Gut microbes are increasingly being recognized as critical contributors to nutrient accessibility in animals. The globally distributed omnivorous American cockroach (Periplaneta americana) harbors many bacterial phyla (e.g., Bacteroidetes) that are abundant in vertebrates. P. americana thrives on a highly diverse plant-enriched diet, making this insect a rich potential source of uncharacterized polysaccharolytic bacteria. We have cultivated, completely sequenced, and functionally characterized several novel Bacteroidetes species that are endemic to the P. americana gut, and many of these isolates can degrade simple and complex polysaccharides. Cultivation and genomic characterization of these Bacteroidetes isolates further enable deeper insight into how these taxa participate in polysaccharide metabolism and, more broadly, how they affect animal health and development.
KEYWORDS: Bacteroidetes, PULs, polysaccharolysis
ABSTRACT
Beneficial gut microbes can facilitate insect growth on diverse diets. The omnivorous American cockroach, Periplaneta americana (Insecta: Blattodea), thrives on a diet rich in plant polysaccharides and harbors a species-rich gut microbiota responsive to host diet. Bacteroidetes are among the most abundant taxa in P. americana and other cockroaches, based on cultivation-independent gut community profiling, and these potentially polysaccharolytic bacteria may contribute to host diet processing. Eleven Bacteroidetes isolates were cultivated from P. americana digestive tracts, and phylogenomic analyses suggest that they were new Bacteroides, Dysgonomonas, Paludibacter, and Parabacteroides species distinct from those previously isolated from other insects, humans, and environmental sources. In addition, complete genomes were generated for each isolate, and polysaccharide utilization loci (PULs) and several non-PUL-associated carbohydrate-active enzyme (CAZyme)-coding genes that putatively target starch, pectin, and/or cellulose were annotated in each of the isolate genomes. Type IX secretion system (T9SS)- and CAZyme-coding genes tagged with the corresponding T9SS recognition and export C-terminal domain were observed in some isolates, suggesting that these CAZymes were deployed via non-PUL outer membrane translocons. Additionally, single-substrate growth and enzymatic assays confirmed genomic predictions that a subset of the Bacteroides and Dysgonomonas isolates could degrade starch, pectin, and/or cellulose and grow in the presence of these substrates as a single sugar source. Plant polysaccharides enrich P. americana diets, and many of these gut isolates are well equipped to exploit host dietary inputs and potentially contribute to gut community and host nutrient accessibility.
IMPORTANCE Gut microbes are increasingly being recognized as critical contributors to nutrient accessibility in animals. The globally distributed omnivorous American cockroach (Periplaneta americana) harbors many bacterial phyla (e.g., Bacteroidetes) that are abundant in vertebrates. P. americana thrives on a highly diverse plant-enriched diet, making this insect a rich potential source of uncharacterized polysaccharolytic bacteria. We have cultivated, completely sequenced, and functionally characterized several novel Bacteroidetes species that are endemic to the P. americana gut, and many of these isolates can degrade simple and complex polysaccharides. Cultivation and genomic characterization of these Bacteroidetes isolates further enable deeper insight into how these taxa participate in polysaccharide metabolism and, more broadly, how they affect animal health and development.
INTRODUCTION
Complex polysaccharides are an abundant and valuable raw material for metabolite and energy production in animals, provided that they generate or have access to the enzymes required to cleave polysaccharides into assimilable monosaccharides (1, 2). Glycoside hydrolases (GH) and polysaccharide lyases (PL) constitute a broad class of enzymes that break glycosidic bonds within polysaccharides (3). Although invertebrates and vertebrates produce some GH and PL de novo, typically gut microbial symbionts supply the bulk of the polysaccharolytic enzymes that degrade dietary polysaccharides into assimilable monosaccharides (4–8). Frequently, these microbes produce enzymes that release monosaccharides that are not absorbed by the producing organism but remain available as “public goods” for the wider microbial community or host (9, 10). Liberated monosaccharides can be directly absorbed by the host or fermented by gut microbes to yield short-chain fatty acids that can contribute to both host and microbial metabolism (6, 11–13). The Bacteroidetes phylum includes species that are commonly abundant in gut bacterial microbiomes of many omnivorous and xylophagous invertebrates and vertebrates, and many of these bacteria can degrade different types of polysaccharides, including complex carbohydrates from plants found in their host’s diets (14, 15).
Bacteroidetes can use carbohydrate-active enzymes (CAZymes) (e.g., GH, carboxyl esterase [CE], carbohydrate binding molecules [CBM], and PL) that hydrolyze carbohydrate substrates into oligo- and monosaccharides. Genes encoding CAZymes in Bacteroidetes can be found as individual or multiple loci or in coregulated, substrate-specific gene clusters called polysaccharide utilization loci (PULs) (16) that detect, transport, and hydrolyze complex carbohydrates into assimilable monosaccharides (2). In addition to CAZymes, PULs encode the following: (i) SusD cell surface glycan binding proteins, (ii) SusC-like/TonB-dependent transporters, and (iii) transcriptional regulators that include extracytoplasmic (ECT) sigma/anti-sigma factors as well as hybrid and conventional two-component systems (2). PULs responsive to plant-based and host-derived glycans have been experimentally characterized in Bacteroides thetaiotaomicron, Bacteroides ovatus, Bacteroides cellulosilyticus, Bacteroides xylanisolvens, and other Bacteroidetes that are endemic in humans (8, 10, 17–20). However, PULs detected in Bacteroides spp. and Bacteroidetes genera that reside in other animals, namely, insects, that consume largely plant-based diets remain underdescribed.
Termites and cockroaches (Insecta: Blattodea) are globally distributed and conspicuously effective consumers of largely to strictly plant-based diets, and several CAZymes have been detected in the relatively few genomes available for symbiotic, gut-residing Bacteroidetes species (21–23). While strict wood feeding is common among lower termites, omnivorous cockroaches, including the American cockroach (Periplaneta americana), comprise the lineages basal to the Blattodea (24). The P. americana diet is comprised of scavenged plant- and animal-based detritus that includes feces and exuvia (25). P. americana does not synthesize the enzymes required for the complete hydrolysis of some complex dietary polysaccharides (e.g., pectin) into monomeric sugars (4, 11, 25, 26), yet it endogenously produces both amylases and β-endoglucanases (27, 28). Like many animals, P. americana likely relies upon gut microbes to produce many of the necessary diet-degradative enzymes (11). Metagenomics and 16S rRNA gene community profiling have shown that Bacteroidetes and Firmicutes, which include many polysaccharolytic bacterial species, are abundant (>0.1% of the total community) in the gut microbial communities of P. americana (29–31). Although Bacteroidetes in P. americana include well-characterized bacterial genera (30, 31) with known complex carbohydrate-degrading abilities (17, 32–34), the particular polysaccharolytic capabilities of bacteria endemic to these insect digestive tracts are not well understood.
Eleven Bacteroidetes species were successfully isolated from P. americana with the aim of characterizing the polysaccharolytic capabilities of cockroach-associated Bacteroidetes and inferring their contributions to host nutrient accessibility. Comprehensive genome sequencing and comparative genomic approaches were used to identify and annotate CAZy-coding genes arranged in either PUL or non-PUL clusters and determine their relatedness to other Bacteroidetes species detected in distinct habitats and within diverse ecological niches. Detailed genome annotation revealed a subset of non-PUL CAZymes that could use type IX secretion systems (T9SS) to be extracellularly exported, which could further enable polysaccharide utilization. In vivo and in vitro assays were used to experimentally verify predicted polysaccharolytic activities inferred from genome annotations.
RESULTS
Genomic analysis of P. americana Bacteroidetes isolates.
Near-complete (97.1% to 99.8% complete) genome assemblies were obtained for each of the eleven P. americana Bacteroidetes isolates, and their sizes ranged between 3.01 to 6.18 Mbp (Table 1), with most isolates being greater than the average (3.66-Mbp) size of 557 complete, publicly available Bacteroidetes genomes (NCBI genome database, accessed 15 May 2019). Genomes PAB51 (6.18 Mbp) and PAD521 (5.24 Mbp) were the largest and bear the greatest number of coding DNA sequences (CDS) (4,815 and 4,557, respectively). The percent G+C contents (G+C%) for all genomes ranged from 37.38 to 43.04% (see Fig. S1A in the supplemental material), with genomes PAD511, PAD521, and PAD25 having higher G+C% than the genomes of previously sequenced Dysgonomonas (22) and genomes PAB519, PAB224, and PAR221 having slightly lower G+C% than previously sequenced Bacteroides and Paludibacter genomes (35, 36). All isolate genomes encoded the minimum complement of tRNAs sufficient to translate all of the proteinogenic amino acids. Only the PAD521 genome appeared to harbor a 31.29-kb plasmid with 30 predicted protein-coding regions, and several CDS on this contig were annotated as orthologues of proteins involved in plasmid replication (Hin recombinase, superfamily II DNA-RNA helicases, and plasmid stabilization system proteins). Finally, the relative proportions of clusters of orthologous groups (COGs) encoded by P. americana Bacteroidetes isolate genomes were insignificantly different from those for Bacteroidetes isolates that inhabit other invertebrates, animals, and environmental systems (see Fig. S1B in the supplemental material).
TABLE 1.
General genomic features from P. americana Bacteroidetes isolates
| Parameter |
Dysgonomonas |
Bacteroides |
Paludibacter | Parabacteroides | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PAD511 | PAD520 | PAD521 | PAD25 | PAD216 | PAB51 | PAB519 | PAB214 | PAB224 | PAR221 | PAP52 | |
| No. of scaffolds | 159 | 70 | 263 | 59 | 54 | 350 | 163 | 53 | 128 | 103 | 61 |
| Estimated genome size (Mb) | 3.78 | 3.91 | 5.24 | 3.75 | 3.18 | 6.18 | 5.55 | 3.39 | 4.48 | 3.11 | 3.01 |
| Coverage per base (×) | 312 | 447 | 199 | 304 | 430 | 159 | 99 | 144 | 122 | 434 | 419 |
| N50 | 106,899 | 141,582 | 51,575 | 315,951 | 141,940 | 80,268 | 89,892 | 225,770 | 87,256 | 51,319 | 136,473 |
| G+C content (%) | 41.72 | 38.92 | 40.76 | 42.54 | 37.38 | 42.77 | 39.34 | 41.37 | 39.42 | 39.89 | 43.04 |
| No. of: | |||||||||||
| CDS genes | 3,109 | 3,337 | 4,557 | 3,285 | 2,679 | 4,815 | 4,434 | 2,711 | 3,757 | 2,603 | 2,413 |
| rRNAsa (16S, 5.8S, 23S) | 4 (1, 2, 1) | 3 (1, 1, 1) | 3 (1, 1, 1) | 4 (1, 2,1) | 3 (1, 1, 1) | 3 (1, 1, 1) | 6 (1, 3, 2) | 3 (1, 1, 1) | 4 (1, 2, 1) | 3 (1, 1, 1) | 3 (1, 1, 1) |
| tRNAs | 39 | 47 | 44 | 44 | 50 | 50 | 54 | 53 | 51 | 42 | 54 |
| Noncoding RNAs | 11 | 12 | 29 | 15 | 9 | 39 | 21 | 7 | 16 | 10 | 9 |
| CDS-coding genome (%) | 88.1 | 88.3 | 84.2 | 89.6 | 90.1 | 89.7 | 90 | 91.7 | 90.6 | 86.1 | 89.6 |
| Genome completeness (%)b | 99.3 | 99.1 | 99.1 | 98.8 | 99.5 | 99.3 | 99.8 | 98.5 | 99.8 | 97.1 | 98.6 |
| Closest cultivated organism (% identity)c | Dysgonomonas gadei (79.2) | Dysgonomonas capnocytophagoides (69) | Dysgonomonas gadei (81.8) | Dysgonomonas gadei (71.3) | Dysgonomonas macrotermitis (69.2) | Bacteroides fragilis (79) | Bacteroides nordii (79.4) | Bacteroides nordii (76.5) | Bacteroides fragilis (78.8) | Paludibacter propionicigenes (70) | Parabacteroides gordonii (78.7) |
Presence is scored if complete ribosomal gene is found in the corresponding genome contigs.
Completeness based on BUSCO analysis using the Bacteroidetes BUSCO database.
Based on average amino acid identity (AAI).
