Jiuqu starter provides enzymes to the simultaneous saccharification and fermentation process of baijiu (Chinese liquor) production; however, the key saccharifying enzymes associated with alcoholic fermentation from Jiuqu and their effects on ethanol production remain unclear. We confirmed that Jiuqu was the main source of carbohydrate hydrolases for baijiu fermentation and identified two types of saccharifying enzymes from multiple microbes as the key enzymes associated with alcoholic fermentation. Moreover, a proper combination of multiple saccharifying enzymes could enhance ethanol production in baijiu fermentation. This combination provides an approach to optimize the profile of saccharifying enzymes for enhancing ethanol production in baijiu and other food fermentations.
KEYWORDS: Jiuqu, saccharifying enzyme, synergistic effect, ethanol, baijiu, Chinese liquor
ABSTRACT
Chinese Jiuqu (fermentation starter) provides saccharifying enzymes for baijiu (Chinese liquor) fermentation, which undergoes a simultaneous saccharification and fermentation process. However, the key saccharifying enzymes associated with alcoholic fermentation from Jiuqu and their effects on ethanol production remain poorly understood. In this study, we identified 51 carbohydrate hydrolases in baijiu fermentation by metaproteomics analysis. Through source-tracking analysis, approximately 80% of carbohydrate hydrolases in the baijiu fermentation were provided by Jiuqu. Among these enzymes, alpha-amylase (EC 3.2.1.1) and glucoamylase (EC 3.2.1.3), from Aspergillus, Rhizomucor, and Rhizopus, were positively related to starch hydrolysis and ethanol production, indicating that they were the key saccharifying enzymes associated with alcoholic fermentation in the baijiu fermentation. Moreover, a combined mixture of alpha-amylase and glucoamylase (in a ratio of 1:6, wt/wt) enhanced ethanol production in a simulative baijiu fermentation under laboratory conditions. This result revealed a synergistic effect of multiple saccharifying enzymes on ethanol production in baijiu fermentation. Our study provides a potential approach to improve the efficiency of saccharification and alcoholic fermentation by optimizing the profile of saccharifying enzymes for fermentation of baijiu and other beverages.
IMPORTANCE Jiuqu starter provides enzymes to the simultaneous saccharification and fermentation process of baijiu (Chinese liquor) production; however, the key saccharifying enzymes associated with alcoholic fermentation from Jiuqu and their effects on ethanol production remain unclear. We confirmed that Jiuqu was the main source of carbohydrate hydrolases for baijiu fermentation and identified two types of saccharifying enzymes from multiple microbes as the key enzymes associated with alcoholic fermentation. Moreover, a proper combination of multiple saccharifying enzymes could enhance ethanol production in baijiu fermentation. This combination provides an approach to optimize the profile of saccharifying enzymes for enhancing ethanol production in baijiu and other food fermentations.
INTRODUCTION
Baijiu (Chinese liquor), one of the oldest distilled liquors in the world, is generated by simultaneous saccharification and fermentation of cereals (sorghum, wheat, rice, corn, etc.) (1–3). This unique, complex process always selects Jiuqu (a mixed starter including Daqu, Xiaoqu, and Fuqu) as the saccharifying and fermenting agent (4). Jiuqu, composed of abundant microorganisms and enzymes, determines the hydrolysis of cereal-derived macromolecular nutrients in baijiu fermentation (1–3). This hydrolytic process determines the acquisition of available nutrients (like fermentable sugars) and subsequently influences microbial metabolism and the formation of flavor in baijiu (5–10). Therefore, it is important to identify the key saccharifying enzymes associated with alcoholic fermentation in Jiuqu and study their effects on ethanol production for improving baijiu fermentation (11, 12).
Jiuqu provides microorganisms and enzymes to baijiu fermentation and significantly contributes to ethanol and flavor compound generation (13, 14). Many studies revealed the structure and composition of microbial communities in Jiuqu (15–20). Furthermore, source-tracking analysis indicates that Jiuqu provides approximately 10% to 20% of the bacterial communities and 60% to 80% of the fungal communities to the baijiu fermentation (5, 21). Meanwhile, Jiuqu also provides abundant enzymes (starch-hydrolyzing enzymes, proteinases, cellulases, phases, and so on), accumulated from Jiuqu preparations, to the baijiu fermentation (15–20). However, the key saccharifying enzymes associated with alcoholic fermentation from Jiuqu remain unclear. In addition, starch hydrolysis determines the acquisition of fermentable sugars in baijiu fermentation and requires the sequential catalysis of multiple enzymes as a result of complex physical structure and modifications (22). Previous studies revealed the profile of saccharifying enzymes in Jiuqu and further indicate that the saccharifying activity of Jiuqu influences starch hydrolysis and metabolite formation in baijiu fermentation (6, 7, 19). However, the most efficient mode of saccharifying enzymes for enhancing ethanol production remains poorly understood. As a result, it is important to reveal the key saccharifying enzymes and study their effects on ethanol production for improving baijiu fermentation.
Here, we selected a typical Jiuqu (classified as Xiaoqu) as the subject and used enzymatic source-tracking analysis and multivariate statistical analysis to identify the key saccharifying enzymes associated with alcoholic fermentation from Jiuqu. Moreover, we revealed the metabolic features of the key saccharifying enzymes on ethanol production in a simulative baijiu fermentation under laboratory conditions. Our study sheds new light on the effects of multiple saccharifying enzymes on ethanol production. Furthermore, our study also provides a potential approach to optimize the profile of saccharifying enzymes for enhancing ethanol production in baijiu fermentation in particular and food fermentations in general.
RESULTS
Enzyme profile in baijiu fermentation.
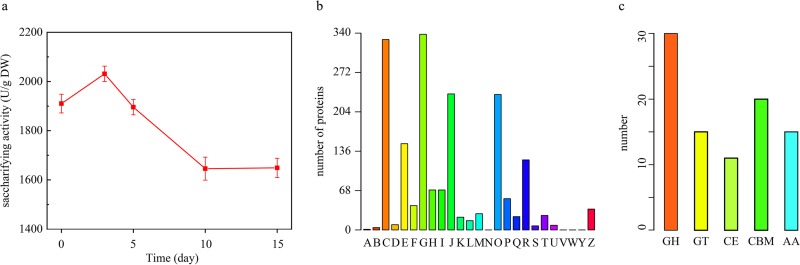
Saccharifying activity increased from 1,910 ± 58.02 to 2,031 ± 51.18 U/g dry weight (DW) in the baijiu fermentation from days 0 to 3 and then decreased to 1,649 ± 69.20 U/g DW at the end of fermentation. (Fig. 1a).
FIG 1.
Enzyme profile in baijiu fermentation. (a) Dynamic of saccharifying activity in baijiu fermentation. (b) Identified protein functions in COG database annotation. A, RNA processing and modification; B, chromatin structure and dynamics; C, energy production and conversion; D, cell cycle control, cell division, and chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation, ribosomal structure, and biogenesis; K, transcription; L, replication, recombination, and repair; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, posttranslational modification, protein turnover, and chaperon functions; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms; U, intracellular trafficking, secretion, and vesicular transport; V, defense mechanisms; W, extracellular structures; Y, nuclear structure; Z, cytoskeleton. (c) Category of identified carbohydrate hydrolases in the baijiu fermentation. GH, glycoside hydrolase families; GT, glycosyltransferase families; CE, carbohydrate esterase families; AA, auxiliary activity families; CBM, carbohydrate-binding module families. Bar, numbers of proteins.
