first identified putative DAG substrates by selecting lipids that showed >15% reduction in cellular levels in recombinant DGK- versus mock-transfected lipidomes (log2 fold change of −0.23, Fig. 2b). We compared whether these DAG substrates were unique, shared within subtype, or shared across subtypes based on fatty acyl chain length and unsaturation. Generally, we discovered minimal overlap in DAG substrates across the DGK superfamily. A few exceptions were noted including overlap in DAG substrates between type 3 and 5 DGKs (C18:1_C20:3 DAG for DGKε and DGKθ) as well as DAGs shared across type 3, 4, and 5 DGKs (C18:0_C22:6 and C16:0_C22:6 for DGKε, DGKζ2, and DGKθ, Fig. 2b). In support of distinct DGK specificities, we identified DAG lipids uniquely regulated by DGKθ (C16:1_C18:1), DGKη (C16:0_C20:2), and DGKε (C16:0_C20:4) that exhibited diversity in fatty acyl chain length and unsaturation (Fig. 2b). We could not detect depletions in any detected DAGs in DGKδ1- and DGKι-transfected cells suggesting that further optimization of conditions are needed to study these isoforms (Fig. 2a).
Figure 2. Assigning DAG substrate specificity to the DGK superfamily.
a) Heat map displaying live cell alterations in the DAG lipidome (log2 fold change) in recombinant human DGK overexpressed- compared with mock non-transfected-HEK293T cells (n=5 biological samples). All 10 isoforms from the DGK superfamily were analyzed by targeted lipidomics. b) Heat map highlighting altered DAGs that were unique and/or shared across DGK isoforms that showed at least a 15% change (log2 fold change of −0.23) in cellular lipid abundance (n=5 biological samples). Note that C18:0_C22:6 (DGKζ2) and C16:0_C22:6 (DGKε) DAGs showed changes at the set cutoff (~0.23). All data shown are representative of two experiments (n=2 biologically independent experiments).
The DAG specificity observed across DGK subtypes was also observed between isoforms within members of type 1 DGKs. Specifically, DGKα and DGKγ each appeared to regulate a mutually exclusive set of DAG substrates that differed in terms of fatty acid makeup. DGKα substrates were enriched for long-chain saturated or monounsaturated fatty acids while DGKγ substrates contained more polyunsaturated fatty acid species with multiple DAGs containing linoleoyl FAs (Fig. 2b). We did not detect any unique DAG substrates for DGKβ given that the sole DGKβ substrate (C18:1_C22:1) was also shared with DGKα (Fig. 2b). We did not identify any DAG substrates that were shared between DGKα and DGKγ or across all 3 type 1 DGKs.
Hierarchical clustering of our targeted metabolomics results revealed interesting relationships with regard to the overall lipid changes when comparing DGK isoforms and the fatty acyl compositions of DAGs (Supplementary Fig. 8 and 9). For example, the type 1 and 2 DGK isoforms appeared to cluster within their respective subtypes as one would expect based on domain topology and sequence homology. However, DGKζ2 shared higher similarity with DGKε and DGKθ compared with type 4 DGKι as determined by DAG substrate profiles (Fig. 2b and Supplementary Fig. 8). We also measured cellular changes in lipid PA product in recombinant DGK- compared with mock-transfected cells (Supplementary Fig. 10). We identified cellular alterations in several DAG and PA lipid pairs with matching fatty acyl compositions that supports our gain-of-function lipidomics approach to capture DGK metabolic activity in live cells (i.e. DAG depletion and PA production, Supplementary Fig. 11). For several isoforms (e.g. DGKγ, DGKκ, and DGKι), we did not detect clear enhancements in cellular PA levels in response to DGK overexpression (Supplementary Fig. 10), which suggests further metabolism of this lipid intermediate as was observed for DGKε (i.e. C16:0_C20:4 PI, Fig. 1b).
Collectively, our findings show that DGK isoforms exhibit unique DAG fatty acyl specificity both within and across DGK subtypes in live cells, and for several of these DAG substrates we provide evidence of direct DAG to PA metabolism using our metabolomics strategy (Fig. 2 and Supplementary Fig. 11).
Chemoproteomic profiling of type 1 DGK active sites
We implemented a LC-MS/MS quantitative chemical proteomic assay using ATP acyl phosphates to probe ligand binding of DGKs as previously described7, 41. Our goal was to identify functional differences in type 1 DGK active sites in order to understand how isoforms with high sequence homology (Supplementary Fig. 12) are capable of exhibiting DAG fatty acyl specificity. Recombinant rat DGKα, DGKβ and DGKγ were transiently transfected in isotopically light and heavy amino acid-labeled HEK293T cells through stable isotope labelling by amino acids in cell culture (SILAC)42. Light and heavy recombinant lysates were then treated with dimethyl sulfoxide (DMSO) vehicle or free ATP (1 mM), prior to addition of ATP acyl phosphate probe to label active site lysine residues. Light and heavy proteomes were combined in a 1:1 ratio, digested with protease, and desthiobiotin-modified tryptic peptides enriched using avidin affinity chromatography. Samples were then analyzed by LC-MS/MS to identify and quantify isotopically-labeled active-site peptides as previously described7, 41. See Methods for additional details.
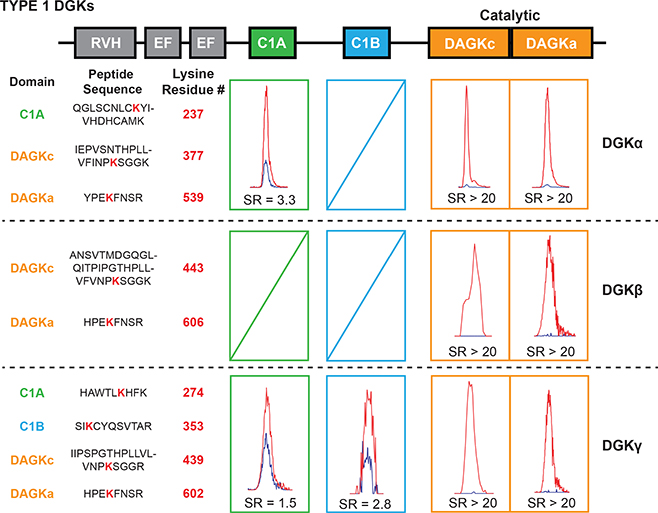
Using our quantitative chemical proteomic approach, we identified probe-modified sites in the lipid kinase domain (DAGKc/DAGKa region) of DGKβ and DGKγ that were highly competed by ATP as determined by SILAC ratios (SR) of MS1 chromatographic peak areas >5 in DMSO/ATP comparisons (Fig. 3). In contrast to similar ATP binding profiles, we observed marked differences in probe modification of C1 domains across type 1 DGKs. Consistent with our previous results7, we found a single probe-modified site in C1A for DGKα (K237, Fig. 3). Extending our studies to the other type 1 isoforms revealed probe modifications located in C1A (K274) and C1B (K353) of DGKγ (Fig. 3). We could not identify a probe modification site in either C1 domains of DGKβ (Fig. 3). Pre-treatment with free ATP (1 mM) resulted in mild competition of DGKγ C1A (SR = 1.5) and C1B (SR = 2.8), which is comparable with the ATP sensitivity profile of DGKα C1A observed previously7 (SR = 3.3, Fig. 3). The differences in ATP probe binding across type 1 DGK C1s were not due solely to availability of lysines given that these lysine sites are well conserved in type 1 DGKs and good tryptic peptide sequence overlap exists for likely LC-MS/MS detection (Supplementary Fig. 12).
Figure 3. Activity-based profiling of functional binding sites in type 1 DGK active sites.