P. americana harbors several previously uncultivated Bacteroidetes isolates.
Phylogenetic reconstructions using 16S rRNA gene sequences derived from bacterial genomes isolated from P. americana indicated that five (PAD511, PAD520, PAD521, PAD25, and PAD216) clustered within the Dysgonomonas genus and four (PAB214, PAB224, PAB51, and PAB519) within the Bacteroides genus. Two remaining isolates, PAR221 and PAP52, grouped within the Paludibacter and Parabacteroides clades, respectively (Fig. 1a). Most isolates formed distinct, new clades that, in 16S rRNA gene-based phylogenies, were sister to uncultivated Bacteroidetes taxa detected in cockroaches (Shelfordella lateralis), termites (Coptotermes curvignathus), and beetles (Pachnoda ephippiata). Following complete genome sequencing and annotation of these genomes, concatenated protein phylogenetic analysis confirmed cladogenesis within the Bacteroidetes (Fig. 1b), with all the Bacteroides isolates forming a new monophyletic cluster within the tree. Meanwhile, Dysgonomonas, Parabacteroides, and Paludibacter isolates were distributed within their corresponding generic clades. In general, average amino acid identity (AAI) values for the isolate genomes were 65 to 80% relative to publicly available Bacteroidetes genomes (Table 1; see Data Set S1 in the supplemental material), which suggests that the following P. americana isolates represented new species among their respective genera: Bacteroides sp. PAB519, Bacteroides sp. PAB214, Bacteroides sp. PAB224, Bacteroides sp. PAB51, Dysgonomonas sp. PAD511, Dysgonomonas sp. PAD520, Dysgonomonas sp. PAD521, Dysgonomonas sp. PAD25, Dysgonomonas sp. PAD216, Parabacteroides sp. PAP52, and Paludibacter sp. PAR221 (hereafter all isolates will be referred to by their strain designation, e.g., PAR221 for Paludibacter sp. PAR221).
FIG 1.
Cladogenesis of P. americana Bacteroidetes species. Maximum-likelihood trees of 16S rRNA genes (a) and 14 concatenated single-copy gene markers (b) are shown. Barnesiella intestinihominis was used as an outgroup. Scale bars indicate 20% and 5% estimated sequence divergence, respectively. See Data Set S2 and Table 5 for the accession numbers of sequences used in these analyses.
P. americana Bacteroidetes isolates were detected by two cultivation-independent approaches in several P. americana individuals. Using isolate-specific 16S rRNA primers (Table 2), all of the P. americana Bacteroidetes, with the exception of PAD25 and PAR221, were detected in, on average, 85% of unpooled gut DNA extracts collected from ten adult P. americana individuals (see Table S1A in the supplemental material). Moreover, PAB519, PAB224, and PAP52 were detected in more than 98% of the individuals tested (see Table S1A in the supplemental material). In a separate experiment, nearly all P. americana Bacteroidetes isolates (i.e., 9 out of 11) were detected in over half of 16S rRNA gene community surveys of 121 P. americana feces samples collected weekly over 8 weeks from 21 P. americana individuals that were lab reared on three distinct diets (see Table S1B in the supplemental material). Consistent detection of these isolates across multiple individuals in both digestive tissues and frass, and over time, supports the hypothesis that they are endemic to P. americana.
TABLE 2.
P. americana Bacteroidetes species-specific 16S rRNA primer sequences
| Taxon | Isolate | Primer direction | Primer sequence (5′→3′) | Tm (°C) | Amplicon length (bp) |
|---|---|---|---|---|---|
| Bacteroides | PAB224 | Forward | TCAAGAAGCCGAGCCGTAAG | 60.1 | 206 |
| Reverse | GCTGAGCTAATCCCCCGATAAG | 60.4 | 206 | ||
| PAB51 | Forward | CACGTGTGGAGTTTTGTATGT | 56.8 | 203 | |
| Reverse | ACCTCCACTATACTCAAGACGCC | 61.5 | 203 | ||
| PAB214 | Forward | ACGTGTAGTGTTTTGCATGTACCG | 62.1 | 210 | |
| Reverse | TCCGCCTACCTCTACTGTACTCA | 61.7 | 210 | ||
| PAB519 | Forward | TGGGAATAAAGTGACGTACGTGT | 60 | 231 | |
| Reverse | CGAATTCCGCCAACCTTTACTTTAC | 60.9 | 231 | ||
| Dysgonomonas | PAD25 | Forward | CTCCGAATGGGTACAAGGGTCAT | 62 | 218 |
| Reverse | TCCATGCAGAACCACTCGACTAG | 62 | 218 | ||
| PAD521 | Forward | CATTACGTGTAGTGTATTGCATGTACTG | 60.7 | 194 | |
| Reverse | CTCAAGGCTACCAGTTTCAACGG | 61.7 | 194 | ||
| PAD511 | Forward | TACGTGTAGTATATTGCATGTACCATATG | 59.3 | 209 | |
| Reverse | CCGCCTACTTCATCTATACTCAAGAAAC | 61.4 | 209 | ||
| PAD216 | Forward | GTACTAGGGTAAAACAGGGGACGT | 61.6 | 203 | |
| Reverse | AACCCAGTTTCAACGGCAATTTTAAG | 61.4 | 203 | ||
| PAD520 | Forward | GTGCTAGGGTAAAACATATCACGAGTG | 61.8 | 209 | |
| Reverse | AAGTCTTCCAGTTTCAACGGCAA | 61.2 | 209 | ||
| Parabacteroides | PAP52 | Forward | CTTCTTTTATTGGGGAATAACGGCAG | 60.7 | 220 |
| Reverse | TCAAGACTAACAGTTTCAACGGCA | 60.9 | 220 | ||
| Paludibacter | PAR221 | Forward | TTGTATGTACTTTACGAATAAGCATCGG | 60.1 | 190 |
| Reverse | GCCTCTACTGCACTCAAGAACAC | 61.4 | 190 |
P. americana Bacteroidetes have diverse CAZyme and PUL repertoires.
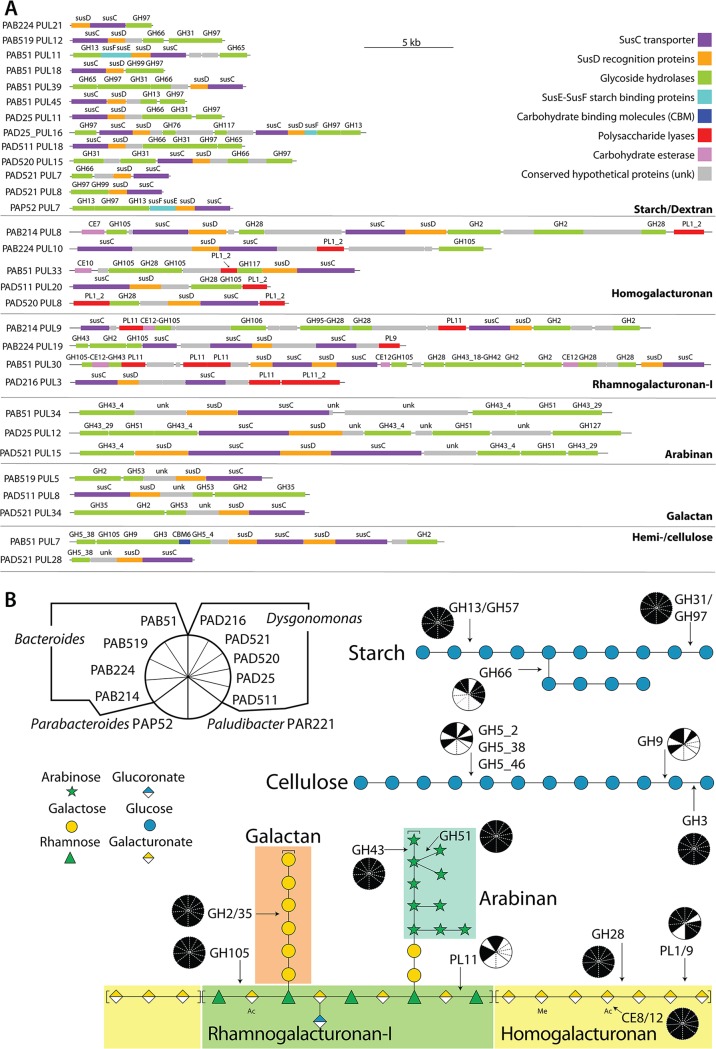
CAZy domain-encoding genes were most abundant in Bacteroides sp. isolate genomes, intermediate among the Dysgonomonas spp., and Paludibacter sp., and least abundant in the Parabacteroides sp. isolate genome (Table 3), with only a subset located in PUL gene clusters (Table 3; see Table S2 and Data Set S2 in the supplemental material). In comparison to previously annotated Bacteroides genomes, the PAB51 genome encoded ∼2× the PUL-associated CAZymes present in the closely related Bacteroides ihuae and Bacteroides graminisolvens genomes (Table 3; see Table S2 in the supplemental material). Additionally, PAB51 and PAB519 had the greatest number of complete PULs (67 and 42, respectively), while PAD216 and PAR221 maintained the fewest, with only three PULs in either genome (Table 3). A comparative analysis of all cultivated Bacteroidetes used in this study revealed isolate-specific PUL repertoires (see Fig. S2 in the supplemental material), and yet a single GH32-anchored PUL was common and highly syntenic across all P. americana Bacteroides isolates and other Bacteroides associated with either humans or insects (see Fig. S3 in the supplemental material). GH32 enzymes hydrolyze sucrose and are involved in the metabolism of fructan and levan polymers (37). The presence of GH32 in this syntenic PUL suggests a sucrose catabolic capability that is common to Bacteroides species. Across the examined isolates, starch, dextran, pectin, and cellulose were the predicted putative target substrates for some of the CAZy domain-encoding genes present in complete PULs (Fig. 2A).
TABLE 3.
Comparison of polysaccharide utilization loci in P. americana Bacteroidetes isolates and near-neighbor Bacteroidetes from other habitats
| Species (genome size, Mbp) | No. of: |
|||
|---|---|---|---|---|
| Polysaccharide utilization CAZy domains | PULs |
|||
| Total | Complete | Incomplete | ||
| Bacteroides graminisolvens | 190 | 31 | 26 | 5 |
| Bacteroides ihuae | 218 | 39 | 32 | 7 |
| Bacteroides sp. PAB51 (6.18) | 415 | 75 | 67 | 8 |
| Bacteroides sp. PAB224 (4.48) | 162 | 21 | 15 | 6 |
| Bacteroides sp. PAB519 (5.55) | 234 | 60 | 42 | 18 |
| Bacteroides sp. PAB214 (3.39) | 160 | 17 | 13 | 4 |
| Dysgonomonas capnocytophagoides | 211 | 37 | 26 | 11 |
| Dysgonomonas gadei | 282 | 58 | 44 | 14 |
| Dysgonomonas macrotermitis | 230 | 29 | 26 | 3 |
| Dysgonomonas sp. PAD511 (3.78) | 162 | 26 | 22 | 4 |
| Dysgonomonas sp. PAD520 (3.91) | 158 | 19 | 16 | 3 |
| Dysgonomonas sp. PAD521 (5.24) | 187 | 34 | 26 | 8 |
| Dysgonomonas sp. PAD25 (3.75) | 118 | 18 | 14 | 4 |
| Dysgonomonas sp. PAD216 (3.18) | 120 | 4 | 3 | 1 |
| Paludibacter propionicigenes | 145 | 17 | 11 | 6 |
| Paludibacter sp. PAR221 (3.11) | 66 | 8 | 3 | 5 |
| Parabacteroides gordonii | 244 | 88 | 62 | 26 |
| Parabacteroides sp. PAP52 (3.01) | 76 | 18 | 13 | 5 |
FIG 2.
P. americana Bacteroidetes isolates encode starch-, cellulose-, and pectin-targeting CAZymes. (A) CAZymes in predicted polysaccharide utilization loci (PUL) are grouped by predicted substrate targets, and labels indicate strain and PUL numbering based on PULpy pipeline annotations (see Data Sets S2 and S5 in the supplemental material for a complete list of all PULs annotated in each P. americana Bacteroidetes genome). (B) Substrate models for starch, cellulose, and pectin were constructed using the Symbol Nomenclature for Glycans system (https://www.ncbi.nlm.nih.gov/glycans/snfg.html), and pie charts indicate if the relevant CAZyme predicted to act on the indicated (arrow) linkage was (filled wedge) or was not (open wedge) annotated in the P. americana Bacteroidetes genomes.
Starch- and dextran-targeting PULs.
α-1,4 (amylose) and α-1,6 (amylopectin and dextran) glycosidic bonds that link α-d-glucoside units in starch and dextran are depolymerized and metabolized by bacterial α-glycosidases (38–40), and 13 PULs in the P. americana Bacteroidetes genomes included at least one GH13, GH66, or GH97 α-glycosidase gene (Fig. 2A and B). These PULs were distributed across seven P. americana Bacteroidetes genomes in several conformations, including a GH13 duplication in a single PUL (PAP52) or across two PULs (PAB51), GH97 in a single PUL (PAB51), and a GH66-GH97 pairing in a single PUL (PAB519, PAD25, PAD511, PAD520, and PAB51) or across two PULs (PAD521) (Fig. 2A). In B. thetaiotaomicron, GH66 has been described as an endo-dextranase that converts dextran into cycloisomaltooligosaccharides (41), and the presence of GH66-coding genes suggests that these P. americana Bacteroidetes can use dextran. In addition to GH13-GH97, the susEF genes, encoding the multidomain starch binding proteins SusEF, were associated with PULs in PAB51, PAP52, and PAD25 (Fig. 2A). In general, these α-glycosidic PULs in the P. americana isolates appear to be well conserved in Bacteroidetes, as they were highly syntenic with α-glycosidic PULs in B. thetaiotaomicron and other human-associated Bacteroidetes (B. xylanisolvens, B. cellulosilyticus, B. ihuae, and Dysgonomonas gadeii) (see Fig. S4A in the supplemental material).