We used a label-free quantitative proteomics approach to study the enzyme profile in the baijiu fermentation (Jiuqu; samples from the baijiu fermentation at days 0 and 3). Among 499,255 spectra obtained, 97,198 (19.47%) identified spectra were assigned to 15,228 peptides and 5,311 proteins. After filtering, we retained 2,373 potential nonredundant protein groups (see Tables S1 and S2 in the supplemental material).
According to Clusters of Orthologous Groups (COG) annotations, 1,819 proteins were classified into 21 categories, covering information storage and processing, cellular processes and signaling, metabolism, and so on (Fig. 1b and Table S3). Approximately 60% of all proteins were related to metabolism, including carbohydrate transport and metabolism, energy production and conversion, amino acid transport and metabolism, and so on. Three hundred thirty-eight proteins were related to carbohydrate transport and metabolism, the most active category in the baijiu fermentation (Fig. 1b and Table S3). Furthermore, we identified 89 carbohydrate hydrolases in Jiuqu and the baijiu fermentation, including 30 glycoside hydrolases (GHs), 15 glycosyltransferases (GTs), 11 carbohydrate esterases (CEs), 15 auxiliary activity enzymes (AAs), and 18 carbohydrate-binding module enzymes (CBMs) (Fig. 1c and Table S4).
Source-tracking analysis of carbohydrate hydrolases and their affiliated microbes in baijiu fermentation.
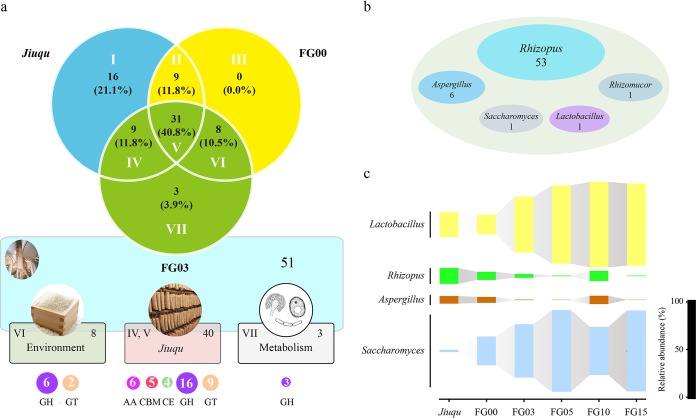
We identified 51 carbohydrate hydrolases in the baijiu fermentation at day 3 (Fig. 2a and Table S4). Among them, up to 40 enzymes appeared in both Jiuqu and the baijiu fermentation, including 6 AAs, 5 CBMs, 4 CEs, 16 GHs, and 9 GTs. Eight enzymes (six glycoside hydrolase and two glycosyltransferase families) appeared only in the baijiu fermentation, and the other three glycoside hydrolase families appeared only in the baijiu fermentation at day 3 (Fig. 2a). These enzymes were mainly affiliated with five genera, including Lactobacillus, Aspergillus, Rhizomucor, Rhizopus, and Saccharomyces (Fig. 2b and Table S4).
FIG 2.
Overview of carbohydrate hydrolases in baijiu fermentation. (a) Source of carbohydrate hydrolases in the baijiu fermentation at day 3. Part I, numbers of proteins only in Jiuqu; part II, numbers of proteins in Jiuqu and fermented samples at day 0; part III, numbers of proteins only in fermented samples at day 0; part IV, numbers of proteins in Jiuqu and fermented samples at day 3; part V, numbers of proteins in Jiuqu and fermented samples at days 0 and 3; part VI, numbers of proteins in fermented samples at days 0 and 3; part VII, numbers of proteins only in fermented samples at day 3. Part VI of the fermented samples at day 3 was from the local environment (raw materials and others). Parts IV and V of fermented samples at day 3 was from Jiuqu. Part VII of fermented samples at day 3 was from microbial metabolism. (b) Microbial source of carbohydrate hydrolases. Numbers are statistics of proteins from specific genera. (c) Dynamic of microbes across Jiuqu and baijiu fermentation. Bar, relative abundance of specific genus across samples.
Furthermore, we used high-throughput sequencing analysis to trace the key microbes (identified as the main microbial source of enzymes by metaproteomics analysis) in the baijiu fermentation. We obtained 450,408 and 535,304 high-quality reads from bacterial 16S rRNA gene sequences and fungal internal transcribed spacer (ITS) regions in all 18 samples. We found an average of 25,023 reads for bacteria and 29,739 reads for fungi per sample. Bacterial and fungal diversity was analyzed with the sequences normalized to 14,624 and 18,753 reads (Table S5). Chao1 richness and Shannon index showed that bacterial and fungal diversity decreased gradually in the baijiu fermentation (Table 1). Furthermore, eight bacterial and seven fungal genera were abundant in Jiuqu or the baijiu fermentation (with relative abundance above 1%) (Table S6). Seven bacterial genera (Acetobacter, Acinetobacter, Klebsiella, Lactobacillus, Pantoea, Pediococcus, and Weissella) and six fungal genera (Aspergillus, Candida, Pichia, Rhizopus, Saccharomyces, and Saccharomycopsis) were dominant in the baijiu fermentation. Among them, Lactobacillus, Aspergillus, Rhizomucor, Rhizopus, and Saccharomyces, which were the main microbial sources of enzymes in the metaproteomics analysis, were also dominant in Jiuqu (Fig. 2c and Table S6).
TABLE 1.
Bacterial and fungal microbiota diversity index based on 16S rRNA and ITS amplicon sequencing across samples
| Sample | No. of clean reads | P value | No. of OTUs | P value | Goods’ coverage | P value | Chao1 richness | P value | Shannon diversity index | P value |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | ||||||||||
| Jiuqu | 33,649 ± 2,301 | 0.07 | 236 ± 6 | <0.001 | 0.9942 ± 0.0002 | 0.04 | 312.00 ± 8.99 | <0.001 | 3.56 ± 0.25 | <0.001 |
| FG00 | 29,540 ± 8,984 | 236 ± 11 | 0.9943 ± 0.0004 | 299.76 ± 8.67 | 4.08 ± 0.03 | |||||
| FG03 | 25,931 ± 11,564 | 130 ± 32 | 0.9956 ± 0.0012 | 213.1 ± 30.28 | 3 ± 0.14 | |||||
| FG05 | 18,131 ± 1,901 | 133 ± 2 | 0.9959 ± 0.0004 | 207.24 ± 41.49 | 2.1 ± 0.08 | |||||
| FG10 | 19,965 ± 1,478 | 117 ± 14 | 0.9962 ± 0.0004 | 171.32 ± 18.26 | 1.69 ± 0.12 | |||||
| FG15 | 22,921 ± 2,524 | 188 ± 63 | 0.9939 ± 0.0018 | 312.69 ± 49.05 | 2.51 ± 0.36 | |||||
| Fungi | ||||||||||
| Jiuqu | 41,605 ± 2,501 | 0.02 | 57 ± 8 | <0.001 | 0.999 ± 0.0001 | <0.001 | 80.28 ± 15.3 | <0.001 | 2.22 ± 0.09 | <0.001 |
| FG00 | 29,513 ± 2,957 | 79 ± 8 | 0.9989 ± 0.0002 | 92.91 ± 11.73 | 2.75 ± 0.27 | |||||
| FG03 | 29,845 ± 3,834 | 46 ± 5 | 0.9991 ± 0.0002 | 70.8 ± 16.07 | 1.51 ± 0.07 | |||||
| FG05 | 30,491 ± 11,378 | 32 ± 5 | 0.9995 ± 0 | 37.3 ± 3.72 | 0.66 ± 0.08 | |||||
| FG10 | 24,952 ± 2,830 | 54 ± 16 | 0.9992 ± 0.0001 | 66.8 ± 10.61 | 2.25 ± 0.12 | |||||
| FG15 | 22,029 ± 4,779 | 37 ± 2 | 0.9992 ± 0 | 52.63 ± 5.21 | 0.71 ± 0.11 |
Identification of key carbohydrate hydrolases in baijiu fermentation.