Chemical proteomics with an ATP acyl phosphate probe was performed to map probe-binding sites of rat type 1 DGKs (DGKα, DGKβ and DGKγ). Competitive binding was measured between probe and free ATP (1 mM). MS1 extracted ion chromatograms of DGK probe-modified active site peptides are shown with corresponding SILAC light/heavy peptide abundance ratios (SR) from vehicle- (light, red peak) and free ATP-treated (heavy, blue peak) proteomes. Probe-modified sites highly competed by free ATP (SR >5) correspond to the ATP substrate binding sites of DGKs. Probe-modified sites that were not sensitive to ATP competition (SR <5) represent binding sites for alternative substrates and/or ligands. Data shown are representative of two experiments (n=2 biologically independent experiments).
In summary, our chemical proteomic studies revealed that type 1 DGK active sites differed principally in probe binding at C1A and C1B domains. Combined with the lack of potent ATP competition at C1 domains, our data suggest that alternative ligands and potentially substrates (e.g. lipids) are bound at these regions of type 1 DGK active sites.
C1 domains are necessary for DGKα metabolic function
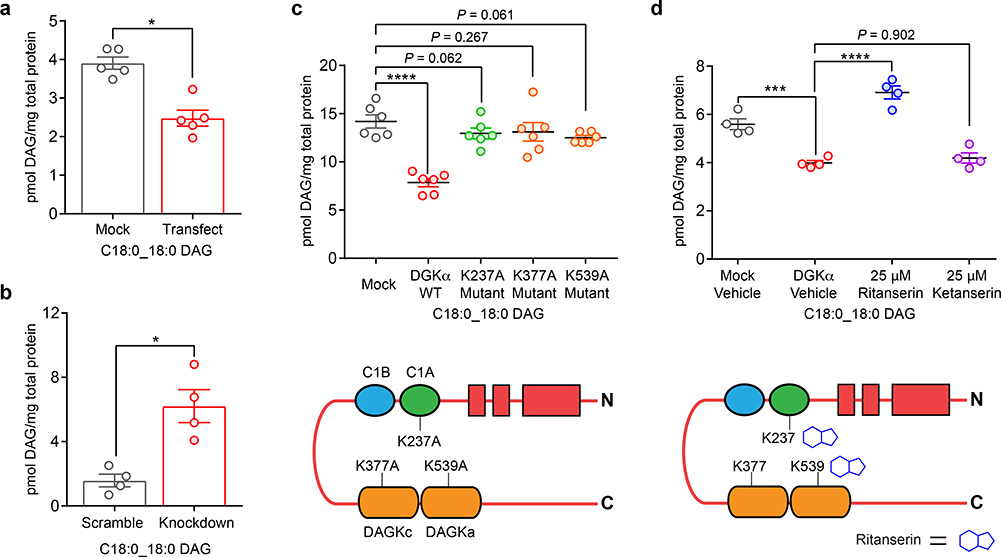
Next, we tested whether C1 domains were necessary for DGK catalyzed metabolism of DAGs in live cells. We focused our efforts on DGKα because the C1A domain of this isoform has been shown to be a critical inhibitor-binding site from previous chemical proteomic studies7, 43. We used LC-MS/MS metabolomics to follow metabolism of the DGKα substrate C18:0_C18:0 DAG as a lipid biomarker of DGKα metabolic activity (Fig. 4a). We confirmed C18:0_C18:0 DAG is an authentic substrate for native human DGKα by performing short hairpin RNA (shRNA) knockdown of endogenous DGKα in A549 cells. Doxycycline-inducible shRNA knockdown resulted in >80% reduction of endogenous DGKα that was accompanied by a significant accumulation of cellular C18:0_C18:0 DAG in A549 cells (Fig. 4b and Supplementary Fig. 13). We identified additional DAG lipids that showed cellular accumulation or reduction upon DGKα knockdown or recombinant overexpression, respectively, which further supports our gain of function approach for mapping authentic lipid substrates of DGKs (Supplementary Fig. 14 and Supplementary Table 3).
Figure 4. C1 domains are necessary and a druggable site for modulating DGKα metabolic function.
a) Overexpression of recombinant human DGKα resulted in metabolism of C18:0_C18:0 DAG (n=5 biological samples). Statistical significance was determined using a Benjamini-Hochberg correction following a two-sided binomial test (*Q < 0.05). b) shRNA-mediated knockdown of endogenous DGKα in A549 cells (0.2 μg/mL doxycycline, 24 hours, n=4 biological samples) resulted in accumulation of cellular C18:0_C18:0 DAG. Statistical significance was assessed using a Benjamini-Hochberg correction following a two-sided binomial test (*Q < 0.05). c) Site-directed mutagenesis (lysine to alanine) of ligand binding sites of rat DGKα (see Fig. 3) resulted in perturbation of metabolic activity in live cells and supports a role for these domains in DGKα metabolic function (n=6 biological samples). Statistical significance was determined using a Dunnett multiple comparison correction following a one-way ANOVA test (****P < 0.0001). d) Treatment of rat DGKα-overexpressing HEK293T cells with ritanserin (blocks C1 and DAGKa sites7) but not the negative control molecule ketanserin disrupted DGKα metabolic activity as evaluated by the C18:0_C18:0 DAG biomarker. Compounds were treated at 25 μM final concentration for 1 hour (n=4 biological samples). Statistical significance was determined using a Tukey’s multiple comparison correction following a one-way ANOVA test (***P < 0.001, ****P < 0.0001). Data shown represent mean ± s.e.m. All data shown are representative of two experiments (n=2 biologically independent experiments).
Our chemoproteomic results (Fig. 3) identified probe-modified lysines in several domains implicated in the catalytic activity of DGKs. We performed site-directed mutagenesis (lysine to alanine) on DGKα to test the importance of these probe-modified sites for metabolic activity. Mutation of C1A (K237A) resulted in loss of DGKα metabolic activity as evidenced by cellular DAG levels that were comparable with mock controls (Fig. 4c and Supplementary Fig. 15). As expected, mutation of individual DAGKc (K377A) or DAGKa (K539A) ATP binding sites also impaired DAG metabolic activity of DGKα (Fig. 4c). We also showed treatment with a DGKα inhibitor ritanserin that binds to C1A7, 44 can block DGKα metabolic activity. Pretreatment of recombinant DGKα-HEK293T cells with ritanserin but not the DGKα-inactive control molecule ketanserin resulted in blockade of C18:0_C18:0 DAG metabolism (Fig. 4d and Supplementary Fig. 16).
In summary, our mutagenesis findings support C1 domains as a critical region necessary for DGKα metabolic function and C1-targeted inhibitors can be used to block DGKα-mediated metabolism in live cells.
Engineering DGK fatty acyl specificity with C1 chimeras
Here, we deployed a protein engineering strategy with C1 domain swapping to test whether 1) atypical (DGKα) and typical (DGKβ, DGKγ) C1s are functionally interchangeable between type 1 DGKs, and 2) C1s regulate DAG fatty acyl specificity of type 1 DGKs in live cells. Our approach is supported by evidence for DGK C1 involvement in ligand recognition and metabolism (Fig 3 and 4) and literature precedence for C1-DAG interactions of other proteins (e.g. PKC38).
We analyzed protein sequences of rat DGKα, DGKβ, and DGKγ to identify homologous regions before and after C1A and C1B, respectively, in order to swap the entire C1A/C1B unit (~150 amino acids) between type 1 isoforms (Supplementary Fig. 17). The result is production of hybrid DGK enzymes (dubbed DGK[α,β,γ]C1[α,β,γ] chimeras) containing native regulatory motifs (EF hands and RVH domain) and catalytic domains (DAGKc/DAGKa) but engrafted with C1 domains transplanted from a separate isoform (Fig. 5a). First, we expressed the recombinant DGK chimeric proteins in HEK293T cells and found that their molecular weights matched their native DGK isoform counterparts with small deviations depending on the length of the exchanged C1 domains (Supplementary Fig. 18). We confirmed that the domain swap of typical and atypical C1s was functionally tolerated by demonstrating catalytic activity of DGK chimeric proteins using a radiolabeled ATP substrate assay in DAG liposomes44 (Supplementary Fig. 19).