HG- and RG-targeting PULs.
Pectic carbohydrate-targeting PULs were detected in P. americana Bacteroidetes isolates PAB214, PAB224, PAB51, PAB519, PAD216, PAD25, PAD511, PAD521, and PAD520 (Fig. 2A). Comparative genome analysis showed high homology among pectinolytic-protein-coding genes in these PULs and those in human (B. cellulosilyticus and D. capnocytophagoides)- and termite (B. reticulotermitis)-associated Bacteroidetes, and the PULs in PAD511 and PAD520 were syntenic with D. capnocytophagoides PUL24 and B. reticulotermitis PUL8 (see Fig. S4B in the supplemental material). In B. thetaiotaomicron, PULs predicted to target homogalacturonan (HG) encoded polysaccharide lyases PL1 and generally a copy of polygalacturonase GH28, as well as rhamnogalacturonyl hydrolase GH105 (8). Similar encoding genes and distribution were found in PULs of PAB214, PAB224, PAB51, PAD511, and PAD520 (Fig. 2A). This indicates that PULs with putative catalytic activity for HG hydrolysis are present in a subset of P. americana Bacteroidetes genomes. Additionally, genes encoding proteins that were annotated as carbohydrate esterase CE7 and CE10 family enzymes were detected in these PULs in the PAB214 and PAB51 genomes, respectively. PL9 and PL11 enzymes were predicted to contribute to rhamnogalacturonan I (RGI) catabolism (42), and PULs in PAB214, PAB51, PAD216, and PAB224 encoded these polysaccharide lyases (Fig. 2A and B). Additionally, RGI catabolic PULs of B. thetaiotaomicron exhibit β-galactosidase (GH2)-, rhamnogalacturonyl hydrolase (GH105)-, and rhamnosidase (GH106)-coding genes (8). Similar genes (GH2, GH105, and GH106) were detected in PUL9, -19, and -30 of PAB214, PAB224, and PAB51, respectively, suggesting their putative role in RGI catabolic activity. Arabinofuranosidase GH43 and GH51 genes have also been detected in pectinolytic PULs from B. thetaiotaomicron and B. xylanisolvens (8, 43), and similarly, genes coding for GH43 and GH51 have been detected in PAB51 PUL34, PAD25 PUL12, and PAD521 PUL15 (Fig. 2A). In particular, GH43 subfamily 4 (GH43_4), classified as α-1,5-arabinase in B. thetaiotaomicron (8), was present in all the putative arabinolytic PULs from P. americana Bacteroidetes (Fig. 2A). Together with arabinan, galactan is commonly found as decoration of rhamnogalacturonan I chains (44). Putative galactan-targeting PULs, exhibiting β-galactosidase GH2- and GH35-coding genes, are present in the PAB519, PAD511, and PAD521 genomes (Fig. 2A and B). Likewise, galactan PULs showing GH2- and GH35-encoding genes have been reported in B. thetaiotaomicron and B. ovatus (8). The above results support the idea of arabinolytic and galactanolytic PULs being present in P. americana Bacteroidetes.
Finally, single and multiple copies of genes encoding carbohydrate esterase CE12 enzymes were also included in the putative RGI hydrolytic PULs in PAB214 and PAB51 (Fig. 2A and B). CE12 has been described within PULs acting on citrus pectin in B. xylanisolvens (43). All of this evidence further supports the inference that these Bacteroides spp. can metabolize pectic carbohydrates.
Putative hemicellulose/cellulose catalytic PULs detected in PAB51 and PAD521 genomes.
Cellulolytic PULs that contain GH3, GH5, GH9, and GH94 CAZy-encoding genes have been reported within Bacteroidetes genomes present in rumen metagenomes (45, 46). Similarly, genes coding for GH3 and GH9 were found in the PUL7 from PAB51 (Fig. 2A). In addition, GH5 subfamily 38 (GH5_38) genes were annotated in PUL7 of PAB51 and PUL28 of PAD521 (Fig. 2A and B). Interestingly, GH5_38 has been classified as cellulase capable of hydrolyzing carboxymethyl cellulose (CMC) and Avicel (47), which suggests a putative cellulolytic activity of PAB51 and PAD521 PULs. However, in contrast to bacterial rumen cellulolytic PULs, genes coding for rhamnogalacturonyl hydrolase GH105, β-galactosidase GH2, and xyloglucanse GH5_4 were also found in PAB51 PUL7 (Fig. 2A). As GH2, GH105, and GH5_4 have been associated primarily with homogalacturonan, galactan, and xyloglucan hydrolysis (8, 47), PAB51 PUL 7 could only be classified as being putatively hemicellulolytic.
Non-PUL-associated CAZy genes.
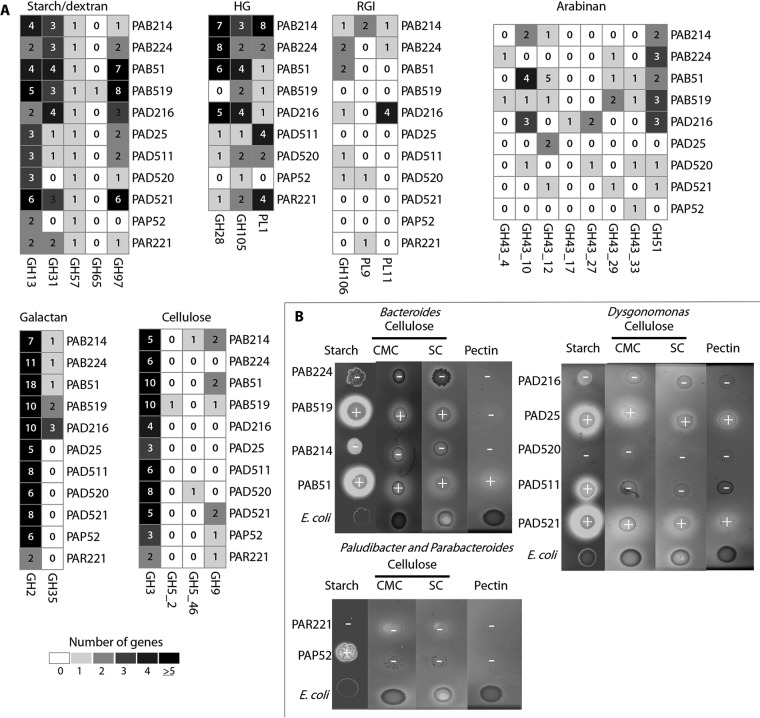
Several CAZy domain-encoding genes, ranging from 79 to 187 per genome, were annotated but not present within complete PULs (see Data Sets S2 and S3 in the supplemental material). Among these non-PUL-associated CAZy genes, amylolytic enzyme (GH13 and GH57)-coding genes were observed in all P. americana Bacteroidetes genomes (Fig. 2B and Fig. 3A). Furthermore, non-PUL α-glycosidase GH97 genes were also present in all except the PAP52 genome (Fig. 2B and 3A). Interestingly, non-PUL GH13 and GH97 in PAB214, PAB224, PAB519, PAB51, and PAD25 contained lipoprotein signal peptide sequences that indicated that they could be cleaved by type II signal peptidase SPII (see Data Set S3 in the supplemental material), suggesting an extracytoplasmic localization of these proteins. All non-PUL GH97-coding genes in the PAD216, PAP52, and PAR221 genomes lacked signal peptides and thus were classified as putative cytoplasmic proteins by both SignalP and LipoP analyses (see Data Set S3 in the supplemental material). Non-PUL dextranases (GH31) were also detected in nearly all of the P. americana Bacteroidetes genomes (Fig. 3A), and most were classified as putative extracytoplasmic SPII lipoproteins (see Data Set S3 in the supplemental material).
FIG 3.
Non-PUL CAZy genes and polysaccharide metabolism exhibited by Bacteroidetes from P. americana. (A) Heat maps showing the number of non-PUL CAZy genes. Numbers in each cell indicate the number of coding genes. (B) Different polysaccharide degradation activity by Bacteroidetes isolates. Isolates were arrayed and grown on solid MTY medium containing starch, CMC, Sigmacell (SC), or pectin. A clear halo after iodine addition surrounded the bacterial colony denoted polysaccharolytic activity. Hydrolysis was scored as positive (+) or negative (−) after RPA analysis. E. coli TOP10 was used as a negative polysaccharolytic bacteria; control.
Among pectinolytic non-PUL CAZy genes annotated in P. americana Bacteroidetes genomes, rhamnogalacturonyl hydrolase GH105 was present in all genomes; polygalacturonase GH28 and polysaccharide lyases PL1 were detected in nearly all of the genomes (Fig. 2B and 3A). SPII lipoprotein signal peptide sequences were associated with these non-PUL GH105, GH28, and PL1 genes in PAB214, PAB519, PAB51, PAD216, PAD511, PAD520, and PAR221, suggesting their extracytoplasmic localization (see Data Set S3 in the supplemental material). Additionally, non-PUL α-l-arabinofuranosidase GH51 was encoded in all Bacteroides genomes as well as in PAD216, PAD520, and PAD521. At least one subfamily of GH43 (GH43_4, _10, _12, _17, _27, _29, and _33) associated with arabinofuranose catabolism (48) was present in each genome (Fig. 3A). Most of the non-PUL GH51 and GH43 proteins, with the exception of a GH51 on PAD216, displayed either type I or type II signal peptidase cleavage motifs, suggesting an extracytoplasmic localization of these enzymes (see Data Set S3 in the supplemental material). Finally, non-PUL β-galactosidase GH2 was present in all P. americana Bacteroidetes isolate genomes. Likewise, GH35 was present in all Bacteroides spp. as well as in PAD216 (Fig. 2B and 3A) and harbored SPI or SPII signal peptides for extracytoplasmic export (see Data Set S3 in the supplemental material).
Among putative non-PUL cellulolytic CAZy genes, β-glucosidase GH3 was abundant in all P. americana Bacteroidetes genomes (Fig. 2B and 3A), with GH3 classified as a possible extracytoplasmic lipoprotein in PAB214, PAB224, PAB519, PAB51, PAD25, PAD511, and PAR221 (see Data Set S3 in the supplemental material). Furthermore, non-PUL GH5_2 and GH5_46, previously classified as cellulolytic GH5 enzymes (47), were present in PAB519, PAB214, and PAD520 (Fig. 2B and 3A), and they included internal SPI and SPII lipoprotein signal peptide sequences. Finally, genes encoding non-PUL-associated GH9, with SPI cleavage motifs, were also annotated in the PAB214, PAB51 PAB519, PAD521, PAP52, and PAR221 genomes (Fig. 3A; see Data Set S3 in the supplemental material).
T9SS.
Type IX secretion systems (T9SS) can contribute to CAZyme secretion in Bacteroidetes (46, 49), and genes encoding all necessary components of the canonical T9SS assembly (gldJ, gldK, gldL, gldM, gldN, sprA, sprE, sprT, porQ, and porV) were detected in the PAB214, PAD511, PAD521, PAD25, and PAR221 P. americana Bacteroidetes genomes (Table 4). Notably, PAB214 is unique among publicly available Bacteroides genomes in that it encodes all components of the canonical T9SS assembly (Table 4). CAZy and other proteins require a conserved carboxy-terminal domain (CTD) with two sequential motifs (PxGxYVV and KxxxK) to be secreted by T9SS (50), and genes encoding CAZy domains with CTDs were detected in nearly all the P. americana Bacteroidetes isolate genomes (see Data Set S4 in the supplemental material). Most of the CAZyme genes with conserved T9SS CTDs in PAB214, PAD216, PAD520, and PAR221 were annotated as polysaccharide lyases PL1, PL9, and PL11 (see Data Set S4 in the supplemental material), which suggests that these isolates potentially use the T9SS to assist with the metabolization of complex polysaccharides (i.e., pectin).
TABLE 4.