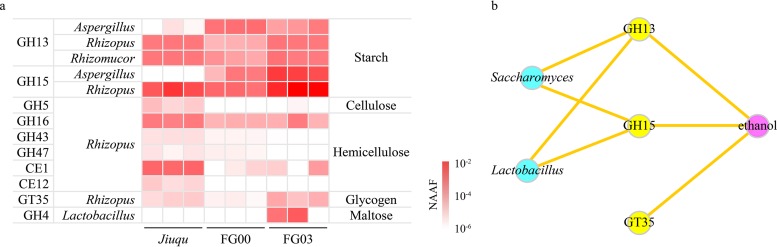
Thirty glycoside hydrolases were classified into GH4, GH5, GH13, GH15, GH16, GH17, GH43, GH47, and GH109 families and were related to polysaccharide hydrolysis (Table S4). GH13 (alpha-amylase, EC 3.2.1.1) and GH15 (glucoamylase, EC 3.2.1.3) were related to starch hydrolysis and affiliated with Rhizopus, Aspergillus, and Rhizomucor. GH5 (cellulase, EC 3.2.1.4) was related to cellulose hydrolysis and affiliated with Rhizopus. GH16 [endo-1,3(4)-beta-glucanase, EC 3.2.1.6], GH43 (beta-xylosidase, EC 3.2.1.37), and GH47 (alpha-mannosidase, EC 3.2.1.113), affiliated with Rhizopus, were related to hemicellulose hydrolysis (23–25). Besides extracellular glycoside hydrolases, several intracellular glycosyltransferases were also related to polysaccharide hydrolysis. For example, GT35 (alpha-1,4 glucan phosphorylase, EC 2.4.1.1) was related to stored glycogen or oligosaccharide hydrolysis and was affiliated with Rhizopus. In addition, we also found several intracellular enzymes related to oligosaccharide or disaccharide hydrolysis. For example, GH4 (maltose-6-phosphate hydrolase, EC 3.2.1.122) was related to maltose hydrolysis and affiliated with Lactobacillus (Fig. 3a and Table S4).
FIG 3.
Identification of key carbohydrate hydrolases in baijiu fermentation. (a) Carbohydrate hydrolases associated with saccharides hydrolysis and their microbial sources. Heatmap indicates the expression of proteins across samples. (b) Relationships between enzymes and microbes or metabolites, based on the connection for a significant (P < 0.05) and strong correlation (Spearman’s rho of >0.6). Yellow spots, enzymes; blue spots, microbes; purple spots, metabolites. Line, strong correlations between objects.
We identified the key carbohydrate hydrolases in the baijiu fermentation, combined with the interactions among enzymes, microbes, and metabolites (based on Spearman’s rank correlations of >0.6, P < 0.05). Here, GH13 and GH15 were positively correlated with the growth of Lactobacillus and Saccharomyces (Fig. 3b). Meanwhile, GH13, GH15, and GT35 were also positively correlated with ethanol production (Fig. 3b and Table S7).
Enzyme profile of isolated strains.
We isolated functional strains from samples and studied their enzyme profiles under culture-dependent conditions (Fig. S4). A Rhizopus microsporus strain grew from 6.00 ± 0.75 to 8.81 ± 0.13 log10 (CFU/g DW), and the saccharifying activity increased from 1,962 ± 112.8 to 20,031 ± 498.2 U/g DW in the 8-day fermentation (Fig. S4a and b). A Lactobacillus fermentum strain grew from 6.06 ± 0.11 to 9.00 ± 0.16 log10 (CFU/ml), whereas a Lactobacillus helveticus strain grew from 6.17 ± 0.11 to 9.00 ± 0.15 log10 (CFU/ml) in the 72-h fermentation (Fig. S4c). The pH decreased from 6.21 ± 0.05 to 5.00 ± 0.04 in the fermentation of L. fermentum, whereas it decreased from 6.35 ± 0.05 to 5.02 ± 0.04 in the fermentation of L. helveticus (Fig. S4d).
Effects of key carbohydrate hydrolases on ethanol production.
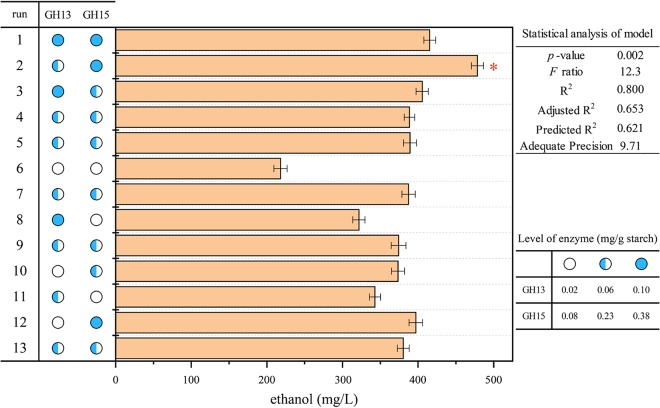
We assessed the effects of two starch hydrolases (GH13 and GH15) on ethanol production by statistical analysis. The two-component experiment had 13 enzyme mixtures according to a central composite design model (Fig. 4 and Tables S8 and S9). Analysis of variance (ANOVA) revealed that the quadratic mixture model was significant (F = 12.3, P < 0.01). A predicted R2 of 0.62 was reasonable, with an adjusted R2 of 0.65. Adequate precision was 9.71, and it indicated an adequate signal (Fig. 4) (26). As a result, GH15 was the most important factor influencing ethanol production (P < 0.01). The best enzyme profile (combination 2) produced 478.5 ± 7.98 mg/liter ethanol (Fig. 4, Table S8). A mixture of GH13 and GH15 (in a ratio of 1:6, wt/wt) had the highest predicted ethanol yield, corresponding to 439.9 mg/liter. To validate the predictive model, we combined GH13 and GH15 at a ratio of 1:6 (wt/wt) and achieved 435.8 ± 6.80 mg/liter ethanol (Table S9). This result indicated that a proper combination of multiple starch hydrolases enhances ethanol production.