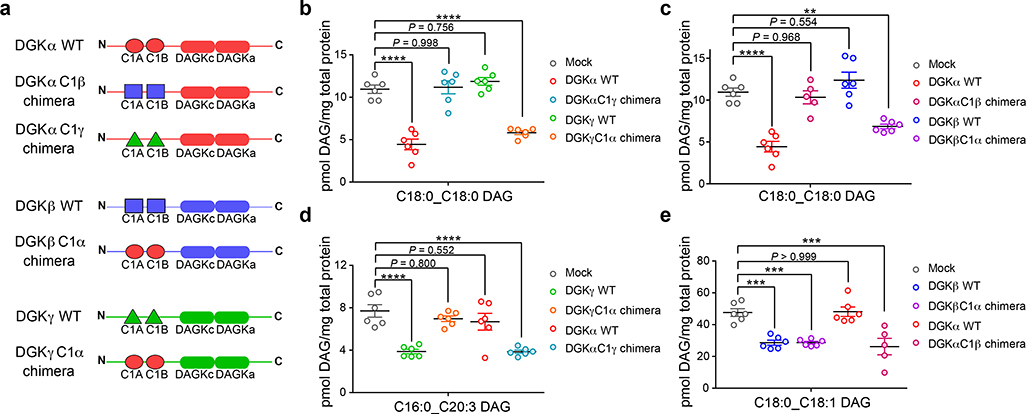
Figure 5. Programming DAG substrate specificity of DGKs in live cells by C1 domain engineering.
a) Schematic of recombinant rat DGK wild-type (WT) and chimeric proteins used for metabolomics analyses. See Supplementary Note for plasmid construct sequences and Supplementary Fig. 18 and 19 for validation of chimeric protein expression and activity. Targeted lipidomics demonstrated that DAG fatty acyl specificity of type 1 DGKs can be engineered via C1 domain swapping: b) DGKα C18:0_C18:0 DAG specificity transferred to DGKγ; c) DGKα C18:0_C18:0 DAG specificity transferred to DGKβ; d) DGKγ C16:0_C20:3 DAG specificity transferred to DGKα; e) DGKβ C18:0_C18:1 DAG specificity transferred to DGKα. Significance of lipid species alterations was determined using a Tukey’s multiple comparison correction following a one-way ANOVA test (**P < 0.01, ***P < 0.001, ****P < 0.0001). All data show represents mean ± s.e.m.; (n=5–6 biological samples). All data shown are representative of two experiments (n=2 biologically independent experiments).
We performed targeted metabolomics profiling of rat WT DGKs to identify DAG substrates that were specific for each respective type 1 isoform: C18:0_C18:0 DAG, DGKα; C18:0_C18:1 DAG, DGKβ; C16:0_C20:3 DAG, DGKγ (Supplementary Fig. 20). We used these DAGs as lipid biomarkers to probe how C1 domain exchange affects DGK lipid metabolism in live cells. Remarkably, we observed both loss and gain of DAG fatty acyl specificity for DAG biomarkers depending on C1 identity of the corresponding DGK chimeric protein (Fig. 5b–e). Specifically, exchange of native C1 domains of DGKα with C1 domains from either DGKγ (DGKαC1γ chimera) or DGKβ (DGKαC1β chimera) resulted in loss of cellular metabolism for C18:0_18:0 DAG (Fig. 5b and c). In parallel, we observed a gain in DAG fatty acyl specificity for DGKα, which exhibited negligible activity towards C16:0_C20:3 DAG but gained this metabolic specificity upon engraftment of DGKγ C1s (DGKαC1γ chimera, Fig. 5d).
We observed a similar metabolic effect for DGKγ that is characterized by gain of C18:0_C18:0 DAG specificity encoded by the transplanted DGKα C1s (Fig. 5b) that is mirrored by loss of specificity for its C16:0_C20:3 DAG substrate (Fig. 5d). In contrast, the exchange of C1 domains in DGKβ resulted in a different effect compared with DGKα and DGKγ. Specifically, DGKβ containing DGKα C1s (DGKβC1α chimera) gained specificity for C18:0_C18:0 DAG (Fig. 5c) without loss of activity for its preferred C18:0_18:1 DAG substrate (Fig. 5e). This result could possibly be due to other structural features inherent to DGKβ, which in addition to C1 domains can modulate DAG fatty acyl specificity of this isoform.
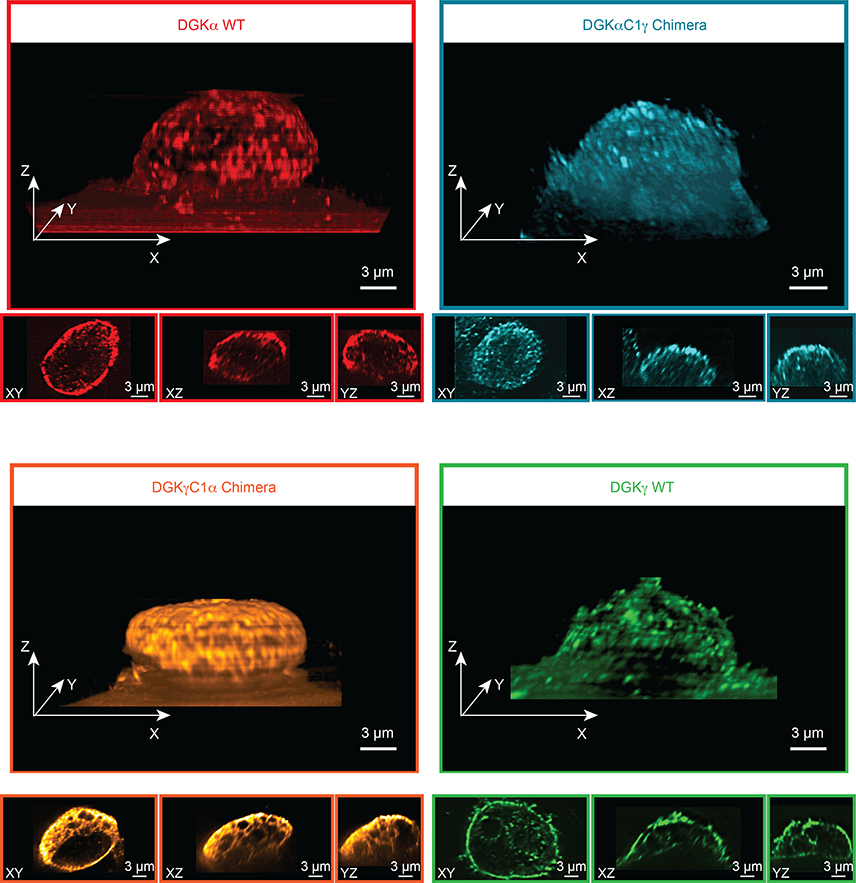
Given that C1 domains regulate membrane association38, we performed lattice-light sheet microscopy45, 46 to determine whether the altered DAG specificity observed for DGK chimeric proteins was due to changes in subcellular localization. Cells were fixed using 4% formaldehyde following 48 hours of transfection to recapitulate the expression and activity profile used for our metabolomic analyses (Fig. 5b–e). We found that the DGKγC1α chimera showed distinct localization at the perimeter of what we suspect to be intracellular organelles, given the well-defined voids in the 3D fluorescence intensity distribution (Fig. 6). In contrast, the DGKαC1γ chimera displayed scattered localization throughout the cell and was not enriched for a particular subcellular locale. Neither chimera showed localization that was overtly akin to the behavior of WT DGKα or DGKγ. Our microscopy data shows subcellular localization of the C1 chimeras are different from each other and from wild-type C1 donor DGKs and supports a localization-independent mechanism for the engineered DAG substrate specificity observed by metabolomics (Fig. 6 and Supplementary Fig. 21–23).
Figure 6. Lattice-light sheet microscopy shows DGK chimeras are not localized based on C1 identity.