Comparison of type IX secretion system genes in P. americana Bacteroidetes isolates and near-neighbor Bacteroidetes from other habitats
| Species | No. of type IX secretion system domains |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| gldJ | gldK | gldL | gldM | gldN | sprA | sprE | sprT | porQ | porV | |
| Bacteroides graminisolvens | 0 | 0 | 0 | 1 | 0 | 0 | 7 | 3 | 0 | 0 |
| Bacteroides ihuae | 1 | 0 | 1 | 0 | 0 | 0 | 7 | 3 | 0 | 1 |
| Bacteroides sp. PAB51 | 4 | 3 | 0 | 1 | 0 | 1 | 4 | 1 | 0 | 0 |
| Bacteroides sp. PAB224 | 1 | 1 | 1 | 1 | 0 | 0 | 10 | 2 | 0 | 0 |
| Bacteroides sp. PAB519 | 4 | 4 | 2 | 1 | 0 | 2 | 9 | 2 | 1 | 1 |
| Bacteroides sp. PAB214 | 4 | 4 | 2 | 1 | 1 | 4 | 8 | 3 | 2 | 2 |
| Dysgonomonas capnocytophagoides | 1 | 1 | 1 | 1 | 1 | 2 | 5 | 9 | 2 | 2 |
| Dysgonomonas gadei | 2 | 2 | 4 | 1 | 2 | 2 | 7 | 8 | 2 | 2 |
| Dysgonomonas macrotermitis | 1 | 1 | 2 | 2 | 1 | 2 | 4 | 4 | 2 | 2 |
| Dysgonomonas sp. PAD511 | 2 | 2 | 1 | 2 | 2 | 3 | 5 | 3 | 1 | 2 |
| Dysgonomonas sp. PAD520 | 3 | 3 | 2 | 1 | 1 | 3 | 5 | 0 | 3 | 2 |
| Dysgonomonas sp. PAD521 | 2 | 2 | 2 | 2 | 2 | 3 | 9 | 5 | 1 | 1 |
| Dysgonomonas sp. PAD25 | 1 | 1 | 1 | 2 | 1 | 3 | 11 | 3 | 1 | 1 |
| Dysgonomonas sp. PAD216 | 2 | 2 | 2 | 4 | 2 | 3 | 8 | 0 | 2 | 2 |
| Paludibacter propionicigenes | 3 | 2 | 1 | 1 | 1 | 2 | 6 | 1 | 2 | 3 |
| Paludibacter sp. PAR221 | 5 | 5 | 3 | 2 | 1 | 2 | 7 | 3 | 2 | 3 |
| Parabacteroides gordonii | 9 | 9 | 3 | 1 | 1 | 2 | 13 | 5 | 3 | 3 |
| Parabacteroides sp. PAP52 | 2 | 2 | 1 | 1 | 1 | 3 | 5 | 0 | 2 | 2 |
In vivo pectin degradation.
PAB214, PAB224, PAP52, and PAR221 culture supernatants were capable of liberating significant amounts of reducing sugars equivalent to d-galacturonic acid following citrus peel pectin hydrolysis (analysis of variance [ANOVA] with Dunnett test, P < 0.05) (Fig. 4; see Table S3 in the supplemental material). Interestingly, isolates PAB214 and PAR221 harbored genes for complete T9SS assembly and pectinolytic-coding genes tagged with T9SS CTD signal (Table 4; see Data Set S4 in the supplemental material). Cultivation of these isolates in MTY broth plus pectin resulted in isolate growth and the release of 20 to 46 μg ml−1 of d-galacturonic acid equivalents into the medium (Fig. 4; see Table S4 in the supplemental material).
FIG 4.

Quantification of reducing sugars (d-galacturonic acid equivalents) in the supernatant medium produced after pectin hydrolysis by Bacteroidetes from P. americana. The red line shows the mean of d-galacturonic acid equivalents present in the supernatant of the noninoculated control medium. Values are shown as means ± standard errors of the means (SEM) from five independent experiments. Asterisks indicate significant differences in reducing sugars in the supernatants in comparison with the noninoculated control medium (**, P < 0.05; ***, P < 0.001).
Plate-based assays of genome-inferred polysaccharide hydrolytic activity.
Genomic predictions of starch, pectin, and cellulose substrate hydrolysis by the P. americana Bacteroidetes isolates were tested by in vivo growth assays on MTY solid medium amended with a single polysaccharide substrate. Hydrolysis of all substrates under anaerobic conditions by at least one isolate was observed (Fig. 3B). Overall, the isolates exhibited basal to substantial growth on solid MTY medium with glucose, starch, CMC, microcrystalline Sigmacell cellulose (Sigmacell), and/or pectin (Fig. 3B; see Fig. S5 in the supplemental material). Despite basal colony formation by all isolates, substantial polysaccharide hydrolysis was detected in only a few isolates, where visible zones of clearing on polysaccharide-containing plates stained with iodine were observed (Fig. 3B). Visual polysaccharolytic activity and relative polysaccharolytic activity (RPA) analysis showed that PAD521, PAD25, and PAB51 exhibited zones of clearing surrounding colonies on all polysaccharide substrates (Fig. 3B; see Table S4 in the supplemental material). PAB519 exhibited positive hydrolysis of starch, CMC, and Sigmacell but not pectin (Fig. 3B). PAD511 and PAP52 exhibited slight starch hydrolysis, but no visible halo was detected with the other polysaccharides tested (Fig. 3B). PAB224, PAB214, PAD216, PAD520, and PAR221 did not exhibit halo production on any polysaccharide tested. Finally, no significant halo was observed when glucose was used as a carbon source (see Fig. S5 in the supplemental material).
Cellulose-based liquid medium growth assays.
Dysgonomonas and Bacteroides isolates that exhibited zones of clearing on solid MTY medium containing both Sigmacell and CMC (PAD521, PAD25, PAB51, and PAB519) reached the stationary phase in liquid MTY supplemented with CMC (Fig. 5A and B). In particular, PAD521 showed a higher cell density and a higher growth rate (by around 0.2 unit of optical density at 600 nm [OD600] h−1) when CMC was present than in control non-CMC medium (Tables S5 and S6 in the supplemental material). PAB51 and PAD25 exhibited a slight but significant improvement in growth rate and total growth when CMC was present in comparison with those in non-CMC control medium (Fig. 5A and B; see Tables S5 and S6 in the supplemental material). Although PAB519 showed higher cell density and growth rate when CMC was added to the medium (Fig. 5B), there was no significant difference between CMC and no-substrate MTY controls (see Tables S5 and S6 in the supplemental material), suggesting that this isolate did not rely upon CMC for growth. Lastly, in vitro cellulolytic activity was quantified using cell-free lysates prepared from PAD521, PAD25, PAB519, and PAB51, and they exhibited significantly greater (P < 0.05) CMC degradation and release of d-glucose equivalents than Escherichia coli (Fig. 5C). Although PAD521 and PAD25 showed significantly higher CMC hydrolysis activity than PAB519 and PAB51, no significant intraspecies differences were observed (Fig. 5C).
FIG 5.

Polysaccharide growth and in vitro degradation assays. (A and B) Growth curves of Dysgonomonas spp. (A) and Bacteroides spp. (B) isolated from P. americana in MTY medium with CMC. MTY medium without CMC was used as a noncellulose substrate medium control. Values are shown as means ± SEM from three independent experiments. (C) DNS test showing the hydrolysis of CMC to d-glucose equivalents (reducing sugars) by cleared bacterial lysates of P. americana Bacteroidetes isolates. E. coli TOP10 was used as a noncellulolytic negative control. Values are shown as means ± SEM from three independent experiments. All values were normalized using boiled bacterial lysate per each treatment. Asterisks indicate significant differences in comparison with E. coli (P < 0.05).
DISCUSSION
P. americana harbors new cultivable Bacteroidetes species.
P. americana harbors a species-rich gut bacterial community that includes many previously detected (30, 31) but uncultivated taxa. Among the many isolates obtained by anaerobic cultivation approaches in this work, eleven were assigned to the Bacteroidetes, and phylogenetic/phylogenomic analyses and other comparative methods suggested that all represented new species. Cladogenesis was frequently observed for these Bacteroides, Dysgonomonas, Paludibacter, and Parabacteroides spp., with nearest neighbors being uncultivated taxa detected in the digestive tracts of other cockroach species (Fig. 1a and b), which suggests that the P. americana Bacteroidetes isolates maintain persistent associations with cockroaches and were not recently taken up from the environment or diet. Maintenance of specific bacterial species in the digestive tract is common in other insects, such as flies (51), mosquitoes (52), and beetles (53). Consistent detection of all Bacteroidetes isolates in the digestive tracts and frass of several P. americana individuals (see Table S1A and B in the supplemental material) further supports the hypothesis that these bacteria are endemic to the digestive tracts of this host species and are likely intergenerationally transmitted via coprophagy (25).
Polysaccharide catabolism by P. americana Bacteroidetes.
Little is known about the role and influence of Bacteroidetes in polysaccharide metabolism in Blattarian insects. Metagenomic loci assigned to Bacteroidetes residing within the green banana cockroach (Panchlora sp.) were annotated as encoding CAZymes that putatively target cellulose, hemicellulose and pectin (23), but they were not functionally characterized. Annotations of the P. americana Bacteroidetes isolate genomes predicted several genes encoding CAZy domain-containing proteins with some integrated into PUL like operons (Fig. 2A). These annotations accurately predicted the growth of several isolates on starch, cellulose and pectin (Fig. 3B). While previously identified Bacteroidetes PULs were predicted to target polysaccharides like starch and pectin (2, 8, 54–56), recent metagenomic analyses of mammalian rumen microbiota have yielded a Bacteroidales-linked PUL that putatively targets cellulose (45, 57). Unique PULs putatively targeting at least one of these polysaccharides were detected in P. americana Bacteroidetes isolate genomes (Fig. 2A).
Starch, pectin, and cellulose metabolism.
Starch degradation and catabolism in the human-associated Bacteroides thetaiotaomicron is mediated by the starch utilization system (Sus) PUL, which encode a transcriptional regulator (SusR), outer membrane binding lipoproteins (SusDEF), a TonB-dependent transporter (SusC), and the GH13-GH97 α-glycoside hydrolases (56). PAB51 PUL11, PAP52 PUL7, and PAD25 PUL16 (Fig. 2A) were among the B. thetaiotaomicron starch-like PULs detected in P. americana Bacteroidetes isolate genomes, highlighting a starch metabolic system spanning Bacteroides inhabiting distinct hosts.
Although endogenous amylases and cellulases in P. americana have been reported (11, 26, 28, 58), plant fibers are also largely comprised of hemicellulose and pectin (59), and no polygalacturonases, polysaccharide lyases, or rhamnogalacturonan hydrolases have been described within this insect. However, PULs with genes encoding polygalacturonases (GH28), pectin acetylesterases (CE12), and other pectinases (i.e., polysaccharide lyases and rhamnogalacturonyl hydrolases [GH105]) were found in most of the P. americana Bacteroides isolate genomes (Fig. 2A and B and 3A). Given that P. americana lacks endogenous pectinases, pectinolytic gut symbionts may be relied upon to supply the enzymes necessary for deconstructing this abundant dietary component, as observed in honey bees (Apis mellifera) and scale insects (Dactylopius coccus) (60, 61). In B. thetaiotaomicron, dietary pectic glycans are degraded by PULs catabolizing galactan, arabinan, homogalacturonan (HG), and rhamnogalacturonan I (RGI) (8). Similarly, P. americana Bacteroides PAB214, PAB224, and PAB51 genomes carry PUL products putatively active on homogalacturonan and rhamnogalacturonan I, and the latter isolate also harbors a putative arabinan-targeting PUL (Fig. 2A). Although orthologous galactan PULs were not detected in strains PAB51, PAB214, and PAB224, multiple non-PUL-associated β-galactosidases (GH2 to GH35) that may target this substrate were observed (Fig. 3A; see Data Set S3 in the supplemental material) and thus may be metabolized by these strains. Among these strains, only PAB51 grew on and degraded pectin on solid medium (Fig. 3B), while PAB214 and PAB224 exhibited little to no growth or substrate degradation on these plates (Fig. 3B), which suggests that although pectinolytic PULs were predicted in these strains, the pectin solid medium was not suitable for growing these strains.
Rumen metagenomes have revealed putative Bacteroidetes PULs for cellulose metabolism (45, 62, 63) in which endoglucanases GH5 and GH9 yield oligomers (cellobiose) from cellulose matrix which are then phosphorylated into assimilable glucose 1-phosphate by GH94 (45, 63). Similarly, P. americana Bacteroidetes PAB51 PUL7 and PAD521 PUL28 encode GH5 subfamily 38, which has been characterized as a cellulase in the Bacteroidetes species Prevotella ruminicola showing catalytic activity on CMC and Avicel (47). Comparative analysis using all Bacteroidetes genomes highlighted the uniqueness of the PAB51 PUL7 and PAD521 PUL28 structure in that no other cultivated P. americana Bacteroidetes genomes harbored similar PUL architecture (see Fig. S2 in the supplemental material). Although the PAB51 PUL7 gene cluster encodes canonical cellulolytic protein-coding genes (GH3, GH5_38, and GH9), rhamnogalacturonanolytic GH105-, galactolytic GH2-, and xylanolytic GH5_4 (Fig. 2A)-encoding genes are also present. The presence of these “noncellulolytic” genes suggests that this PUL may not exclusively target cellulose substrates but also degrades hemicellulose. Nonetheless, cloning and functional characterization of these genes is required to confirm the catalytic activity of PAB51 PUL7.