FIG 4.
Statistical analysis of the predictive model for ethanol production in a similar baijiu fermentation system under laboratory conditions. Bar, ethanol concentrations in every experiment. Statistical analysis of the model indicates the evaluation of the central composite design. Circles indicate the levels of factors.
DISCUSSION
Our study identified the key saccharifying enzymes associated with alcoholic fermentation from Jiuqu in baijiu production. The proper combination or profile of the key saccharifying enzymes could enhance ethanol production. Our study shed light on the effects of multiple saccharifying enzymes on ethanol production. Furthermore, our study also provides an approach to optimize the profile of saccharifying enzymes for enhancing ethanol production in baijiu fermentation in particular and other food fermentations in general.
Jiuqu was the main source of saccharifying enzymes associated with alcoholic fermentation in baijiu production.
Saccharifying activity reached a peak, and 51 carbohydrate hydrolases were identified in the baijiu fermentation at day 3 (Fig. 1a and 2a). Approximately 80% of 51 enzymes were provided by Jiuqu, and they were affiliated mainly with Aspergillus, Rhizomucor, and Rhizopus (Fig. 2a and b). These microbes in the baijiu fermentation were also provided by Jiuqu (Fig. 2c).
In addition, the microbial compositions of Jiuqu (classified as Xiaoqu) in this study were different from the previous reports of Daqu, including bacteria such as Bacillus, Enterobacter, and lactic acid bacteria (5, 21). The universal Jiuqu starter can be classified as Daqu, Xiaoqu, and Fuqu, and their industrial characteristics were different in raw materials, inoculations, and culture conditions (1). The Jiuqu starter studied here is always made of stewed rice and soy beans and inoculated with strains in the initial preparation. Its preparation undergoes a back-slopped fermentation process under a lower temperature (30 to 40°C) (see Fig. S1 in the supplemental material), whereas Daqu is always made of raw wheat, barley, or pea and prepared in a spontaneous fermentation process under a higher temperature (40 to 70°C) (1). The industrial and host-associated stress responses in food microbes may drive the microbial community assembly in the back-slopped or spontaneous fermentation of various Jiuqu starters (27). According to the lifestyle of microbes in food fermentations, the microbiota of back-slopped Jiuqu starters may be shaped by the selection of the most competitive microorganisms, whereas the community assembly of Daqu may depend on microbial dispersals from plant or local environmental sources (28, 29). As a result, the selected and competitive microbes were dominated in back-slopped Jiuqu starters, like the vertebrate- or host-adapted lactic acid bacteria (e.g., L. helveticus), whereas the plant commensal microbes, like Bacillus, Enterobacteriaceae, and plant-associated lactic acid bacteria, were dominant in spontaneously fermented Daqu (5, 21). In addition, the microbial adaptation to environmental factors (e.g., temperature, acidity, and moisture) also may influence the community assembly in the preparation of Jiuqu starters (30, 31). In sum, the microbial stress responses and adaptation contribute to the community assembly in the back-slopped or spontaneous fermentation of Jiuqu starters.
Multiple saccharifying enzymes from multiple microbes contributed to alcoholic fermentation in baijiu production.
Starch hydrolysis in cereals requires a combination of multiple endo- and exoactive enzymes, including alpha-amylase (EC 3.2.1.1), beta-amylase (EC 3.2.1.2), isoamylase (EC 3.2.1.68), glucoamylase (EC 3.2.1.3), and others (32). We combined the metaproteomics, culture-dependent, and multivariate statistical analyses to identify the key saccharifying enzymes from Jiuqu. Alpha-amylase (GH13) and glucoamylase (GH15) were identified as two key saccharifying enzymes and were positively related to exogenous cereal-derived starch hydrolysis and alcoholic fermentation in the baijiu fermentation (Fig. 5). Alpha-amylase (GH13) plays a central role in the endohydrolytic action of starch at alpha-1,4 bonds to release a mixture of soluble linear and branched glucans. The linear and branched glucans are hydrolyzed to release short malto-oligosaccharides or maltose by alpha-amylase, isoamylase, and beta-amylase (32). Glucoamylase (GH15) takes part in the exohydrolytic action of starch at alpha-1,4(6) bonds to release glucose (32). Furthermore, glucoamylase can act synergistically with alpha-amylase to release glucose directly from the starch granule surface (33). In this study, these alpha-amylases (GH13) and glucoamylases (GH15) were mainly secreted by Aspergillus, Rhizomucor, and Rhizopus in the baijiu fermentation (Fig. 3a and 5). This finding can be distinguished from a single strain being responsible for enzyme secretion and starch hydrolysis in the production of other beverages or foods (34, 35). This typical saccharification of multiple enzymes from multiple microbes contributes to the diversity of microbial communities and abundant flavor compounds in the simultaneous saccharification fermentation process of Chinese baijiu (1).
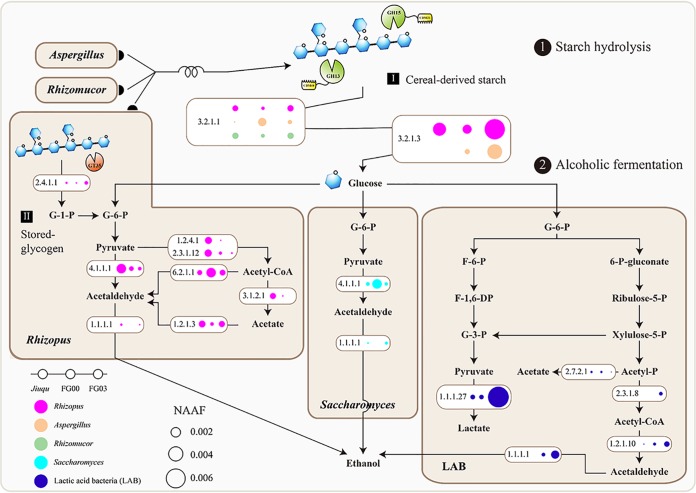
FIG 5.
Functional model for polysaccharide hydrolysis and ethanol production in Chinese baijiu fermentation. G-1-P, glucose 1-phosphate; G-6-P, glucose 6-phosphate; F-6-P, fructose-6-phosphate; F-1,6-DP, fructose-1,6-diphosphate; G-3-P, 3-phospho glyceraldehyde; 6-P-gluconate, 6-phosphate-gluconate; ribulose-5-P, ribulose-5-phosphate; xylulose-5-P, xylulose-5-phosphate; acetyl-P, acetyl-phosphate. Enzymes: EC 3.2.1.1, alpha-amylase; EC 3.2.1.3, glucoamylase; EC 2.4.1.1, alpha-1,4 glucan phosphorylase; EC 4.1.1.1, pyruvate decarboxylase; EC 1.1.1.1, aldehyde-alcohol dehydrogenase; EC 1.2.4.1, pyruvate dehydrogenase; EC 2.3.1.12, acetyltransferase; EC 6.2.1.1, acetyl-coenzyme A (CoA) synthetase; EC 3.1.2.1, acetyl-CoA hydrolase; EC 1.2.1.3, aldehyde dehydrogenase; EC 1.1.1.27, l-lactate dehydrogenase; EC 2.7.2.1, acetate kinase; EC 2.3.1.8, phosphotransacetylase; EC 1.2.1.10, aldehyde-alcohol dehydrogenase.