Recombinant rat DGK isoforms were imaged by immunofluorescence using an anti-FLAG primary and DyLight 550 secondary antibody staining. Images were acquired at 561 nm excitation using an illumination intensity of the sample at 1 W/cm2. The raw 3D image data was de-skewed and deconvolved as described previously45, 46 and 3D images were rendered using the 3D Viewer plugin in Fiji (see Methods for details). The reconstructed voxel size of the 3D image is 100 × 100 × 100 nm. For each 3D image, three orthogonal cross sections through the cell center are also displayed. Microscopy results reveal that subcellular localization of the C1 chimeras are different from each other and from wild-type C1 donor DGKs. Data shown are representative of two experiments (n=2 biologically independent experiments).
In summary, our protein engineering, LC-MS/MS metabolomics, and microscopy data provide evidence that C1 domains directly mediate DAG fatty acyl specificity of type 1 DGKs in live cells and the programmable nature of the tandem C1s is not simply due to protein localization.
DISCUSSION
DGKs are specialized multi-domain lipid kinases responsible for metabolizing lipid secondary messengers involved in cellular activation. Metabolic specificity of DGKs is believed to arise from diverse regulatory domains outside of the conserved catalytic core to control when and where these enzymes are active. Despite this widely accepted model, biochemical studies have shown that a minimal DGK architecture is sufficient for attaining DAG substrate specificity despite lack of elaborate regulatory domains33, 34, 35. Here, we applied chemical proteomics, metabolomics, and protein engineering to annotate a direct role for C1 domains in mediating DAG fatty acyl specificity of DGKs.
Given the variable expression of native DGKs in cell and tissues, we used a gain of function approach combined with LC-MS/MS metabolomics to enable assignment of lipid substrate specificity for all 10 mammalian enzymes in live cells. We validated lipids identified from recombinant overexpression as authentic substrates by showing that knockdown of endogenous DGKs resulted in accumulation of DAGs with identical fatty acyl composition (Supplementary Fig. 14 and Supplementary Table 3). Our lipidomics approach was crucial for identifying isoform-specific biomarkers to track activity of individual DGK isoforms. Our initial proof of concept studies used DGKε to validate the capability of our method for quantitating changes in DAGs with fatty acyl chains that matched previous findings33, 34, 35. We observed depletion of arachidonic acid-containing DAG and accumulation of PI species with identical fatty acyl composition (C16:0 and C20:4 fatty acids, Fig. 1b and d) that are in agreement with previous reports of regulation of the PI cycle by DGKε34, 35.
Our live cell lipidomics findings revealed several interesting and unexpected features of DGK specificity. First, we observed minimal overlap in DAG acyl specificity across isoforms (Fig. 2a). Specifically, only a handful of identified DAGs were shared between isoforms from two or more subtypes (Fig. 2b). Several isoforms including DGKδ1, DGKκ, and DGKι did not show appreciable changes in cellular DAGs in our recombinant system (Fig. 2a). Interestingly, the latter two isoforms showed a paradoxical depletion of cellular PA when overexpressed in cells (Supplementary Fig. 10). It is tempting to speculate that DGKκ may not function solely as a DAG kinase based on our metabolomic findings and its unique catalytic domain topology (i.e. peptide region separating DAGKc and DAGKa regions47, 48, 49, Supplementary Fig. 1). Whether this effect is due to feedback regulation or an alternative metabolic function of DGKκ is a subject that will be explored in future studies.
The DAG fatty acyl specificity observed between type 1 DGKs was surprising and prompted us to explore ligand binding in their active sites to gain insights into how molecular specificity could be achieved. As expected, the ATP binding regions of DGKα, DGKβ, and DGKγ mapped to homologous regions including conserved probe-modified lysine sites (DAGKc and DAGKa sites, Fig. 3). In contrast, we observed differential probe labeling in C1A and C1B of type 1 DGKs that correlated with the DAG acyl specificity observed by metabolomics. Specifically, DGK isoforms (DGKα and DGKγ) that showed programmable DAG specificity also contained C1 domains sensitive to covalent reaction with ATP acyl phosphate probe (Fig. 3, Fig. 5b and d). Interestingly, the absence of probe binding at DGKβ C1s was correlated with a lesser effect of these domains in mediating DAG acyl specificity for this isoform (Fig. 3 and Fig. 5e).
Although our metabolomics and proteomics data support C1 involvement in DGK specificity, we used protein engineering to provide direct evidence for this model. We generated DGK C1 chimeric proteins to demonstrate that the DAG fatty acyl specificity of DGKs could be programmed by exchanging C1 domains in live cells (Fig. 5). Our studies suggest that C1s are a component of the DGK active site to help select lipid substrates and this hypothesis is supported by previous chemical proteomic findings of a putative DGKα interdomain active site composed of C1A and the catalytic domains43. Our previous and current findings present a working model whereby the regulatory domains could be involved in formation of DGK active sites (Supplementary Fig. 1). This hypothesis would provide a framework for future studies to map additional ligand binding sites outside of the catalytic domain using existing as well as DGK-tailored activity-based probes to gain deeper understanding of mechanisms regulating DGK specificity in biological systems.
While our studies provide key insights into DGK lipid specificity, we recognize that recombinant overexpression systems can alter cellular localization, structure, and regulation of DGK isoforms, which could affect our lipid specificity assessments. We also acknowledge the potential for compensatory effects (e.g. feedback regulation) in our live cell lipidomics studies. Together, these limitations could explain the increases in DAGs with overexpression of certain DGKs (Fig. 2a), the limited examples of direct DAG to PA conversion (Supplementary Fig. 11), and some differences in DGKε specificity (e.g. broader substrate profile and a preference for C16:0_C20:4 over C18:0_C20:4 DAG, Fig. 2a) compared with previous findings33, 40. Another caveat is overexpression of one recombinant DGK could affect expression of other native DGKs as well as proteins outside of the DGK superfamily. Despite validation that native DGKα expression was not affected (Supplementary Fig. 4), additional studies evaluating proteome-wide changes (including additional endogenous DGK isoforms) are needed to rule out potential non-specific effects from our recombinant system. Although our current findings support DGK C1 domains as a principal determinant of DAG fatty acyl specificity, future studies aimed at integrating C1 biology with additional regulatory mechanisms including, e.g. subcellular localization and membrane curvature50 are needed to decipher DGK function in vivo.
In summary, the DGK superfamily plays multifaceted roles in mammalian physiology and disease, and efforts are underway to target individual isoforms for therapeutic purposes. The identification of C1 domains as a key site for directing specificity of DGK activity is an important step toward advancing basic understanding of lipid signaling networks as well as the identification of ligand binding sites for developing isoform-selective inhibitors.
ONLINE METHODS
Reagents
Unless otherwise specified, all reagents were purchased from Fisher Scientific. Polyethyleneimine (Polysciences Inc., Cat# 24765), ritanserin ≥99% by HPLC (Tocris Bioscience, Cat# 1955), ketanserin tartrate ≥97% by HPLC (Tocris Bioscience, Cat# 0908), 1-stearoyl-2-arachidonoyl-d8-sn-glycerol (SAG-d8; Cayman Chemical Company, Cat# 10009872), and arachidonic acid-d8 (AA-d8; Cayman Chemical Company, Cat# 390010).