Even though Bacteroides cellulosilyticus was reported to be capable of cellulose hydrolysis in vitro (64), subsequent growth and kinetic studies showed that B. cellulosilyticus was unable to grow using cellulose as a sole carbon source (65). Likewise, Dysgonomonas termitidis has been reported to exhibit weak carboxymethyl cellulose hydrolytic activity (66); however, the analyses described in this report uncovered no complete cellulolytic PUL in the D. termitidis genome. PAB51 and PAD521 grew on solid medium containing either carboxymethyl cellulose or microcrystalline Sigmacell cellulose and appeared to hydrolyze the substrates in the medium through an apparent extracellular mechanism (Fig. 3B). Furthermore, growth kinetic assays revealed that carboxymethyl cellulose in the liquid medium significantly improved growth of PAB51 and PAD521 isolates (Fig. 5A and B). Taken together, these results suggest that a subset of P. americana Bacteroidetes isolates could use both cellulose species, perhaps due to the endo- and exocellulolytic GHs encoded in PULs (Fig. 2A) or to non-PUL-associated hydrolases (Fig. 3A).
P. americana Bacteroidetes deploy “selfish” and “public goods” polysaccharolytic strategies.
Bacterial polysaccharide degradative strategies can theoretically impact individual fitness, community dynamics, and nutrients available for host uptake. The PUL mechanism allows for a “selfish” strategy of carbohydrate utilization where internalization of polysaccharides prior to hydrolysis ensures maximum access to released sugars and avoids loss to the host and gut microbes. Alternatively, extracellular polysaccharide hydrolysis may reflect a “public goods” strategy, where hydrolyzed products are accessible to the host and other microbial community members (2, 10, 19). Most of the non-PUL-associated CAZymes detected in P. americana Bacteroidetes genomes were classified as lipoprotein extracytoplasmic enzymes (see Data Set S3 in the supplemental material). This suggests that CAZymes in P. americana Bacteroidetes can be located in the outer bacterial membrane or be exported outside the periplasmic space by secretion systems (46, 50) and therefore hydrolyze different substrates in the exterior milieu. The presence of zones of clearance around some P. americana Bacteroidetes isolates (i.e., PAB519, PAB51, PAD25, and PAD521) growing on solid medium plates containing different carbon substrates provides evidence of extracellular polysaccharide hydrolysis (Fig. 3B). Particularly, these isolates have non-PUL extracytoplasmic CAZy genes for starch (GH13), pectin (PL1, GH28, and GH105), and cellulose (GH5_2, GH5_46, and GH9) hydrolysis (Fig. 3A).
In other Bacteroidetes (i.e., Cytophaga hutchinsonii, Flavobacterium johnsoniae, and “Candidatus Paraporphyromonas polyenzymogenes”), it has been shown that non-PUL mechanisms are employed for polysaccharide metabolism where glycoside hydrolases are secreted into the medium by the type IX secretion system (T9SS) for breaking down polysaccharide chains extracellularly (46, 49, 67, 68). Genes for the complete assembly of T9SS were annotated in the PAB214, PAD511, PAD521, PAD25, and PAR221 genomes (Table 4). In addition to a functional T9SS, a CTD tag in the proteins is required for being recognized and secreted by this system (50). Correspondingly, the PAB214 and PAR221 genomes encode multiple non-PUL polysaccharide lyases (PL1, PL9, and PL11), rhamnogalacturonyl hydrolases (GH105), and β-galactosidases (GH2), associated with pectin metabolism in other Bacteroidetes (8), that have integrated CTD tags (see Data Set S4 in the supplemental material). Furthermore, PAB214 and PAR221 were capable of releasing significant amounts of d-galacturonic acid equivalents and of growth in MTY broth with pectin (Fig. 4). In B. thetaiotaomicron, PL1, PL11, and GH105 are responsible for liberating d-galacturonic acid units from homogalacturonan and rhamnogalacturonan I in the periplasmic space (8), and coding sequences for these CAZymes in the PAB214 and PAR221 genomes possess signal peptides for extracytoplasmic export (see Data Set S3 in the supplemental material). Additionally, pectinases in these isolates also encode T9SS CTD recognition domains (see Data Set S4 in the supplemental material). This, along with the d-galacturonic acid release evidence, supports the hypothesis that pectinases in PAB214 and PAR221 are released into the medium, possibly by T9SS, and hydrolyze pectin components extracellularly. T9SS has been implicated in secretion of hydrolytic enzymes for chitin, cellulose, and hemicellulose metabolism (46, 49, 67). T9SS-enabled secretion of pectinolytic enzymes has not been reported for Bacteroidetes, including B. thetaiotaomicron, and thus further experimental work is needed to test this hypothesis and possibly a new role of T9SS. It is worth noting that although pectin hydrolysis in liquid medium was observed for PAB214 or PAR221, both exhibited poor growth and no halo on solid MTY medium with pectin (Fig. 3B). Further characterization of these strains is necessary to determine if the assay conditions were not favorable for pectin-stimulated growth or eliciting pectinase activity.
Strictly PUL-based polysaccharide degradation may limit direct contributions of monosaccharides to community-available resources and host metabolism but may increase contributions of important products like vitamins and short-chain fatty acids produced by these bacteria. In contrast, extracellular degradation of polysaccharides may exact a slight metabolic cost for the secretion of degradative enzymes and loss of exclusive access to liberated nutrients, yet this “noble” act may facilitate cross-feeding of degradative products, increasing the overall availability of community- or host-assimilable carbohydrates, and provide a basis for synergistic interactions within the bacterial community. Further detailing of the polysaccharolytic abilities of these isolates will help to uncover the many ways in that members of the Bacteroidetes participate in and impact the microbial communities of invertebrates and vertebrates.
MATERIALS AND METHODS
Isolate cultivation.
Bacteria were isolated from adult Periplaneta americana insects reared on dog food and maintained in the insectary at The Ohio State University. Following euthanization, insect bodies were surface sterilized with a 70% ethanol–10% bleach solution for 30 s and aseptically dissected under a sterile atmosphere. Gut tissues were suspended in 1 ml modified TY plus glucose (MTYG) medium (MgSO4·7H2O, 0.25 g liter−1; CaCl2·H2O, 15 mg liter−1; tryptone, 10 g liter−1; yeast extract, 5 g liter−1; NaCl, 2 g liter−1; l-cysteine, 0.25 g liter−1; trace element solution [69], 1 ml liter−1, 3.6 g liter−1 d-glucose; 1 M potassium phosphate [pH 6.7], 10 ml; Pfennig vitamin mixture [70], 1 ml; 1 M NaHCO3, 10 ml; 0.25% hemin in 1 N NaOH, 0.4 ml; 1% vitamin K1 in absolute ethanol, 0.4 ml) and homogenized with pestles. Suspensions were serially diluted to 10−9 in MTYG liquid medium, and 0.1 ml of each dilution was spread on solid MTYG plates (18 g liter−1 agar was added) and incubated under anaerobic conditions (90% N2, 5% CO2, and 5% H2) at 30°C in the dark for up to 24 days. Colonies representing distinct morphologies were picked, restreaked on solid MTYG medium, and incubated anaerobically (30°C) for 3 to 14 days; this was repeated at least two times to obtain pure isolates. Liquid cultures were prepared by inoculating a single colony into MTYG medium (5 ml) and incubating at 30°C under anaerobic conditions until cultures became turbid. Cultures were examined for purity under a photomicroscope, and those that contained cells with two or more distinct morphologies were restreaked for isolation on MTYG plates. Cultures containing cells with a uniform morphology were considered pure and were harvested in 10% dimethyl sulfoxide for storage at –80°C. Genomic DNA was extracted from isolate cultures using the DNeasy blood and tissue kit (Qiagen) following the manufacturer’s instructions for Gram-negative bacteria. Bacterial DNA was used as a template to amplify near-full-length 16S rRNA genes with 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′) primers. Amplicons were produced using the following thermocycler parameters: 1 cycle of 97°C for 5 min; 40 cycles of 97°C for 30 s, 40 to 55°C for 30 s, and 68°C for 2 min; and 1 cycle of 68°C for 15 min. Amplification of ∼1.5-kbp products was confirmed by gel electrophoresis, and the products were purified using the DNA Clean & Concentrator kit according to manufacturer’s instructions (Zymo Research, Irvine, CA). 16S rRNA gene amplicons were sequenced by Sanger capillary chemistry and manually checked to ensure a single read per base position in >90% of the sequence; sequences meeting this criterion were deemed pure. These sequences were matched to their closest bacterial relative by BLASTN (71) searches, and the results of the analysis of isolates classified within Bacteroides, Dysgonomonas, Parabacteroides, and Paludibacter genera were described.
Genome sequencing, assembly, and annotation.
All bacterial genomes were sequenced on an Illumina HiSeq platform using the Illumina Nextera XT Library kit. Read quality trimming and adaptor removal were performed (TrimGalore 0.4.1 parameters: -q 30 –phred 33 –illumina –paired; https://github.com/FelixKrueger/TrimGalore) prior to assembly of high-quality reads in SPAdes 3.7 with default parameters (72) or IDBA-UD 1.1 assemblers (parameters: –pre_correction –step 15) (73). Scaffolds were retrieved by SSPACE 1.3 (74) with BWA (75) as an aligner (parameters: -x 0 -z 0 -k 0 -g 0 -a 0.70 -n 15 -T 30 -p 0). deBruijn graphs were visualized with Bandage 0.8.0 (76) to identify possible plasmids as circular contigs. For annotation, open reading frames (ORFs) and protein-coding sequences (CDS) from genomes were predicted using Prodigal 2.6 with default parameters (77). Completeness of genome sampling as represented by the scaffolds was estimated by the BUSCO 3.2.0 pipeline (78) using the “Bacteroidetes/Chlorobi group” orthologue database, with translations of predicted ORFs. PROKKA version 1.12 (79) was used for gene annotation with SignalP (44) for Gram-negative signal peptides and Infernal (80) for noncoding RNA annotation (–gram -/neg –rfam parameters, respectively). Manual curation of hypothetical genes was performed using the NCBI “nr” database (downloaded on September 2017) via BLASTP (71). Metabolic pathways were predicted using the GhostKoala tool from KEGG (81) and manually curated using BioCyc (82). Additionally, all genomes were annotated using the NCBI Prokaryotic Genome Annotation Pipeline PGAP-4.6 (83). Correlations between PROKKA locus identifiers and PGAP locus identifiers of each genome are shown in Data Set S2 in the supplemental material.
Phylogenetic and comparative genomic analyses.
Near-full-length (>1,400-nucleotide [nt]) 16S rRNA gene sequences were retrieved from isolate genome annotations and compared to the NCBI “nt” (downloaded September 2018) and Arb-SILVA (accessed in January 2019) databases using BLASTN to identify the nearest neighbors (see Data Set S2 in the supplemental material) for phylogenetic analyses. MAFFT 7.3.10 was used to align all sequences (84), and a maximum-likelihood-based (ML) phylogenetic tree based on the general time-reversible (GTR) model, as recommended by JModelTest 2.1.10 (85), was generated using PhyML 3.1 (86) with the Shimodaira-Hasegawa-like (SH) procedure for internal branch support (87).
A total of 148 Bacteroidetes genomes (Table 5) that were complete or nearly complete (based on the presence of >90% of 224 “Bacteroidetes/Chlorobi group” single-copy orthologues following a BUSCO analysis; see Data Set S2 in the supplemental material) were downloaded from the NCBI “assembly” database (www.ncbi.nlm.nih.gov/assembly/) and used for phylogenomic and comparative analyses. A concatenated protein-based phylogeny was constructed using 14 orthologous genes (rplA, rplC, rplD, rplE, rplF, rplL, rplP, rplS, rpmA, rpoB, rpsC, rpsE, pgk, and infC). These were identified by the “hmmsarch” tool (parameters: hmmsearch -Z 5000 -E 0.005 –domE 0.005 –domtblout .hmmsearch -o/dev/null $ref_dir”.”markers.hmm $input_seq) from HMMER 3.1b2 (88) using hidden Markov model (HMM) profiles of these markers from the AMPHORA2 pipeline database (89). These markers were single-copy unique and conserved across all the Bacteroidetes genomes used in this analysis. Protein sequences were concatenated and aligned using the multigenome2blocks pipeline developed for this work (https://github.com/avera1988/multigenome2blocks). Prottest3 3.4.2 (90) was used to select the Le-Gascuel (LG + I + G) substitution model. ML phylogeny was constructed using PhyML 3.1 (86) with the SH procedure for internal branch support (87).
TABLE 5.