We also applied a culture-dependent method to emphasize the metaproteomics analysis. We isolated a Rhizopus microsporus strain from Jiuqu samples, which was the most important microbe for secreting saccharifying enzymes. We found the saccharifying activity increased as R. microsporus grew in the simulative Jiuqu preparation under laboratory conditions (Fig. S4a and b). Moreover, Aspergillus and Rhizomucor species are also identified as secreted saccharifying enzymes under culture-dependent conditions (20, 36), which corresponds to the metaproteomics analysis (Fig. 5). Furthermore, Aspergillus, Rhizomucor, and Rhizopus species can be selected as candidates to improve the quality of Jiuqu by adjusting the structure of microbial communities, fortifying functional microbes, or developing pure cultures (37–39). However, a proper combination of saccharifying enzymes should be identified for enhancing ethanol production, orienting it to adjust the microbial communities of Jiuqu.
In addition, several intracellular phosphorylases were related to stored glycogen or oligosaccharide hydrolysis and also contributed to ethanol production in the baijiu fermentation (Fig. 5). Alpha-1,4 glucan phosphorylase (GT35) will cleave glycogen to generate glucose-1-phosphate by consuming inorganic phosphate, and glucose-1-phosphate can be converted to glucose-6-phosphate without ATP by phosphoglucomutase (40, 41). Glucose-6-phosphate then can be metabolized to generate ethanol in cells. Furthermore, the bacterial phosphorylases can select extracellular maltodextrin as a source of carbon and energy when maltodextrin is available in the environment (42). The fungal phosphorylases should get more attention in the utilization of extracellular polysaccharides. We also identified an intracellular maltose-6-phosphate hydrolase, affiliated with Lactobacillus species, in the metaproteomics analysis (Fig. 3a). Meanwhile, we isolated Lactobacillus fermentum and Lactobacillus helveticus strains from the baijiu fermentation. L. fermentum and L. helveticus then were cultured using the manual medium that selected maltose as the only carbon source, and we found the pH decreased as the cells grew in the fermentation (Fig. S4c and d). This finding indicated that L. fermentum and L. helveticus can utilize maltose (43, 44). These results corresponded to those from the metaproteomics analysis (Fig. 3a). Furthermore, these Lactobacillus species were dominant across Jiuqu and baijiu fermentation according to the high-throughput sequencing analysis (Fig. S5). Here, we focused on the role of glycoside hydrolases in polysaccharide hydrolysis and ethanol production in food fermentations (45, 46). Additionally, the stored glycogen or oligosaccharide hydrolysis, catalyzed by phosphorylases (starch, maltose, or other phosphorylases), also should contribute to ethanol production. These catalytic pathways are energy efficient in intracellular systems and should not be overlooked (18, 19, 47, 48). Furthermore, various phosphorylases (starch, maltose, or other phosphorylases) are widely exploited in industry because of the economic role of their glycosyltransferase reactions. Meanwhile, as starch is inexpensive and generally distributed in the local environment, multiple phosphorylases have broad applications in food, cosmetic, plastic, and pharmaceutical industries (49–51).
A proper combination of saccharifying enzymes could enhance ethanol production.
The effects of two glycoside hydrolases (GH13 and GH15) were positively related to starch hydrolysis and ethanol production in baijiu fermentation (Fig. 5). Additionally, we isolated a Saccharomyces cerevisiae strain from samples that was generally responsible for ethanol metabolism in baijiu fermentation (52). Therefore, we combined the mixtures of two saccharifying enzymes and studied their effects on ethanol production in an in vitro system. As a result, we acquired a proper combination of GH13 and GH15 for enhancing ethanol production, and we obtained the responsible strains from Jiuqu samples. This method provides a potential approach to selecting the responsible strains to adjust microbial assembly and metabolism in the preparation of Jiuqu or to develop pure cultures for obtaining the optimized enzyme profile of starters to enhance ethanol production in baijiu fermentation.
In conclusion, we identified the key saccharifying enzymes associated with alcoholic fermentation from Jiuqu in baijiu production. Moreover, we revealed that the proper combination of the key saccharifying enzymes could enhance ethanol production. Our study shed light on the effects of multiple saccharifying enzymes on ethanol production. Furthermore, our study provides a potential approach to optimizing the profile of saccharifying enzymes for enhancing ethanol production in baijiu fermentation in particular and other food fermentations in general.
MATERIALS AND METHODS
Sample collection.
Samples were collected in July 2017 from a famous baijiu distillery in Guangzhou Province, China (lat 29.43, long 115.59) (see Fig. S1 and S2 in the supplemental material). In Jiuqu preparations, raw materials (rice and soybean) were stewed and mixed with caky balls (inoculated with strains). Mixed materials were shaped into bricks and transferred into a brewing facility for fermentation. Matured Jiuqu bricks were stored for 2 weeks and smashed to powder for baijiu fermentation. The mixture of Jiuqu powder, raw material (rice), and water (in a ratio of 1:5:16, wt/wt/wt) was put into a fermentation silo (about 50 m3) and then sealed for a 15-day baijiu fermentation (Fig. S1). We tracked 3 Jiuqu brewing facilities on 7, 8, and 9 July 2017 (Fig. S2). Each facility was an independent batch. Three Jiuqu powder samples (about 1,000 g) were collected before being transferred into each fermentation silo. We tracked three silos that used the corresponding Jiuqu samples. Samples were collected from three points (1, 2, and 3) at the upper, middle, and bottom layer (a, b, and c) in every fermentation silo (Fig. S2). One final sample was obtained by pooling samples from different sites. Samples (about 1,000 g) were collected at days 0, 3, 5, 10, and 15 from 3 fermentation silos and named FG00, FG03, FG05, FG10, and FG15. All samples were stored at –20°C until microbial and physicochemical analysis. Separate samples of Jiuqu, FG00, and FG03 (200 g each) were immediately frozen in liquid nitrogen after collection and then stored at –80°C for metaproteomics analysis.
Physicochemical and enzymatic activity analysis.
The moisture of samples was measured by detecting its weight loss after drying 10-g samples at 105°C for 4 h (sufficient to reach constant weight). The saccharifying activity of samples was measured as previously described (53, 54). A unit of saccharifying activity was defined as the amount (in micrograms) of glucose converted from starch by 1 g of sample per minute under the assay conditions.
Protein preparation and mass spectrometry analyses.
To achieve adequate predictive power for the enzyme profile, samples of Jiuqu, fermentation samples at day 0 and day 3, were applied to a label-free quantitative proteomics analysis. The proteomic samples were prepared as described earlier (19, 55). A ProteoExtract abundant protein extraction kit (Merck KGaA, Darmstadt, Germany) was used to remove the high-abundance proteins from the proteomic samples.