Cell culture
HEK293T cells were purchased from ATCC. DGKA Knockdown A549 cells were generated by Dr. Kwon-Sik Park Lab at University of Virginia. HEK293T cells were cultured in DMEM with 10% FBS (U.S. Source, Omega Scientific) and 1% L-glutamine (Thermo Fisher Scientific) in 10 cm2 plates. Media in cell cultures were switched to serum-free DMEM 4 hrs prior to harvesting for all lipid analyses or 1 hr for compound treatments. SILAC HEK293T cells were cultured in DMEM for SILAC (Fisher Scientific) supplemented with 10% dialyzed FBS (Omega Scientific) in 10 cm2 plates. Light medium was supplemented with 100 μg ml−1 L-arginine and 100 μg ml−1 L-lysine. Heavy medium was supplemented with 100 μg ml−1 [13C615N4]L-arginine and 100 μg ml−1 [13C615N2]L-lysine. Light or heavy amino acids were incorporated for at least 5 passages prior to utilizing SILAC HEK293T cells for experiments. All cells were grown to ∼80% confluency in a 37 °C incubator with 5% CO2. Cells were switched to serum-free DMEM media prior to compound treatments
Transient transfection
Recombinant DGK proteins were produced by transient transfection of HEK293T cells with recombinant DNA. pDONR223-DGKK was a gift from William Hahn and David Root (Addgene plasmid # 23487). pCSF107mT-GATEWAY-3’-FLAG was a gift from Todd Stukenberg (Addgene plasmid # 67619). pCSF107mT-DGKK-FLAG construct was generated by recombination of the Addgene plasmids using the Gateway cloning system (Invitrogen). All other vectors were either developed by Dr. Thurl Harris (University of Virginia, School of Medicine) or gifted to Dr. Kevin Lynch (University of Virginia, School of Medicine) by Dr. Kaoru Goto (Yamagata University, School of Medicine) and Dr. Fumio Sakane (Chiba University) and were kindly shared for these studies: pcDNA3-FLAG-DGKA (rat), pcDNA3-FLAG-DGKB (rat), pcDNA3-FLAG-DGKG (rat), pcDNA3.1-FLAG-DGKH (human), pCMV-Tag2B-FLAG-DGKQ (human), pGC-FLAG-DGKA (human), pGC-FLAG-DGKB (human), pGC-FLAG-DGKG (human), pcDNA3-DGKE-3xFlag (human), pCMV-HA-DGKI (human), and pCMV-SPORT6-HA-DGKZ (human). A549 cells with doxycycline-inducible knockdown of DGKA were developed and provided by Dr. Kwon-Sik Park (University of Virginia, School of Medicine). HEK293T cells were seeded at a density of 440,000 cells in complete DMEM and grown to 50–60% confluency. A polyethyleneimine (PEI) stock solution was prepared (1 mg/mL, pH 7.4) and filter sterilized. Serum-free DMEM (600 μL) was mixed gently with 20 μL of sterile PEI (1 mg/mL, and pH 7.4), and 2.6 μg DNA in a sterile microfuge tube. Mixtures were incubated for 30 min at 25 °C. The mixture was then added drop-wise to each 10 cm2 plate, rocked back and forth to mix, and placed back in the 37 °C incubator. Cell pellets were harvested after two full days of growth, snap-frozen in liquid N2, and stored at −80 °C until use. Recombinant proteins were produced by transient transfection in SILAC HEK293T cells using the procedure described in ‘Cell culture’ above, except that cells were plated at a concentration of 1 × 106 cells per 10 cm2 plate and grown to ∼70% confluency prior to introducing transfection mixture.
Western blot analysis of recombinant protein expression
The following antibodies were purchased for western blot studies: HA Epitope Tag Monoclonal Antibody (Thermo Fisher Scientific) Cat# PI26183; Anti-FLAG antibody produced in rabbit (Sigma-Aldrich) Cat# F7425; Goat anti-mouse DyLight 650 (Thermo Fisher Scientific) Cat# 84545; Goat anti-rabbit DyLight 550 (Thermo Fisher Scientific) Cat# 84541; DGKA Antibody Rabbit Polyclonal (Proteintech) Cat# 11547. Cell lysates were separated via ultracentrifugation at 100,000 x g for 45 min at 4 °C. Proteins separated by SDS-PAGE (7.5% polyacrylamide, TGX Stain-Free Mini Gel) at 150 V for 55 min. Gel transfers were performed using the Bio-Rad Trans-Blot Turbo RTA Midi Nitrocellulose Transfer Kit with a Bio-Rad Trans-Blot Turbo Transfer System (25V, 10 min). The nitrocellulose blot was then incubated in blocking solution (30 mL, 3% BSA in Tris-buffered saline and Tween 20 (TBS-T) (1.5 M NaCl, 0.25 M Tris, 0.1% Tween 20 in double distilled H2O (ddH2O), pH 7.4) for 1 hr at 25 °C with gentle shaking. The blot was then transferred immediately to primary antibody solution (1:1,000 anti-FLAG, 1:10,000 anti-HA or 1:1,000 anti-DGKα in TBS-T) and incubated overnight at 4 °C with gentle shaking. The blot was then rinsed 5 times for 5 min in TBS-T, transferred immediately into secondary antibody solution (1:10,000 anti-species DyLight 550 or DyLight 650 in TBS-T), and incubated for 1 hr at 25 °C with gentle shaking. The blot was then rinsed 5 times for 5 min in TBS-T, transferred into ddH2O, and imaged by in-blot fluorescence scanning on a ChemiDoc MP Imaging System. Each lane displayed in western blots represents an individual biological replicate of that overexpression/treatment condition.
Doxycycline-induced shRNA knockdown of DGKα in A549 cells
A549 cells were infected with lentiviral shRNAs targeting human DGKA gene in Tet-pLKO-puro plasmid (a gift from Dmitri Wiederschain; Addgene #21915). Oligonucleotide sequences for the target are as follows: shDGKA-1, 5′-CGGATTGACCCTGTTCCTAAC-3′, for shDGKA-2 5′-CGGCCAGAAGACAAGTTAGAA-3’. Lentiviral particles were produced by co-transfecting the lentiviral plasmid with 2 packaging plasmids (psPAX2 and pMD2.G) in HEK293T cells using PEI (Sigma, 408727). Supernatants containing the lentiviral particles were harvested 48 and 72 hrs after the transfection and filtered through 0.45 μm PVDF membrane. Infections were performed with viral supernatant in the presence of 5 μg/ml polybrene (Sigma, H9268). Puromycin (2 μg ml−1; A1113803, Thermo Fisher Scientific) was used to select stably transduced cells. DGKA knockdown in the selected cells was induced by adding doxycycline to culture media (Sigma, D9891) and was validated by immunoblot. A549 cells were cultured in a similar manner to HEK293T cells until they reached 80% confluency. Cells were treated with 10 μL of a 0.2 μg/μL doxycycline stock diluted in DMSO for 48 hrs. Following treatment, cells were starved, harvested, and analyzed through either LC-MS/MS of lipid extracts or through western blot analysis of endogenous DGKα expression. DGKα knockdown was confirmed by western blot.
Site-directed mutagenesis
Primers for type 1 DGK lysine-alanine mutants were generated and purchased from Integrated DNA Technologies (IDT). The mutagenesis reaction was performed by combining the following in a PCR tube: MilliQ H2O (34.8 μL), PfuUltra enzyme (1 μL), 10X PfuUltra buffer (5 μL), dNTP mix (1 μL), and template plasmid (5 μL). Primers were diluted to 10 μM in MilliQ H2O and 1.6 μL of each (forward and reverse) were added to the mutagenesis master mix to bring the final volume to 50 μL. The mixture was placed in a thermocycler under the following protocol after being brought up to 98 °C: 98 °C (10 secs), 55 °C (30 secs), 72 °C (30 secs) for 35 cycles then left at 4 °C overnight. Following PCR DpnI restriction enzyme (1 μL) was added, mixed, and incubated at 37°C for 1 hr. This product was then transformed into electrocompetent E.coli cells (XL-1 from Agilent) and grown to obtain isolated mutated plasmid.