Bacteroidetes genomes used for comparative genomics and phylogenomics
| GenBank assembly accession no. | Organism |
|---|---|
| GCF_000374585.1 | Bacteroides barnesiae |
| GCF_000273725.1 | Bacteroides caccae |
| GCF_001688725.2 | Bacteroides caecimuris |
| GCF_002221665.1 | Bacteroides caecimuris |
| GCF_001318345.1 | Bacteroides cellulosilyticus |
| GCF_000195615.1 | Bacteroides clarus |
| GCF_000154845.1 | Bacteroides coprocola |
| GCF_000157915.1 | Bacteroides coprophilus |
| GCF_000212915.1 | Bacteroides coprosuis |
| GCF_001640865.1 | Bacteroides dorei |
| GCF_000155815.1 | Bacteroides eggerthii |
| GCF_000226135.1 | Bacteroides faecis |
| GCF_000156195.1 | Bacteroides finegoldii |
| GCF_000195635.1 | Bacteroides fluxus |
| GCF_000009925.1 | Bacteroides fragilis |
| GCF_000210835.1 | Bacteroides fragilis |
| GCF_000374365.1 | Bacteroides gallinarum |
| GCF_000428125.1 | Bacteroides graminisolvens |
| GCF_000186225.1 | Bacteroides helcogenes |
| GCF_900104585.1 | Bacteroides ihuae |
| GCF_001578635.1 | Bacteroides intestinalis |
| GCF_900128455.1 | Bacteroides mediterraneensis |
| GCF_000499785.1 | Bacteroides neonati |
| GCF_000273175.1 | Bacteroides nordii |
| GCF_000315485.1 | Bacteroides oleiciplenus |
| GCF_001314995.1 | Bacteroides ovatus |
| GCF_000187895.1 | Bacteroides plebeius |
| GCF_000375405.1 | Bacteroides propionicifaciens |
| GCF_000190575.1 | Bacteroides salanitronis |
| GCF_000381365.1 | Bacteroides salyersiae |
| GCF_000403195.1 | Bacteroides sartorii |
| GCF_000218365.1 | Bacteroides sp. |
| GCF_000159875.2 | Bacteroides sp. |
| GCF_000526555.1 | Bacteroides sp. |
| GCF_000162155.1 | Bacteroides sp. |
| GCF_000162175.1 | Bacteroides sp. |
| GCF_000157055.1 | Bacteroides sp. |
| GCF_001185845.1 | Bacteroides sp. |
| GCF_000163655.1 | Bacteroides sp. |
| GCF_000162195.1 | Bacteroides sp. |
| GCF_000185585.1 | Bacteroides sp. |
| GCF_000158515.2 | Bacteroides sp. |
| GCF_000157075.2 | Bacteroides sp. |
| GCF_002159755.1 | Bacteroides sp. |
| GCF_002161765.1 | Bacteroides sp. |
| GCF_002161565.1 | Bacteroides sp. |
| GCF_000157095.2 | Bacteroides sp. |
| GCF_000162215.1 | Bacteroides sp. |
| GCF_000163675.1 | Bacteroides sp. |
| GCF_000702285.1 | Bacteroides sp. |
| GCF_000702225.1 | Bacteroides sp. |
| GCF_001811695.1 | Bacteroides sp. |
| GCF_001815255.1 | Bacteroides sp. |
| GCF_000382465.1 | Bacteroides sp. |
| GCF_900155865.1 | Bacteroides sp. |
| GCF_900108345.1 | Bacteroides ndongoniae |
| GCF_000785025.1 | Bacteroides sp. |
| GCF_000154525.1 | Bacteroides stercoris |
| GCF_000011065.1 | Bacteroides thetaiotaomicron |
| GCF_000513195.1 | Bacteroides timonensis |
| GCF_000154205.1 | Bacteroides uniformis |
| GCF_000012825.1 | Bacteroides vulgatus |
| GCF_002161115.1 | Bacteroides xylanisolvens |
| GCA_001899125.1 | Dysgonomonas sp. |
| GCA_002409595.1 | Dysgonomonas sp. |
| GCA_002482385.1 | Dysgonomonas sp. |
| GCA_002482615.1 | Dysgonomonas sp. |
| GCA_002483985.1 | Dysgonomonas sp. |
| GCF_000213555.1 | Dysgonomonas gadei |
| GCF_000213575.1 | Dysgonomonas mossii |
| GCF_000296465.1 | Barnesiella intestinihominis |
| GCF_000376405.1 | Dysgonomonas mossii |
| GCF_000426485.1 | Dysgonomonas capnocytophagoides |
| GCF_000473955.1 | Coprobacter fastidiosus |
| GCF_000803105.1 | Coprobacter secundus |
| GCF_001047035.1 | Dysgonomonas macrotermitis |
| GCF_001261715.1 | Dysgonomonas sp. |
| GCF_001261735.1 | Dysgonomonas sp. |
| GCF_001487125.1 | Sanguibacteroides massiliensis |
| GCF_900128985.1 | Dysgonomonas macrotermitis |
| GCA_000699785.1 | Parabacteroides distasonis |
| GCA_001405935.1 | Parabacteroides distasonis |
| GCA_002257605.1 | Parabacteroides sp. |
| GCA_000307455.1 | Parabacteroides distasonis |
| GCA_000699805.1 | Parabacteroides distasonis |
| GCA_002206325.2 | Parabacteroides distasonis |
| GCA_002272965.1 | Parabacteroides sp. |
| GCA_001406015.1 | Parabacteroides distasonis |
| GCA_000307375.1 | Parabacteroides johnsonii |
| GCA_002160095.1 | Parabacteroides distasonis |
| GCA_000012845.1 | Parabacteroides distasonis |
| GCA_000157035.2 | Parabacteroides sp. |
| GCA_000162275.1 | Parabacteroides sp. |
| GCA_000307435.1 | Parabacteroides distasonis |
| GCA_000307475.1 | Parabacteroides sp. |
| GCA_000307495.1 | Parabacteroides merdae |
| GCA_000364265.1 | Parabacteroides sp. |
| GCA_000403825.2 | Parabacteroides goldsteinii |
| GCA_900128505.1 | Parabacteroides timonensis |
| GCA_000699745.1 | Parabacteroides distasonis |
| GCA_000699765.1 | Parabacteroides distasonis |
| GCA_001404395.1 | Parabacteroides distasonis |
| GCA_001405775.1 | Parabacteroides distasonis |
| GCA_000969835.1 | Parabacteroides goldsteinii |
| GCA_000162535.1 | Parabacteroides sp. |
| GCA_000307395.1 | Parabacteroides goldsteinii |
| GCA_000428565.1 | Parabacteroides gordonii |
| GCA_000969825.1 | Parabacteroides gordonii |
| GCA_000969845.1 | Parabacteroides sp. |
| GCA_900186615.1 | Parabacteroides bouchesdurhonensis |
| GCA_002159645.1 | Parabacteroides sp. |
| GCA_002161725.1 | Parabacteroides johnsonii |
| GCA_900108035.1 | Parabacteroides chinchillae |
| GCA_900292045.1 | Parabacteroides pacaensis |
| GCA_001404575.1 | Parabacteroides merdae |
| GCA_000307345.1 | Parabacteroides merdae |
| GCA_000154105.1 | Parabacteroides merdae |
| GCA_001078555.1 | Parabacteroides sp. |
| GCA_900168155.1 | Parabacteroides chartae |
| GCA_900155425.1 | Parabacteroides massiliensis |
| GCA_900162725.1 | Parabacteroides sp. |
| GCA_001039445.1 | Parabacteroides goldsteinii |
| GCA_900232875.1 | Parabacteroides provencensis |
| GCA_000436315.1 | Parabacteroides sp. |
| GCA_000699905.1 | Parabacteroides distasonis |
| GCA_001915675.1 | Parabacteroides sp. |
| GCA_000156495.1 | Parabacteroides johnsonii |
| GCA_002383435.1 | Paludibacter sp. |
| GCA_002427615.1 | Paludibacter sp. |
| GCA_001618385.1 | Paludibacter jiangxiensis |
| GCA_000183135.1 | Paludibacter propionicigenes |
Pan and core genome analyses were conducted using GETHOMOLOGUES 2.0 (91), and OrthoMCL (92) was used for orthologue clustering (parameters: -A -c -t 0 -M -n 35). Core genes corresponding to each genome were parsed from the GETHOMOLOGUES pangenome matrix results using custom Bash and Perl scripts (https://github.com/avera1988/Comparative_genomics), and they were used to calculate the average amino acid identity (AAI) between Dysgonomonas, Bacteroides, Parabacteroides, and Paludibacter genomes using the AAI calculator from the “enveomics” collection tools (93). AAI distance matrices were calculated and visualized by custom Perl and R scripts (deposited in https://github.com/avera1988/Comparative_genomics).
Finally, to compare functional profiles between P. americana and public Bacteroidetes genomes, all clusters of orthologous groups (COG) of proteins from each genome (Table 5) were annotated using the cdd2cog pipeline (https://github.com/aleimba/bac-genomics-scripts). Comparisons between proportions of COG genes from P. americana and non-P. americana Bacteroidetes were performed using custom R scripts (https://github.com/avera1988/Comparative_genomics). Two-tailed ANOVA and Tukey honestly significant difference (HSD) post hoc tests were used to evaluate significant differences in the proportion of genes among COG profiles between P. americana and non-P. americana Bacteroidetes genomes.
Cultivation-independent detection of isolates in wild P. americana cockroaches.
Bacteroidetes isolates were detected in P. americana digestive tracts by strain-specific PCR. Briefly, adult Periplaneta americana insects were obtained from the insectary at The Ohio State University and were habituated to a diet of gamma-irradiated rodent food for 1 week. Ten cockroaches were dissected to remove the full-length gastrointestinal tract, and DNA was extracted from these tissues with the Qiagen DNeasy Blood and Tissue DNA extraction kit. Diagnostic PCR was conducted on each extract using isolate-specific primers (Table 2) and the following thermocycler conditions: 3 min at 95°C, followed by 35 cycles of 1 min at 95°C, 15 s at 58°C, and 1 min at 68°C, followed by 3 min of extension at 68°C. Isolate-specific primers were searched against the Arb-SILVA 16S rRNA gene database and against an in-house 16S rRNA gene database of cockroach gut isolates to ensure specificity. Isolate-specific primers were further tested against closely related species within an in-house isolate collection to ensure amplification specificity. Three independent PCRs were conducted with each gut extract and isolate-specific primer combination, and positive amplification in at least two of three replicates was scored as presence of the isolate in an individual cockroach. Additionally, the presence of P. americana Bacteroidetes isolates was surveyed in feces of other P. americana individuals by 16S rRNA gene amplicon tag community profiling. A complete description of the methodology for the 16S rRNA gene amplicon tag survey is in the supplemental material.
PUL and CAZyme annotation and screening.
Polysaccharide utilization loci (PULs) were identified in the PROKKA general feature files (*.gff) and protein prediction fasta (*.faa) files for the assembled isolate and the publicly available genomes using the PULpy pipeline with the dbCAN database version 7 (downloaded January 2019) for carbohydrate-active enzyme (CAZyme) annotations (94, 95). PULs were annotated as “complete” if they contained a SusC/SusD pair and at least one adjacent CAZy-coding gene in the final PULpy annotation. CAZymes specific to the backbone of each polysaccharide class present in the PULpy summary files were used to manually identify putative amylolytic (GH13, GH31, GH65, GH66, GH97, or GH99) and cellulolytic (GH5, GH9, and GH94) PULs. Because the GH5 family encompasses multiple β-glucosidase subfamilies with diverse carbohydrate substrates, we considered for annotation only those GH5 CAZymes acting on cellulose (i.e., GH5_2, _38, and _46) as described by Aspeborg et al. (47). Similarly, for pectin hydrolysis we looked for homogalacturonan (GH28, GH105, or PL1), rhamnogalacturonan I (GH28, GH42, GH27, GH105, GH106, PL9, PL11, or PL26), galactan (GH2 or GH35), and arabinan (GH43 or GH51) catabolic CAZymes compared with previously pectinolytic PULs described in B. thetaiotaomicron (8). Because GH43 contains multiple subfamilies with diverse carbohydrate substrates, only GH43_1, _4, _5, _6, _10, _11, _12, _16, _21, _26, _27, _29, _33, and _35, described by Mewis et al. (48) as α-l-arabinofuranosidases, were considered in the analysis. GETHOMOLOGUES was used to identify PULs that were either common across or unique to specific isolates and publicly available genomes. Common and unique PUL protein clusters for the Bacteroides, Dysgonomonas, Parabacteroides, and Paludibacter genera used in these analyses were manually extracted from the pangenome_matrix result using custom bash and R scripts. Total gene organization and syntenic comparisons were plotted using genoPlotR (96).