Peptides were analyzed by an EASY-nLC1200 coupled to a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Five microliters (approximately 2.5 μg total peptide) was loaded into a C18 analytical column (75 μm [inner diameter] by 25 cm) (Thermo Fisher Scientific, Waltham, MA) for separation for 240 min. The mobile phase was controlled by solvents A (2% acetonitrile and 0.1% formic acid) and B (80% acetonitrile and 0.1% formic acid) at 300 nl/min, using the following gradients: starting with 5% B for 2 min, 23% B for min 3 to 147, 29% B for min 148 to 183, 48% B for min 184 to 208, 100% B for min 209 to 215, and 0% B until 240 min. Linear trap quadrupole mass spectrometer conditions were set as previously reported (19).
Mass spectrometric data analysis.
Raw data were uploaded to the database and analyzed by Thermo Scientific Proteome Discoverer (PD) 1.4 software connected to an in-house Mascot server (V 2.4.1; Matrix Science, Boston, MA). Proteins were searched on the UniProt database (http://www.uniprot.org). The peptide and protein candidates were generated by a strict filtering based on a false discovery rate (FDR) of 0.01. Label-free quantification (LFQ) was applied by MaxQuant (56). The detailed procedure was described earlier (57). Meanwhile, we constructed the database based on the genomic and proteomic information of the dominant microbial members (identified by the high-throughput sequencing analysis) from the NCBI database (https://www.ncbi.nlm.nih.gov/), including Lactobacillus fermentum (CP002033.1), Lactobacillus helveticus (NC_010080.1), Klebsiella pneumoniae (FO203501.1), Pediococcus pentosaceus (NC_008525.1), Weissella confusa (NZ_CAGH00000000.1), Aspergillus ruber (KK088411.1), Candida glabrata (LMAA01000056.1), Pichia kudriavzevii (NC_042506.1, NC_042507.1, NC_042508.1, NC_042509.1, and NC_042510.1), Rhizopus microsporus (KV921258.1), Saccharomyces cerevisiae (NC_001136.10), Saccharomycopsis fibuligera (CP015978.1, CP015979.1, CP015980.1, CP015981.1, CP015982.1, CP015983.1, and CP015984.1), and others. The reconstructed database was also used for protein research. We identified parent proteins by the highest score for a given peptide mass. Identified proteins were annotated by Gene Ontology (GO), Clusters of Orthologous Groups (COG), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. We determined normalized area abundance factors (NAAF) to estimate protein abundances by chromatographic peak areas from the proteomics data (58).
DNA extraction, amplification, and sequence processing.
Total DNA was extracted from samples by the E.Z.N.A. (easy nucleic acid isolation) soil DNA kit (Omega Bio-Tek, Norcross, GA). The 16S rRNA gene V3-V4 hypervariable region was amplified by universal primers 338F (forward) and 806R (reverse) (59). The internal transcribed spacer (ITS) was amplified by primers ITS1F and ITS2 (60). These primers contained a set of barcode sequences unique to each sample. PCR products were purified by a PCR purification kit, and their concentrations were measured by a Thermo Scientific NanoDrop 8000 UV-visible spectrophotometer (NanoDrop Technologies, Wilmington, DE). The barcoded PCR products were sequenced by a MiSeq benchtop sequencer for 250-bp paired-end sequencing (2 by 250 bp; Illumina, San Diego, CA) at Beijing Auwigene Tech., Ltd. (Beijing, China).
The MiSeq-generated raw sequence data were processed by the QIIME pipeline (v 1.8.0) (61). We removed the sequences with quality scores of <30 for quality trimming, choosing only sequences over 200 bp to analyze. We removed sequences that did not perfectly match the PCR primer. Chimeras were removed by UCHIME software (62). The trimmed sequences then were clustered into operational taxonomic units (OTUs) with a 97% identity threshold by QIIME’s uclust pipeline (63). A single representative sequence from each OTU was aligned to the Greengenes database (v13.8) and the UNITE fungal ITS database (v6.0) (64, 65). Singleton OTUs were removed before further analysis. Chao1 richness and Shannon diversity indexes were calculated by QIIME (v 1.8.0) (66).
Strain isolation and cultivation.
The bacterial and fungal strains were isolated from Jiuqu and fermented samples by using Man-Rogosa-Sharpe medium (MRS), Wallerstein laboratory medium (WL), and potato dextrose agar (PDA). Genomic DNA extraction and identification of the single isolated strains were applied as described in previous studies (5, 19). R. microsporus JJ01 was inoculated into the raw materials used for Jiuqu preparations, and the culture conditions were set as previously described (19). L. fermentum JJ01 and L. helveticus JJ02 were cultured for 72 h at 37°C in manual medium [including 20 g/liter maltose, 10 g/liter tryptone, 10 g/liter beef extract, 5 g/liter yeast extract, 5 g/liter CH3COONa·3H2O, 2 g/liter K2HPO4·3H2O, 2 g/liter (NH4)2HC6H5O7, 0.58 g/liter MgSO4·7H2O, 0.25 g/liter MnSO4·H2O, and 1 ml/liter Tween 80]. pH was determined with a pH detector (INESA Scientific Instrument Co., Ltd., Shanghai, China). Other physicochemical parameters, real-time quantitative PCR, and enzyme activity analysis were applied as previously reported (19).
HPLC analysis.
The extraction of ethanol was done as previously described (5). Ethanol was separated by a column (Aminex HPX-87H; 300 mm by 7.8 mm; Bio-Rad, Hercules, CA) and detected by high-performance liquid chromatography (HPLC; Agilent 1200 HPLC; Agilent Technologies, Santa Clara, CA) with a refractive index detector (WGE GmbH, Bergheim, Germany). The detection conditions were set as previously described (67). The quantitative standard curve was established using ethanol (99.9%, HPLC grade) purchased from J&K Chemical, Ltd. (Shanghai, China).
Optimization of enzyme profile for ethanol production.
Polished rice was soaked in water at 25°C overnight (the ratio of rice to water was 2:3, wt/wt) and steamed at 121°C for 40 min. The solid fraction was suspended in the low-salt buffer (10 mmol/liter Tris-HCl, 0.1 mol/liter NaCl, pH 7.5), and a final 2% (wt/vol) suspension was used for alcoholic fermentation. Alpha-amylase (GH13) and glucoamylase (GH15) were purchased from Anhui Guangyuan Industrial Co., Ltd. (Hefei, China), and used for carbohydrate hydrolysis and alcoholic fermentation. The phylogenetic tree of commercial enzymes and the ones identified in the baijiu fermentation is shown in Fig. S3.