Lipid extraction
Modified Bligh Dyer method (CHCl3:MeOH:H2O/1:2:2; 0.1N HCl) was used to extract lipids from HEK293T cell pellets (~6.0 million cells). Antioxidant BHT (butylated hydroxy toluene) was added at 50 μg/mL during extraction. CHCl3 (1 mL), and MeOH (2 mL) were added with 1.5 mL of ddH2O containing resuspended HEK293T cells in a two-dram vial. 1N HCl (500 μL) was added last to bring the final concentration of acid to 0.1 N. Samples were vortexed, incubated on ice for 20 min, and centrifuged at 2,000 x g for 5 min at 4 °C. Lipid standards (10 pmol each of AA-d8 and SAG-d8) were added to organic solvents prior to mixing and lipid extraction of cells. The organic layer was transferred and aqueous layer extracted with addition of 1.5 mL of 1:2 CHCl3:MeOH solution. The extracted organic layers were combined and dried down under nitrogen stream. Samples were resuspended in 240 μL of 1:1 MeOH:IPA and stored at −80 °C until further analysis.
LC-MS/MS analysis of lipid extracts
The lipid samples were analyzed by LC-MS/MS. A Dionex Ultimate 3000 RS UHPLC system was used with an analytical column (Kinetex® 1.7 μm C18 100 Å, Phenomenex, LC column 100 × 2.1 mm) and reverse phase LC solvents (A: ACN:H2O/50:50, 10 mM NH4HCO2, 0.1% formic acid; B: ACN:IPA:H2O/10:88:2, 10 mM NH4HCO2, 0.1% formic acid) with the following gradient: Flowrate 0.25 mL/min, 0 min 65% A, 4 min 40% A, 12 min 15% A, 21 min 0% A, 24 min, 0% A, 24.1 min 100% A, 27 min 0% A, 30 min 100% A, 33 min 0% A, 35 min 65% A. The eluted lipids were ionized by electrospray using a HESI-II probe into an Orbitrap Q-Exactive Plus mass spectrometer (Thermo Scientific). Data acquisition was performed using both PRM targeted and top10 data-dependent (ddMS2) global analysis methods. PRM targeting of diacylglycerol and phosphatidic acid lipid species was accomplished by filtering for the [M+NH4]+ and [M-H]- adduct ions, respectively and subsequent MS/MS (MS2) detection of expected diagnostic fragment ions (neutral loss of fatty acyl chains in positive mode and loss of the fatty acyl chain along with H in the negative mode). Intensities of the lipid species were measured using TraceFinder™ software, which identified fragment ions across multiple samples based on the MS2 filter defined in a targeted list of lipids and aligned them according to the intensities of the ion species found in each raw file. Lipid identifications and peak alignments were performed using LipidSearch™ software while quantitative analysis of the aligned intensities was exported and analyzed using Prism GraphPad version 7.03. We found use of the acid-modified Bligh Dyer method extraction method resulted in ~140% enhanced lipid coverage across multiple lipid classes (e.g. phospholipids, triacylglycerols, etc.) compared with other lipid extraction methods (e.g. Folch method; Supplementary Table 1). Positive and negative ion annotations for each sample were combined and aligned within a chromatographic time window to allow greater confidence in lipid identifications using appropriate MS2 product ions and neutral losses from the compiled dataset in LipidSearch™ analysis software51. Aligned results were further filtered by retention time constraints, signal to noise ratio, intensity ratio of diagnostic ion, main adduct ion, identification quality, and peak quality specifications. See Supplementary Fig. 5 for an example.
Sample preparation for quantitative LC-MS/MS analysis using ATP acyl phosphates
Samples were prepared as previously described7.
LC-MS/MS analysis of SILAC samples
SILAC samples were analyzed as previously described7.
Immunofluorescence microscopy
HEK293T cells were seeded at 50,000 cells/mL on sterile 22 mm borosilicate glass coverslips (Fisher Scientific) in 6-well dishes with 3 mL of complete DMEM or at 10,000 cells/mL on sterile 5 mm borosilicate glass coverslips in 24-well dishes with 1 mL of complete DMEM. Once cells reached 60% confluency, they were transfected with desired recombinant DGK isoform for 48 hrs. Cells were washed in PBS (2X), fixed with 4% formaldehyde in PBS for 20 mins, washed in PBS (2X), and stained with a 1:2,000 wheat germ agglutinin (WGA) stain conjugated to either an Alexa 350 fluorophore (Thermo Fisher Scientific) or Alexa 488 fluorophore (Biotium) in PBS at 37 °C for 15 mins. Fixed cells were again washed in PBS (2X), permeabilized with 0.125% Triton X-100 in PBS for 5 mins, and subsequently washed in PBS (2X). Cells were incubated with 1:1,000 anti-FLAG antibody from rabbit (Thermo Fisher Scientific) diluted in 3% (w/v) BSA-PBS at 37 °C for 3 hrs. Cells were washed in 0.1% Tween 20-PBS (5X), incubated with 1:10,000 goat anti-rabbit antibody (Thermo Fisher Scientific) diluted in PBS at 37 °C for 1 hr, and washed in 0.1% Tween 20-PBS (5X). Coverslips were mounted (cell side down) to glass slides using mounting media (Thermo Fisher Scientific) and stored at 25 °C until analysis on a Zeiss 780 NLO Confocal microscope or stored in PBS at 4 °C away from light until analysis with a home-built lattice light sheet microscope.
Lattice-light sheet microscopy image acquisition and data processing
Cell samples were imaged with a home-made lattice light sheet microscope. The square lattice pattern was applied to generate the excitation light sheet. The angle between the excitation light sheet and the glass coverslip is 31.8 degrees. The 3D image is produced from a stack of 2D slices by translating the specimen with a piezo stage (Physik Instrumente, P-621.1CD) through the stationary light sheet horizontally in the plane of the specimen coverslip. Typically, 400 2D slices were collected for one data set with a step size of 200 nm. The illumination intensity of the excitation beam (561 nm) at the sample was about 1 W/cm2. Images were acquired with the exposure time of 100 ms. The raw 3D image data was de-skewed and deconvolved as described previously45, 46 by using the Richardson-Lucy algorithm and the reconstructed 3D images were rendered using the 3D Viewer plugin in Fiji52.
Liposomal DAG kinase assay
The liposomal method for measuring DAG kinase activity was modified from previously reported method44. Briefly, lipids were prepared for liposome formation by dissolving dioleoyl phosphatidylcholine (DOPC), dioleoyl phosphatidylserine (DOPS), and the appropriate DAG species in chloroform, combining, and drying in vacuo to remove all solvent. The surface concentration of lipids within the liposomes was 10 mol% DAG, 20 mol% DOPS, and 70 mol% DOPC. The lipids were then hydrated to 10 mM in liposome buffer (50 mM (3-(N-morpholino)propanesulfonic acid) (MOPS), pH 7.5, 100 mM NaCl and 5 mM MgCl2). The hydrated lipids were subjected to five freeze-thaw cycles in liquid nitrogen, followed by extrusion through a 100 nm polycarbonate filter 11 times. The measurement of DGK activity was determined by following the incorporation of the γ-Phosphate from [γ−32P]ATP into DAG to form a radiolabeled PA product. Immediately prior to running assays, [γ−32P]ATP was prepared by adding 0.5 – 2 μL of [γ−32P]ATP (6000 Ci/mmol) to 500 μL of 10 mM ATP. The assays were run as 100 μL reactions and contained liposome buffer, 0.1 mM CaCl2, 1 mM dithiothreitol (DTT), appropriate cell lysate (4.4 μg total protein), 40 μL liposomes (4 mM lipids), and were initiated with the addition of 10 μL prepared 10mM [γ−32P] ATP. The reactions were allowed to proceed for 20 min at 30 °C before being terminated with the addition of 0.5 mL methanol with 0.1 N HCl, followed by 1 mL of ethyl acetate. Phase separation was achieved with the addition of 1 mL 1 M MgCl2, and the organic phase was washed by thoroughly vortexing to remove soluble [γ−32P] ATP. To measure the incorporation of [32P] into DAG, the extract was centrifuged at 200 x G for 1 minute, and 0.5 ml of the organic phase was removed and the radioactivity was measured using a scintillation counter. The activity of each DGK-expressing lysate was normalized to GFP-expressing lysates as background. Activity was calculated as nmol of PA produced per minute per mg of total protein present in the assay.