Non-PUL-associated CAZy genes were identified in each isolate genome annotation by manually searching for loci annotated as either glycoside hydrolase (GH), polysaccharide lyase (PL), carbohydrate esterase (CE), or carbohydrate binding module (CBM), and the results were compared to the PULpy gene results table. All loci not present within the PULpy gene list were classified as non-PUL-associated CAZy genes. Signal peptides for protein secretion in the non-PUL CAZy genes were predicted with SignalP 5.0 (97) and LipoP 1.1 (98), and membrane-spanning regions were predicted with TMHMM 2.0 (99). CAZymes were classified as extracytoplasmic proteins if either the signal peptidase I (SPI) or the signal peptidase II (SPII) sequence was detected in the CAZy-encoding gene by both SignalP and LipoP.
T9SS detection.
PROKKA protein sequence predictions from the isolate genomes were compared to type XI secretion system (T9SS) HMM profiles in the TXSSscan database (100) using the “hmmscan” (parameters: hmmscan –cpu 40 –domtblout) tool from HMMER 3.1b2 (88) to identify genes encoding domains that comprised potential components of type IX secretion systems. Putative T9SS component domains were manually parsed from hmmscan result tables and compared with previous annotations of each new genome. Additionally, the presence of the T9SS C-terminal sorting domain (CTD) was identified using the TIGR04183 and TIGR04131 HMM profiles from TIGRFAM (101).
DNS assay.
Bacterial production of polysaccharolytic enzymes in the presence of pectin was evaluated using the 3,5-dinitrosalicylic acid (DNS) assay. Five replicates of each isolate were cultured anaerobically in MTY base liquid medium plus pectin from citrus peel (Sigma) to an OD600 of 0.4. Uninoculated sterile media were used as negative hydrolysis controls. One milliliter was collected from all samples and centrifuged at 12,000 × g for 10 min to pellet bacterial cells, and 100 μl of supernatant, which contained possible reducing sugars, was removed and incubated with 100 μl of DNS reagent in an Eppendorf thermocycler (one cycle of 10 min at 95°C; held at 2°C) (102). Fifty microliters of reaction mix was diluted in 250 μl of deionized water for absorbance measurements at 540 nm using a FLUOstar Omega plate reader. Standard curves were prepared using d-galacturonic acid to calculate substrate degradation, which was determined by quantifying the amount of reducing sugars released in mg ml−1. One-way ANOVA with Dunnett post hoc tests was performed to evaluate differences between treatments and negative controls. All statistics and plots were done in R version 3.5 using the DescTools v0.99.19 library (https://cran.r-project.org/web/packages/DescTools/index.html).
Polysaccharide degradation and growth assays.
Bacteroidetes isolates were plated on MTY base solid medium with one of the following polysaccharides added: starch (10 g liter−1) (Sigma), carboxymethyl cellulose (CMC) sodium salt (10 g liter−1) (EMD Millipore), Sigmacell cellulose (10 g liter−1) (Sigmacell) or pectin from citrus peel (10 g liter−1) (Sigma). Bacteria were incubated for up to 5 days at 30°C under anaerobic conditions. Following incubation, plates were immersed in 5 ml of 2.5% Lugol’s iodine solution to visualize carbohydrate hydrolysis; halos of cleared polysaccharide surrounding colonies were scored as positive hydrolysis. MTYG and MTY base plates were used as positive nonsaccharolytic and growth controls, respectively. E. coli TOP10 was included as a nonpolysaccharolytic bacterial control. Relative polysaccharolytic activity (RPA) was determined as the diameter of the colony plus the clear zone divided by the diameter of the colony. Final RPA values from Bacteroidetes isolates were subtracted from E. coli RPA values to estimate actual polysaccharolytic activity and eliminate false positives.
CMC growth assay and hydrolysis quantification.
To evaluate if cellulose improves growth of P. americana Bacteroidetes isolates, strains were grown in MTY base liquid anaerobic medium without or with carboxymethyl cellulose (1%, wt/vol) (MTY+CMC). Growth was evaluated by measuring the change OD600 every 12 h for 72 h. Total growth (Tg) was calculated as the difference between the maximum and minimum optical densities observed (Tg = OD600max − OD600min). Growth rates (Gr) were calculated as described previously (65). Total bacterial growth (Tg) was divided by the difference in time (Gr = Tg/Tmax − Tmin), where Tmax and Tmin correspond to time points where OD600max and OD600min were reached, respectively. A paired Student t test was used to identify significant differences in growth rate and total growth between MTY+CMC and MTY (control) kinetics for each isolate. Additionally, in vitro enzymatic CMC hydrolysis was determined using the same DNS microplate method as described above (102). Strains exhibiting growth and CMC hydrolysis on solid media were inoculated on MTY+CMC plates, and after 5 days, all biomass from plates was collected in 1.5 ml 0.1 M potassium phosphate buffer (KH2PO4, 9.5 g liter−1; K2HPO4, 5.25 g liter−1; pH 6.5). Bacterial cells were disrupted by five 30-s ultrasonication cycles on ice, and all samples were centrifuged at 12,000 × g for 10 min to collect supernatant that was transferred to fresh microcentrifuge tubes and stored on ice until use. Total supernatant protein content was quantified (bicinchoninic acid [BCA] protein quantification kit; Thermo Scientific), and 500 μl of supernatant was incubated with 500 μl of 1% CMC–0.1 M potassium phosphate buffer solution for 12 h. Total soluble proteins were prepared from E. coli TOP10 cells in the same manner and used as a cellulolytic negative control. Enzymatic activity was stopped by adding 100 μl of DNS, and total reducing sugars relative to a glucose standard curve were determined. Parallel hydrolysis reactions using heat-killed (95°C for 10 min) total soluble protein were performed as negative controls. Reducing sugars from CMC degradation were determined by subtracting sugars present in heated-killed treatments from reducing sugars present in non-heat-killed treatments, and the amount of reducing sugars was normalized per microgram of protein. Statistical differences in amounts of reducing sugars between the isolates and E. coli treatments were calculated by one-way ANOVA and Tukey HSD post hoc tests that were applied to pairwise comparisons between all treatments.
Data availability.
Isolate genome assemblies were deposited at DDBJ/ENA/GenBank under accession numbers QVMG00000000 to QVMQ00000000. The data have been deposited with links to BioProject accession number PRJNA486189 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/). All relevant data are also available from the authors upon request.
Supplementary Material
ACKNOWLEDGMENTS
A.V.-P.D.L., B.C.J., and Z.L.S. received support from the National Science Foundation (IOS-1656786) and The Ohio State University. A.V.-P.D.L. received support from “Apoyo para estancias posdoctorales en el extranjero del Consejo Nacional de Ciencia y Tecnologia (CONACYT).”
We thank Marie Asao, Mady Herrmann, and Sema Osman for their assistance in the lab. We also thank Phil Pope and Eric Martens for conversations and thoughtful comments on the manuscript. All bioinformatics were performed using The Ohio Supercomputing Center “Owens” and The Ohio State University “Unity” clusters.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Martens EC, Kelly AG, Tauzin AS, Brumer H. 2014. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol 426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grondin JM, Tamura K, Déjean G, Abbott DW, Brumer H. 2017. Polysaccharide utilization loci: fuelling microbial communities. J Bacteriol 199:e00860-16. doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies G, Henrissat B. 1995. Structures and mechanisms of glycosyl hydrolases. Structure 3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe H, Tokuda G. 2010. Cellulolytic systems in insects. Annu Rev Entomol 55:609–632. doi: 10.1146/annurev-ento-112408-085319. [DOI] [PubMed] [Google Scholar]
- 5.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 6.Brune A. 2014. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 7.Brune A, Dietrich C. 2015. The gut microbiota of termites: digesting the diversity in the light of ecology and evolution. Annu Rev Microbiol 69:145–166. doi: 10.1146/annurev-micro-092412-155715. [DOI] [PubMed] [Google Scholar]
- 8.Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Baslé A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schückel J, Venditto I, Ralet MC, Henrissat B, Martens EC, Mosimann SC, Abbott DW, Gilbert HJ. 2018. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 3:210–219. doi: 10.1038/s41564-017-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suen G, Weimer PJ, Stevenson DM, Aylward FO, Boyum J, Deneke J, Drinkwater C, Ivanova NN, Mikhailova N, Chertkov O, Goodwin LA, Currie CR, Mead D, Brumm PJ. 2011. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS One 6:e18814. doi: 10.1371/journal.pone.0018814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rakoff-Nahoum S, Coyne MJ, Comstock LE. 2014. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol 24:40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bignell DE. 1977. An experimental study of cellulose and hemicellulose degradation in the alimentary canal of the American cockroach. Can J Zool 55:579–589. doi: 10.1139/z77-073. [DOI] [Google Scholar]
- 12.Hansen AK, Moran NA. 2014. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol Ecol 23:1473–1496. doi: 10.1111/mec.12421. [DOI] [PubMed] [Google Scholar]
- 13.Makki K, Deehan EC, Walter J, Bäckhed F. 2018. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23:705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Engel P, Moran NA. 2013. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 15.Cockburn DW, Koropatkin NM. 2016. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J Mol Biol 428:3230–3252. doi: 10.1016/j.jmb.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 16.Bjursell MK, Martens EC, Gordon JI. 2006. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem 281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- 17.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Baslé A, Morland C, Day AM, Zheng H, Rogers TE, Thompson P, Hawkins AR, Yadav MP, Henrissat B, Martens EC, Dupree P, Gilbert HJ, Bolam DN. 2015. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun 6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLean R, Hamilton BS, Boraston AB, Cartmell A, Ziemer CJ, Lowe EC, Urs K, Day A, Vervecken W, Martens EC, Abbott DW, Cameron EA, Hakki Z, Peña MJ, Atherly T, Davies GJ, Suits MD, Williams SJ, Zhu Y, Piens K, Chen R, Porter NT, Rogowski A, Gilbert HJ, Thompson AJ, Cuskin F, Munōz-Munōz JL, Pudlo NA, Speciale G, Tolbert TJ, Bracke D, Temple MJ. 2015. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakoff-Nahoum S, Foster KR, Comstock LE. 2016. The evolution of cooperation within the gut microbiota. Nature 533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuki M, Oshima K, Suda W, Sakamoto M, Iida T, Hattori M, Ohkuma M. 2014. Draft genome sequence of Bacteroides reticulotermitis strain JCM 10512T, isolated from the gut of a termite. Genome Announc 2:e00072-14. doi: 10.1128/genomeA.00072-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun X, Yang Y, Zhang N, Shen Y, Ni J. 2015. Draft genome sequence of Dysgonomonas macrotermitis strain JCM 19375T, isolated from the gut of a termite. Genome Announc 3:e00963-15. doi: 10.1128/genomeA.00963-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gontang EA, Aylward FO, Carlos C, Del Rio TG, Chovatia M, Fern A, Lo CC, Malfatti SA, Tringe SG, Currie CR, Kolter R. 2017. Major changes in microbial diversity and community composition across gut sections of a juvenile Panchlora cockroach. PLoS One 12:e0177189-21. doi: 10.1371/journal.pone.0177189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Shi Y, Qiu Z, Che Y, Lo N. 2017. Reconstructing the phylogeny of Blattodea: robust support for interfamilial relationships and major clades. Sci Rep 7:1–8. doi: 10.1038/s41598-017-04243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bignell DE. 1981. Nutrition and digestion, p 57–86. In Bell WJ, Adiyodi KG (ed), The American cockroach. Springer Netherlands, Dordrecht, Netherlands. [Google Scholar]
- 26.Tamaki FK, Pimentel AC, Dias AB, Cardoso C, Ribeiro AF, Ferreira C, Terra WR. 2014. Physiology of digestion and the molecular characterization of the major digestive enzymes from Periplaneta americana. J Insect Physiol 70:22–35. doi: 10.1016/j.jinsphys.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Genta FA, Terra WR, Ferreira C. 2003. Action pattern, specificity, lytic activities, and physiological role of five digestive β-glucanases isolated from Periplaneta americana. Insect Biochem Mol Biol 33:1085–1097. doi: 10.1016/s0965-1748(03)00121-8. [DOI] [PubMed] [Google Scholar]
- 28.Oyebanji O, Soyelu O, Bamigbade A, Okonji R. 2014. Distribution of digestive enzymes in the gut of American cockroach, Periplaneta americana (L.). Int J Sci Res Publ 4:1–5. [Google Scholar]