The experimental design and analysis were implemented by Design-Expert software (Stat-Ease, Inc., Minneapolis, MN) to assess the effect of combined GH13 and GH15 on ethanol production. A central composite design required 13 reactions based on setting the low and high levels of two enzymes. The ranges from minimum to maximum content were set as 0.02 to 0.10 mg/g starch for GH13 and 0.08 to 0.38 mg/g starch for GH15 (19). Each reaction mixture (50 ml, pH 7.5) was incubated at 30°C for a 72-h fermentation. Saccharomyces cerevisiae JJ01 was precultured in yeast extract-peptone-dextrose medium at 30°C and 200 rpm for 24 h. Cells were centrifuged (10,000 × g for 3 min), washed twice with sterile saline (0.90% wt/vol), and then inoculated into each mixture with an initial cell density of 1 × 106 cells/ml. After a 72-h fermentation, 10 ml culture broth was centrifuged at 10,000 × g for 5 min, and the supernatant was collected to measure ethanol. The concentration of ethanol was calculated as previously described (5, 68). All data were analyzed by ANOVA, and to develop a statistically based predictive model, F ratio, P value, R2, adjusted R2, predicted R2, and adequate precision were calculated (26). All experiments were run in triplicate.
Data analysis.
We calculated all possible Spearman’s rank correlations to analyze the relationships among microbes, enzymes, and metabolites. A network was created by Cytoscape (ver 3.7.0) to sort through and visualize correlations.
Data availability.
All generated protein data were submitted to the iProX database (http://www.iprox.org) under the project number IPX0001641000/PXD014221. All sequences generated were submitted to the DDBJ database under the accession numbers DRA008485 and DRA008486. The genomic sequences of the isolated Rhizopus microsporus JJ01, Saccharomyces cerevisiae JJ01, Lactobacillus fermentum JJ01, and Lactobacillus helveticus JJ02 strains were deposited in GenBank under the project numbers MH782032, MK994013, MN841929, and MN841930.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the National Key R&D Program of China (2018YFD0400402 and 2016YFD0400500), National Natural Science Foundation of China (NSFC) (grant 31530055), Jiangsu Province Science and Technology Project (BE2017705), and the National First-Class Discipline Program of Light Industry Technology and Engineering (LITE2018-12).
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Jin G, Zhu Y, Xu Y. 2017. Mystery behind Chinese liquor fermentation. Trends Food Sci Technol 63:18–28. doi: 10.1016/j.tifs.2017.02.016. [DOI] [Google Scholar]
- 2.Liu H, Sun B. 2018. Effect of fermentation processing on the flavor of baijiu. J Agric Food Chem 66:5425–5432. doi: 10.1021/acs.jafc.8b00692. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Huo J, Huang M, Zhao M, Luo X, Sun B. 2017. Structural characterization of a tetrapeptide from sesame flavor-type baijiu and its preventive effects against AAPH-Induced oxidative stress in HepG2 cells. J Agric Food Chem 65:10495–10504. doi: 10.1021/acs.jafc.7b04815. [DOI] [PubMed] [Google Scholar]
- 4.Wang M, Yang J, Zhao Q, Zhang K, Su C. 2019. Research progress on flavor compounds and microorganisms of Maotai flavor baijiu. J Food Sci 84:6–18. doi: 10.1111/1750-3841.14409. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Du H, Zhang Y, Xu Y. 2018. Environmental microbiota drives microbial succession and metabolic profiles during Chinese liquor fermentation. Appl Environ Microbiol 84:e02369-17. doi: 10.1128/AEM.02369-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang P, Wu Q, Jiang X, Wang Z, Tang J, Xu Y. 2017. Bacillus licheniformis affects the microbial community and metabolic profile in the spontaneous fermentation of Daqu starter for Chinese liquor making. Int J Food Microbiol 250:59–67. doi: 10.1016/j.ijfoodmicro.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 7.He G, Huang J, Zhou R, Wu C, Jin Y. 2019. Effect of fortified Daqu on the microbial community and flavor in Chinese strong-flavor liquor brewing process. Front Microbiol 10:56. doi: 10.3389/fmicb.2019.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang G, Hamaker BR. 2017. The nutritional property of endosperm starch and its contribution to the health benefits of whole grain foods. Crit Rev Food Sci Nutr 57:3807–3817. doi: 10.1080/10408398.2015.1130685. [DOI] [PubMed] [Google Scholar]
- 9.Wu Z, He F, Qin D, Li H, Sun J, Sun X, Sun B. 2019. Determination of phenolic compounds in alcoholic fermentation materials and spent grains by ultrasound-assisted alkali alcohol extraction coupled with HPLC. Anal Methods 11:5366–5375. doi: 10.1039/C9AY01739A. [DOI] [Google Scholar]
- 10.Zhao D, Jiang Y, Sun J, Li H, Luo X, Zhao M. 2019. Anti-inflammatory mechanism involved in 4-ethylguaiacol-mediated inhibition of LPS-induced inflammation in THP-1 cells. J Agric Food Chem 67:1230–1243. doi: 10.1021/acs.jafc.8b06263. [DOI] [PubMed] [Google Scholar]
- 11.Tang Q, He G, Huang J, Wu C, Jin Y, Zhou R. 2019. Characterizing relationship of microbial diversity and metabolite in Sichuan Xiaoqu. Front Microbiol 10:696. doi: 10.3389/fmicb.2019.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gan S, Yang F, Sahu SK, Luo R, Liao S, Wang H, Jin T, Wang L, Zhang P, Liu X, Xu J, Xu J, Wang Y, Liu H. 2019. Deciphering the composition and functional profile of the microbial communities in Chinese Moutai liquor starters. Front Microbiol 10:1540. doi: 10.3389/fmicb.2019.01540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Zheng X, Han B, Vervoort J, Nout M. 2009. Characterization of Chinese liquor starter, “Daqu,” by flavor type with 1H NMR-based nontargeted analysis. J Agric Food Chem 57:11354–11359. doi: 10.1021/jf902881p. [DOI] [PubMed] [Google Scholar]
- 14.Zheng X, Tabrizi MR, Nout MJR, Han B. 2011. Daqu—a traditional Chinese liquor fermentation starter. J Inst Brewing 117:82–90. doi: 10.1002/j.2050-0416.2011.tb00447.x. [DOI] [Google Scholar]
- 15.Gao Y, Wang H, Xu Y. 2010. PCR-DGGE analysis of the bacterial community of Chinese liquor high and medium temperature Daqu. Microbiology 37:999–1004. [Google Scholar]
- 16.Wang H, Gao Y, Fan Q, Xu Y. 2011. Characterization and comparison of microbial community of different typical Chinese liquor Daqus by PCR-DGGE. Lett Appl Microbiol 53:134–140. doi: 10.1111/j.1472-765X.2011.03076.x. [DOI] [PubMed] [Google Scholar]