Sequence alignments
Lipid kinase sequences were obtained from Uniprot (https://www.uniprot.org/) and aligned using Clustal Omega53, 54.
Hierarchical clustering
Calculation and generation of hierarchical clustering dendrograms was done using R with the following publicly available packages: ‘cluster’, ‘factoextra’, and ‘tidyverse’. A Euclidean method was used to calculate distance measurements while the ‘ward.D2’ agglomeration method was utilized to calculate the dissimilarity values between observed parameters of the lipid analysis for hierarchical clustering.
Quantification and statistical analysis
For the lipid analyses, the intensities of targeted lipid species and their deuterated counterparts obtained from the LC-MS/MS analysis were used for the semi-quantitative determination of lipid concentrations within a sample. Intensities of lipid species were divided by the intensities of deuterated standards (arachidonic acid-d8 (AA-d8) for negative mode and 1-stearoyl-2-arachidonoyl-sn-glycerol-d8 (SAG-d8) for positive mode; 10 pmol). The calculated lipid amounts were normalized based on total protein concentration of respective samples. Statistical analyses were performed in GraphPad Prism (v7.03). In order to determine statistically significant differences between groups analyzing multiple lipid species, a multiple hypothesis t-test (Dunnett’s test) with Benjamini-Hochberg correction for false discovery rates (do not assume consistent standard deviation, confidence interval of 95%) was applied. Otherwise a one-way ANOVA test was implemented to demonstrate significant differences between multiple groups with respect to individual lipid changes. Statistical significance was set at P < 0.05. The number of experimental biological replicates of cell populations in each sample group are indicated by n and can be found in respective figure legends. Data are shown as mean ± standard error of mean (s.e.m.).
Software and algorithms
The following software and algorithms were used in this report: In-gel/in-blot fluorescence scanning and normalization; Image Lab software (Bio-Rad). MS1 and MS2 file conversion; RawConverter (RawConverter). MS data protein search algorithm; ProLuCID (Integrated Proteomics Applications – IP2). MS data lipid search algorithm; LipidSearch (Thermo Fisher Scientific). MS data acquisition; Xcalibur (Thermo Fisher Scientific). MS data protein analysis; Skyline-daily (MacCoss Lab Software). MS data lipid analysis; TraceFinder (Thermo Fisher Scientific). Super-resolution microscopy image analysis – Fiji (ImageJ). Confocal microscopy image analysis – CZI (Zeiss). Sequence alignment software; Clustal Omega (EMBL-EBI). Statistical analysis calculation; Prism (GraphPad). Hierarchical clustering analysis; R (RStudio).
Data availability
All data produced or analyzed for this study are included in the published article (and its supplementary information files) or are available from the corresponding author on reasonable request.
Code availability
All code is available upon reasonable request from the corresponding author.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Ross and all members of the Hsu Lab for helpful discussions. We thank Sean Campbell for assistance in designing DGK chimera plasmids. This work was supported by the University of Virginia (start-up funds to K.-L.H.), National Institutes of Health Grants (DA035864 and DA043571 to K.-L.H.; grant no. GM801868 to T.B.W.; grant no. GM007055 to C.E.F.), U.S. Department of Defense (Grant W81XWH-17–1-0487 to K.-L.H.), NCI Cancer Center Support Grant (5P30CA044579–27 to K.-L.H. and T.E.H.), and the Schiff Foundation (Brain Tumor Research Grant to K.-L.H. and T.E.H.). Images were acquired using the W.M. Keck Center for Cellular Imaging Zeiss 780 Confocal microscopy system at UVA (NIH OD016446).
Footnotes
COMPETING INTERESTS
The authors declare no competing financial interest.
REFERENCES
- 1.Aimon S, Callan-Jones A, Berthaud A, Pinot M, Toombes GE, Bassereau P. Membrane shape modulates transmembrane protein distribution. Dev Cell 2014, 28(2): 212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443(7112): 651–657. [DOI] [PubMed] [Google Scholar]
- 3.Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell 2005, 122(4): 605–617. [DOI] [PubMed] [Google Scholar]
- 4.Almena M, Mérida I. Shaping up the membrane: diacylglycerol coordinates spatial orientation of signaling. Trends Biochem Sci 2011, 36(11): 593–603. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Su Y, Wang X. Phosphatidic acid-mediated signaling. Adv Exp Med Biol 2013, 991: 159–176. [DOI] [PubMed] [Google Scholar]
- 6.van Blitterswijk WJ, Houssa B. Properties and functions of diacylglycerol kinases. Cell Signal 2000, 12(9–10): 595–605. [DOI] [PubMed] [Google Scholar]
- 7.Franks CE, Campbell ST, Purow BW, Harris TE, Hsu KL. The Ligand Binding Landscape of Diacylglycerol Kinases. Cell Chem Biol 2017, 24(7): 870–880 e875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada K, Sakane F, Matsushima N, Kanoh H. EF-hand motifs of alpha, beta and gamma isoforms of diacylglycerol kinase bind calcium with different affinities and conformational changes. Biochem J 1997, 321: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abramovici H, Hogan AB, Obagi C, Topham MK, Gee SH. Diacylglycerol kinase-zeta localization in skeletal muscle is regulated by phosphorylation and interaction with syntrophins. Mol Biol Cell 2003, 14(11): 4499–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kume A, Kawase K, Komenoi S, Usuki T, Takeshita E, Sakai H, et al. The Pleckstrin Homology Domain of Diacylglycerol Kinase eta Strongly and Selectively Binds to Phosphatidylinositol 4,5-Bisphosphate. J Biol Chem 2016, 291(15): 8150–8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai S, Sakane F, Kanoh H. Phorbol ester-regulated oligomerization of diacylglycerol kinase delta linked to its phosphorylation and translocation. J Biol Chem 2002, 277(38): 35323–35332. [DOI] [PubMed] [Google Scholar]
- 12.Harada BT, Knight MJ, Imai S, Qiao F, Ramachander R, Sawaya MR, et al. Regulation of enzyme localization by polymerization: polymer formation by the SAM domain of diacylglycerol kinase delta1. Structure 2008, 16(3): 380–387. [DOI] [PubMed] [Google Scholar]
- 13.Jing W, Gershan JA, Holzhauer S, Weber J, Palen K, McOlash L, et al. T cells deficient in diacylglycerol kinase-zeta are resistant to PD-1 inhibition and help create persistent host immunity to leukemia. Cancer Res 2017, 77(20): 5676–5686. [DOI] [PubMed] [Google Scholar]
- 14.Olenchock BA, Guo R, Carpenter JH, Jordan M, Topham MK, Koretzky GA, et al. Disruption of diacylglycerol metabolism impairs the induction of T cell anergy. Nat Immunol 2006, 7(11): 1174–1181. [DOI] [PubMed] [Google Scholar]
- 15.Zha Y, Marks R, Ho AW, Peterson AC, Janardhan S, Brown I, et al. T cell anergy is reversed by active Ras and is regulated by diacylglycerol kinase-alpha. Nat Immunol 2006, 7(11): 1166–1173. [DOI] [PubMed] [Google Scholar]
- 16.Merida I, Andrada E, Gharbi SI, Avila-Flores A. Redundant and specialized roles for diacylglycerol kinases alpha and zeta in the control of T cell functions. Sci Signal 2015, 8(374): re6. [DOI] [PubMed] [Google Scholar]
- 17.Prinz PU, Mendler AN, Masouris I, Durner L, Oberneder R, Noessner E. High DGK-alpha and disabled MAPK pathways cause dysfunction of human tumor-infiltrating CD8+ T cells that is reversible by pharmacologic intervention. J Immunol 2012, 188(12): 5990–6000. [DOI] [PubMed] [Google Scholar]