- 29.Schauer C, Thompson CL, Brune A. 2012. The bacterial community in the gut of the cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites. Appl Environ Microbiol 78:2758–2767. doi: 10.1128/AEM.07788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabree ZL, Moran NA. 2014. Host-specific assemblages typify gut microbial communities of related insect species. Springerplus 3:138–111. doi: 10.1186/2193-1801-3-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tinker KA, Ottesen EA. 2016. The core gut microbiome of the american cockroach, Periplaneta americana, is stable and resilient to dietary shifts. Appl Environ Microbiol 82:6603–6610. doi: 10.1128/AEM.01837-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson KL, Salyers AA. 1989. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer-membrane starch-binding sites and periplasmic starch-degrading enzymes. J Bacteriol 171:3192–3198. doi: 10.1128/jb.171.6.3192-3198.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porter NT, Martens EC. 2017. The critical roles of polysaccharides in gut microbial ecology and physiology. Annu Rev Microbiol 71:349–369. doi: 10.1146/annurev-micro-102215-095316. [DOI] [PubMed] [Google Scholar]
- 34.Schwalm ND, Groisman EA. 2017. Navigating the gut buffet: control of polysaccharide utilization in Bacteroides spp. Trends Microbiol 25:1005–1015. doi: 10.1016/j.tim.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 35.Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci U S A 101:14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohde M, Chang Y-J, Han C, Palaniappan K, Pitluck S, Klenk H-P, Detter JC, Jeffries CD, Land M, Lapidus A, Mavromatis K, Hugenholtz P, Woyke T, Tapia R, Nolan M, Gronow S, Eisen JA, Mikhailova N, Kyrpides NC, Bristow J, Munk C, Göker M, Brambilla E, Hammon N, Goodwin L, Deshpande S, Markowitz V, Chen A, Liolios K, Ivanova N, Hauser L, Pati A, Cheng J-F, Lucas S. 2011. Complete genome sequence of Paludibacter propionicigenes type strain (WB4T). Stand Genomic Sci 4:36–44. doi: 10.4056/sigs.1503846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alberto F, Bignon C, Sulzenbacher G, Henrissat B, Czjzek M. 2004. The three-dimensional structure of invertase (β-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J Biol Chem 279:18903–18910. doi: 10.1074/jbc.M313911200. [DOI] [PubMed] [Google Scholar]
- 38.Naumoff DG. 2005. GH97 is a new family of glycoside hydrolases, which is related to the alpha-galactosidase superfamily. BMC Genomics 6:112. doi: 10.1186/1471-2164-6-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ratnavathi CV, Komala VV. 2016. Sorghum grain quality, p 1–61. In Ratnavathi CV, Patil J, Chavan UD (ed), Sorghum biochemistry. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 40.Ravcheev DA, Godzik A, Osterman AL, Rodionov DA. 2013. Polysaccharides utilization in human gut bacterium Bacteroides thetaiotaomicron: comparative genomics reconstruction of metabolic and regulatory networks. BMC Genomics 14:873. doi: 10.1186/1471-2164-14-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim YM, Yamamoto E, Kang MS, Nakai H, Saburi W, Okuyama M, Mori H, Funane K, Momma M, Fujimoto Z, Kobayashi M, Kim D, Kimura A. 2012. Bacteroides thetaiotaomicron VPI-5482 glycoside hydrolase family 66 homolog catalyzes dextranolytic and cyclization reactions. FEBS J 279:3185–3191. doi: 10.1111/j.1742-4658.2012.08698.x. [DOI] [PubMed] [Google Scholar]
- 42.Yapo BM. 2011. Rhamnogalacturonan-I: a structurally puzzling and functionally versatile polysaccharide from plant cell walls and mucilages. Polym Rev 51:391–413. doi: 10.1080/15583724.2011.615962. [DOI] [Google Scholar]
- 43.Despres J, Forano E, Lepercq P, Comtet-Marre S, Jubelin G, Yeoman CJ, Miller MEB, Fields CJ, Terrapon N, Le Bourvellec C, Renard C, Henrissat B, White BA, Mosoni P. 2016. Unraveling the pectinolytic function of Bacteroides xylanisolvens using a RNA-seq approach and mutagenesis. BMC Genomics 17:147. doi: 10.1186/s12864-016-2472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 45.Naas AE, Mackenzie AK, Mravec J, Schückel J, Willats WGT, Eijsink VGH, Pope PB. 2014. Do rumen Bacteroidetes utilize an alternative mechanism for cellulose degradation?. mBio 5:4–9. doi: 10.1128/mBio.01401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naas AE, Solden LM, Norbeck AD, Brewer H, Hagen LH, Heggenes IM, McHardy AC, Mackie RI, Paša-Tolic L, Arntzen M, Eijsink VGH, Koropatkin NM, Hess M, Wrighton KC, Pope PB. 2018. Candidatus Paraporphyromonas polyenzymogenes encodes multi-modular cellulases linked to the type IX secretion system. Microbiome 6:1–13. doi: 10.1186/s40168-018-0421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aspeborg H, Coutinho PM, Wang Y, Brumer H, Henrissat B. 2012. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol Biol 12:186. doi: 10.1186/1471-2148-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mewis K, Lenfant N, Lombard V, Henrissat B. 2016. Dividing the large glycoside hydrolase family 43 into subfamilies: a motivation for detailed enzyme characterization. Appl Environ Microbiol 82:1686–1692. doi: 10.1128/AEM.03453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kharade SS, McBride MJ. 2014. Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J Bacteriol 196:961–970. doi: 10.1128/JB.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lasica AM, Ksiazek M, Madej M, Potempa J. 2017. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol 7:215. doi: 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pais IS, Valente RS, Sporniak M, Teixeira L. 2018. Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol 16:e2005710. doi: 10.1371/journal.pbio.2005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crotti E, Rizzi A, Chouaia B, Ricci I, Favia G, Alma A, Sacchi L, Bourtzis K, Mandrioli M, Cherif A, Bandi C, Daffonchio D. 2010. Acetic acid bacteria, newly emerging symbionts of insects. Appl Environ Microbiol 76:6963–6970. doi: 10.1128/AEM.01336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morales-Jiménez J, Zúñiga G, Ramírez-Saad HC, Hernández-Rodríguez C. 2012. Gut-associated bacteria throughout the life cycle of the bark beetle Dendroctonus rhizophagus Thomas and Bright (Curculionidae: Scolytinae) and their cellulolytic activities. Microb Ecol 64:268–278. doi: 10.1007/s00248-011-9999-0. [DOI] [PubMed] [Google Scholar]
- 54.Arnal G, Bastien G, Monties N, Abot A, Anton Leberre V, Bozonnet S, O'Donohue M, Dumon C. 2015. Investigating the function of an arabinan utilization locus isolated from a termite gut community. Appl Environ Microbiol 81:31–39. doi: 10.1128/AEM.02257-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ndeh D, Rogowski A, Cartmell A, Luis AS, Baslé A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A, Terrapon N, Buffetto F, Nepogodiev S, Xiao Y, Field RA, Zhu Y, O'Neil MA, Urbanowicz BR, York WS, Davies GJ, Abbott DW, Ralet M-C, Martens EC, Henrissat B, Gilbert HJ. 2017. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544:65–70. doi: 10.1038/nature21725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foley MH, Cockburn DW, Koropatkin NM. 2016. The Sus operon: a model system for starch uptake by the human gut Bacteroidetes. Cell Mol Life Sci 73:2603–2617. doi: 10.1007/s00018-016-2242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naas AE, MacKenzie AK, Dalhus B, Eijsink VGH, Pope PB. 2015. Structural features of a Bacteroidetes-affiliated cellulase linked with a polysaccharide utilization locus. Sci Rep 5:11666. doi: 10.1038/srep11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wharton DRA, Wharton ML, Lola JE. 1965. Cellulase in the cockroach, with special reference to Periplaneta americana (L.). J Insect Physiol 11:947–959. doi: 10.1016/0022-1910(65)90198-8. [DOI] [PubMed] [Google Scholar]
- 59.Broxterman SE, Schols HA. 2018. Interactions between pectin and cellulose in primary plant cell walls. Carbohydr Polym 192:263–272. doi: 10.1016/j.carbpol.2018.03.070. [DOI] [PubMed] [Google Scholar]
- 60.Engel P, Martinson VG, Moran NA. 2012. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A 109:11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martínez-Romero E, Vera-Ponce de León A, Rosenblueth M, Martínez-Romero J, Bustamante-Brito R. 2019. Metatranscriptomic analysis of the bacterial symbiont dactylopiibacterium carminicum from the carmine cochineal Dactylopius coccus (Hemiptera: Coccoidea: Dactylopiidae). Life 9:4. doi: 10.3390/life9010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pope PB, Mackenzie AK, Gregor I, Smith W, Sundset MA, McHardy AC, Morrison M, Eijsink V. 2012. Metagenomics of the svalbard reindeer rumen microbiome reveals abundance of polysaccharide utilization loci. PLoS One 7:e38571-10. doi: 10.1371/journal.pone.0038571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mackenzie AK, Naas AE, Kracun SK, Schückel J, Fangel JU, Agger JW, Willats WGT, Eijsink VGH, Pope PB. 2015. A polysaccharide utilization locus from an uncultured bacteroidetes phylotype suggests ecological adaptation and substrate versatility. Appl Environ Microbiol 81:187–195. doi: 10.1128/AEM.02858-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robert C, Chassard C, Lawson PA, Bernalier-Donadille A. 2007. Bacteroides cellulosilyticus sp. nov., a cellulolytic bacterium from the human gut microbial community. Int J Syst Evol Microbiol 57:1516–1520. doi: 10.1099/ijs.0.64998-0. [DOI] [PubMed] [Google Scholar]
- 65.McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, Gordon JI. 2013. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol 11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pramono AK, Sakamoto M, Iino T, Hongoh Y, Ohkuma M. 2015. Dysgonomonas termitidis sp. nov., isolated from the gut of the subterranean termite Reticulitermes speratus. Int J Syst Evol Microbiol 65:681–685. doi: 10.1099/ijs.0.070391-0. [DOI] [PubMed] [Google Scholar]
- 67.Zhu Y, McBride MJ. 2014. Deletion of the Cytophaga hutchinsonii type IX secretion system gene sprP results in defects in gliding motility and cellulose utilization. Appl Microbiol Biotechnol 98:763–775. doi: 10.1007/s00253-013-5355-2. [DOI] [PubMed] [Google Scholar]
- 68.Zhu Y, Kwiatkowski KJ, Yang T, Kharade SS, Bahr CM, Koropatkin NM, Liu W, McBride MJ. 2015. Outer membrane proteins related to SusC and SusD are not required for Cytophaga hutchinsonii cellulose utilization. Appl Microbiol Biotechnol 99:6339–6350. doi: 10.1007/s00253-015-6555-8. [DOI] [PubMed] [Google Scholar]
- 69.Wahlund TM, Woese CR, Castenholz RW, Madigan MT. 1991. A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum sp. nov. Arch Microbiol 156:81–90. doi: 10.1007/BF00290978. [DOI] [Google Scholar]
- 70.Pfennig N. 1978. Rhodocyclus purpureus gen. nov. and sp. nov., a ring-shaped, vitamin B12-requiring member of the family Rhodospirillaceae. Int J Syst Bacteriol 28:283–288. doi: 10.1099/00207713-28-2-283. [DOI] [Google Scholar]
- 71.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kulikov AS, Prjibelski AD, Tesler G, Vyahhi N, Sirotkin AV, Pham S, Dvorkin M, Pevzner PA, Bankevich A, Nikolenko SI, Pyshkin AV, Nurk S, Gurevich AA, Antipov D, Alekseyev MA, Lesin VM. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng Y, Leung HCM, Yiu SM, Chin F. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 74.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 75.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 79.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 80.Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 82.Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher C, Holland T, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller L, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang P, Karp PD. 2014. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res 42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 86.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 87.Shimodaira H, Hasegawa M. 1999. Letter to the editor: multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16:1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- 88.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu M, Scott AJ. 2012. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 90.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rodriguez-R LM, Konstantinidis KT. 2016. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr 4:e1900v1. doi: 10.7287/peerj.preprints.1900v1. [DOI] [Google Scholar]
- 94.Stewart RD, Auffret M, Roehe R, Watson M. 2018. Open prediction of polysaccharide utilisation loci (PUL) in 5414 public Bacteroidetes genomes using PULpy. bioRxiv doi: 10.1101/421024. [DOI]
- 95.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. DbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:445–451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guy L, Roat Kultima J, Andersson S. 2010. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 98.Juncker AS, Willenbrock H, von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175–182. [PubMed] [Google Scholar]
- 100.Abby SS, Cury J, Guglielmini J, Néron B, Touchon M, Rocha E. 2016. Identification of protein secretion systems in bacterial genomes. Sci Rep 6:1–14. doi: 10.1038/srep23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haft DH, Selengut JD, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res 31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gonalves C, Rodriguez-Jasso RM, Gomes N, Teixeira JA, Belo I. 2010. Adaptation of dinitrosalicylic acid method to microtiter plates. Anal Methods 2:2046–2048. doi: 10.1039/c0ay00525h. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Isolate genome assemblies were deposited at DDBJ/ENA/GenBank under accession numbers QVMG00000000 to QVMQ00000000. The data have been deposited with links to BioProject accession number PRJNA486189 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/). All relevant data are also available from the authors upon request.