- 18.Riese MJ, Wang LC, Moon EK, Joshi RP, Ranganathan A, June CH, et al. Enhanced effector responses in activated CD8+ T cells deficient in diacylglycerol kinases. Cancer Res 2013, 73(12): 3566–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo R, Wan CK, Carpenter JH, Mousallem T, Boustany RM, Kuan CT, et al. Synergistic control of T cell development and tumor suppression by diacylglycerol kinase alpha and zeta. Proc Natl Acad Sci U S A 2008, 105(33): 11909–11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pankratz N, Wilk JB, Latourelle JC, DeStefano AL, Halter C, Pugh EW, et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet 2009, 124(6): 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon-Sanchez J, van Hilten JJ, van de Warrenburg B, Post B, Berendse HW, Arepalli S, et al. Genome-wide association study confirms extant PD risk loci among the Dutch. Eur J Hum Genet 2011, 19(6): 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry 2008, 13(2): 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber H, Kittel-Schneider S, Gessner A, Domschke K, Neuner M, Jacob CP, et al. Cross-disorder analysis of bipolar risk genes: further evidence of DGKH as a risk gene for bipolar disorder, but also unipolar depression and adult ADHD. Neuropsychopharmacology 2011, 36(10): 2076–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Squassina A, Manchia M, Congiu D, Severino G, Chillotti C, Ardau R, et al. The diacylglycerol kinase eta gene and bipolar disorder: a replication study in a Sardinian sample. Mol Psychiatry 2009, 14(4): 350–351. [DOI] [PubMed] [Google Scholar]
- 25.Zeng Z, Wang T, Li T, Li Y, Chen P, Zhao Q, et al. Common SNPs and haplotypes in DGKH are associated with bipolar disorder and schizophrenia in the Chinese Han population. Mol Psychiatry 2011, 16(5): 473–475. [DOI] [PubMed] [Google Scholar]
- 26.Moya PR, Murphy DL, McMahon FJ, Wendland JR. Increased gene expression of diacylglycerol kinase eta in bipolar disorder. Int J Neuropsychopharmacol 2010, 13(8): 1127–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melen E, Himes BE, Brehm JM, Boutaoui N, Klanderman BJ, Sylvia JS, et al. Analyses of shared genetic factors between asthma and obesity in children. J Allergy Clin Immunol 2010, 126(3): 631–637 e631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laramie JM, Wilk JB, Williamson SL, Nagle MW, Latourelle JC, Tobin JE, et al. Multiple genes influence BMI on chromosome 7q31–34: the NHLBI Family Heart Study. Obesity (Silver Spring) 2009, 17(12): 2182–2189. [DOI] [PubMed] [Google Scholar]
- 29.Jiang LQ, de Castro Barbosa T, Massart J, Deshmukh AS, Lofgren L, Duque-Guimaraes DE, et al. Diacylglycerol kinase-delta regulates AMPK signaling, lipid metabolism, and skeletal muscle energetics. Am J Physiol Endocrinol Metab 2016, 310(1): E51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowe CE, Zhang Q, Dennis RJ, Aubry EM, O’Rahilly S, Wakelam MJ, et al. Knockdown of diacylglycerol kinase delta inhibits adipocyte differentiation and alters lipid synthesis. Obesity (Silver Spring) 2013, 21(9): 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chibalin AV, Leng Y, Vieira E, Krook A, Bjornholm M, Long YC, et al. Downregulation of diacylglycerol kinase delta contributes to hyperglycemia-induced insulin resistance. Cell 2008, 132(3): 375–386. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Chang GQ, Leibowitz SF. Diacylglycerol kinase zeta in hypothalamus interacts with long form leptin receptor. Relation to dietary fat and body weight regulation. J Biol Chem 2001, 276(8): 5900–5907. [DOI] [PubMed] [Google Scholar]
- 33.Lung M, Shulga YV, Ivanova PT, Myers DS, Milne SB, Brown HA, et al. Diacylglycerol kinase epsilon is selective for both acyl chains of phosphatidic acid or diacylglycerol. J Biol Chem 2009, 284(45): 31062–31073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shulga YV, Topham MK, Epand RM. Study of arachidonoyl specificity in two enzymes of the PI cycle. J Mol Biol 2011, 409(2): 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shulga YV, Topham MK, Epand RM. Substrate specificity of diacylglycerol kinase-epsilon and the phosphatidylinositol cycle. FEBS Lett 2011, 585(24): 4025–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez de Turco EB, Tang W, Topham MK, Sakane F, Marcheselli VL, Chen C, et al. Diacylglycerol kinase epsilon regulates seizure susceptibility and long-term potentiation through arachidonoyl- inositol lipid signaling. Proc Natl Acad Sci U S A 2001, 98(8): 4740–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marquez VE, Blumberg PM. Synthetic diacylglycerols (DAG) and DAG-lactones as activators of protein kinase C (PK-C). Acc Chem Res 2003, 36(6): 434–443. [DOI] [PubMed] [Google Scholar]
- 38.Das J, Rahman GM. C1 domains: structure and ligand-binding properties. Chem Rev 2014, 114(24): 12108–12131. [DOI] [PubMed] [Google Scholar]
- 39.Ware TB, Shin M, Hsu K-L. Metabolomics analysis of lipid metabolizing enzyme activity. Methods Enzymol 2019, 626: 407–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettitt TR, Wakelam MJO. Diacylglycerol kinase epsilon, but not zeta, selectively removes polyunsaturated diacylglycerol, inducing altered protein kinase C distribution in vivo. J Biol Chem 1999, 274(51): 36181–36186. [DOI] [PubMed] [Google Scholar]
- 41.Franks CE, Hsu KL. Activity-Based Kinome Profiling Using Chemical Proteomics and ATP Acyl Phosphates. Curr Protoc Chem Biol 2019, 11(3): e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Functional Mann M. and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol 2006, 7(12): 952–958. [DOI] [PubMed] [Google Scholar]
- 43.McCloud RL, Franks CE, Campbell ST, Purow BW, Harris TE, Hsu KL. Deconstructing Lipid Kinase Inhibitors by Chemical Proteomics. Biochemistry 2018, 57(2): 231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boroda S, Niccum M, Raje V, Purow BW, Harris TE. Dual activities of ritanserin and R59022 as DGKalpha inhibitors and serotonin receptor antagonists. Biochem Pharmacol 2017, 123: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 2014, 346(6208): 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li D, Shao L, Chen BC, Zhang X, Zhang M, Moses B, et al. ADVANCED IMAGING. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 2015, 349(6251): aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imai S, Kai M, Yasuda S, Kanoh H, Sakane F. Identification and characterization of a novel human type II diacylglycerol kinase, DGK kappa. J Biol Chem 2005, 280(48): 39870–39881. [DOI] [PubMed] [Google Scholar]
- 48.Klauck TM, Xu X, Mousseau B, Jaken S. Cloning and characterization of a glucocorticoid-induced diacylglycerol kinase. J Biol Chem 1996, 271(33): 19781–19788. [DOI] [PubMed] [Google Scholar]
- 49.Sakane F, Imai S, Kai M, Wada I, Kanoh H. Molecular cloning of a novel diacylglycerol kinase isozyme with a pleckstrin homology domain and a C-terminal tail similar to those of the EPH family of protein-tyrosine kinases. J Biol Chem 1996, 271(14): 8394–8401. [DOI] [PubMed] [Google Scholar]
- 50.Bozelli JC Jr., Jennings W, Black S, Hou YH, Lameire D, Chatha P, et al. Membrane curvature allosterically regulates the phosphatidylinositol cycle, controlling its rate and acyl-chain composition of its lipid intermediates. J Biol Chem 2018, 293(46): 17780–17791. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES (Methods-only)
- 51.Breitkopf SB, Ricoult SJH, Yuan M, Xu Y, Peake DA, Manning BD, et al. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 2017, 13(3): 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012, 9(7): 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 2010, 38: W695–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011, 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data produced or analyzed for this study are included in the published article (and its supplementary information files) or are available from the corresponding author on reasonable request.