- 17.Wu Q, Ling J, Xu Y. 2014. Starter culture selection for making Chinese sesame-flavored liquor based on microbial metabolic activity in mixed-culture fermentation. Appl Environ Microbiol 80:4450–4459. doi: 10.1128/AEM.00905-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Yi Z, Jin Y, Huang M, He K, Liu D, Luo H, Zhao D, He H, Fang Y, Zhao H. 2017. Metatranscriptomics reveals the functions and enzyme profiles of the microbial community in Chinese nong-flavor liquor starter. Front Microbiol 8:1747. doi: 10.3389/fmicb.2017.01747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, Wu Q, Xu Y, Sun B. 2018. Specific volumetric weight-driven shift in microbiota compositions with saccharifying activity change in starter for Chinese baijiu fermentation. Front Microbiol 9:2349. doi: 10.3389/fmicb.2018.02349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, Wu Q, Xu Y. 2014. Filamentous fungal diversity and community structure associated with the solid state fermentation of Chinese Maotai-flavor liquor. Int J Food Microbiol 179:80–84. doi: 10.1016/j.ijfoodmicro.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Du H, Xu Y. 2017. Source tracking of prokaryotic communities in fermented grain of Chinese strong-flavor liquor. Int J Food Microbiol 244:27–35. doi: 10.1016/j.ijfoodmicro.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 22.Wu Q, Cao S, Xu Y. 2017. Effects of glutinous and nonglutinous sorghums on Saccharomyces cerevisiae fermentation for Chinese liquor making. Int J Food Sci Technol 52:1348–1357. doi: 10.1111/ijfs.13330. [DOI] [Google Scholar]
- 23.Comtet-Marre S, Parisot N, Lepercq P, Chaucheyras-Durand F, Mosoni P, Peyretaillade E, Bayat AR, Shingfield KJ, Peyret P, Forano E. 2017. Metatranscriptomics reveals the active bacterial and eukaryotic fibrolytic communities in the rumen of dairy cow fed a mixed diet. Front Microbiol 8:67. doi: 10.3389/fmicb.2017.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu N, Yang J, Ji L, Liu J, Yang Y, Yuan H. 2016. Metagenomic and metaproteomic analyses of a corn stover-adapted microbial consortium EMSD5 reveal its taxonomic and enzymatic basis for degrading lignocellulose. Biotechnol Biofuels 9:243. doi: 10.1186/s13068-016-0658-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma G, Khatri I, Subramanian S. 2016. Complete genome of the starch-degrading myxobacteria Sandaracinus amylolyticus DSM 53668T. Genome Biol Evol 8:2520–2529. doi: 10.1093/gbe/evw151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Zhang S, He Q, Hu X. 2015. Application of response surface methodology in experiment design and optimization. Res Explor Lab 34:41–45. doi: 10.1155/2016/2349476. [DOI] [Google Scholar]
- 27.Ruiz L, Aertsen A, Nguyen-The C, Gänzle MG, Alvarez-Ordóñez A. 2017. Editorial: industrial and host associated stress responses in food microbes. Implications for food technology and food safety. Front Microbiol 8:1522. doi: 10.3389/fmicb.2017.01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganzle M, Ripari V. 2016. Composition and function of sourdough microbiota: from ecological theory to bread quality. Int J Food Microbiol 239:19–25. doi: 10.1016/j.ijfoodmicro.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Ganzle MG, Zheng J. 2019. Lifestyles of sourdough lactobacilli—do they matter for microbial ecology and bread quality? Int J Food Microbiol 302:15–23. doi: 10.1016/j.ijfoodmicro.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Li P, Lin W, Liu X, Wang X, Luo L. 2016. Environmental factors affecting microbiota dynamics during traditional solid-state fermentation of Chinese Daqu starter. Front Microbiol 7:1237. doi: 10.3389/fmicb.2016.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao C, Lu Z, Zhang X, Wang S, Ao L, Shen C, Shi J, Xu Z. 2017. Bio-heat is a key environmental driver shaping the microbial community of medium-temperature Daqu. Appl Environ Microbiol 83:e01550-17. doi: 10.1128/AEM.01550-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeeman SC, Kossmann J, Smith AM. 2010. Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 61:209–234. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
- 33.Sun Z, Henson CA. 1991. A quantitative assessment of the importance of barley seed α-amylase, β-amylase, debranching enzyme, and α-glucosidase in starch degradation. Arch Biochem Biophys 284:298–305. doi: 10.1016/0003-9861(91)90299-x. [DOI] [PubMed] [Google Scholar]
- 34.Gomi K. 2019. Regulatory mechanisms for amylolytic gene expression in the koji mold Aspergillus oryzae. Biosci Biotechnol Biochem 83:1385–1401. doi: 10.1080/09168451.2019.1625265. [DOI] [PubMed] [Google Scholar]
- 35.Zhu Y, Tramper J. 2013. Koji—where East meets West in fermentation. Biotechnol Adv 31:1448–1457. doi: 10.1016/j.biotechadv.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Ban S, Qiu S. 2018. Analysis of the mould microbiome and exogenous enzyme production in Moutai-flavor Daqu. J Inst Brew 124:91–99. doi: 10.1002/jib.467. [DOI] [Google Scholar]
- 37.Li P, Lin W, Liu X, Wang X, Gan X, Luo L, Lin W. 2017. Effect of bioaugmented inoculation on microbiota dynamics during solid-state fermentation of Daqu starter using autochthonous of Bacillus, Pediococcus, Wickerhamomyces and Saccharomycopsis. Food Microbiol 61:83–92. doi: 10.1016/j.fm.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Qiu S, Li P, Ban S. 2019. Analysis of microbial community structure in traditional and automated Moutai-flavor Daqu. J Am Soc Brew Chem 77:140–146. doi: 10.1080/03610470.2019.1569886. [DOI] [Google Scholar]
- 39.Ma R, Sui L, Zhang J, Hu J, Liu P. 2019. Polyphasic characterization of yeasts and lactic acid bacteria metabolic contribution in semi-solid fermentation of Chinese baijiu (traditional fermented alcoholic drink): towards the design of a tailored starter culture. Microorganisms 7:147. doi: 10.3390/microorganisms7050147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ubiparip Z, Beerens K, Franceus J, Vercauteren R, Desmet T. 2018. Thermostable alpha-glucan phosphorylases: characteristics and industrial applications. Appl Microbiol Biotechnol 102:8187–8202. doi: 10.1007/s00253-018-9233-9. [DOI] [PubMed] [Google Scholar]
- 41.Liu N, Li H, Chevrette MG, Zhang L, Cao L, Zhou H, Zhou X, Zhou Z, Pope PB, Currie CR, Huang Y, Wang Q. 2019. Functional metagenomics reveals abundant polysaccharide-degrading gene clusters and cellobiose utilization pathways within gut microbiota of a wood-feeding higher termite. ISME J 13:104–117. doi: 10.1038/s41396-018-0255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cifuente JO, Comino N, Trastoy B, D'Angelo C, Guerin ME. 2019. Structural basis of glycogen metabolism in bacteria. Biochem J 476:2059–2092. doi: 10.1042/BCJ20170558. [DOI] [PubMed] [Google Scholar]
- 43.Mukisa IM, Byaruhanga YB, Muyanja C, Langsrud T, Narvhus JA. 2017. Production of organic flavor compounds by dominant lactic acid bacteria and yeasts from Obushera, a traditional sorghum malt fermented beverage. Food Sci Nutr 5:702–712. doi: 10.1002/fsn3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Wang F, Zhang X, Bai X, He H. 2019. Optimization of the mixed cultivation conditions for the Lactobacillus helveticus strain and Lactobacillus rhamnosus strain and the preparation of copprobiotic. China Dairy Ind 47:19–30. (In Chinese.) [Google Scholar]
- 45.Liu J, Chen J, Fan Y, Huang X, Han B. 2018. Biochemical characterisation and dominance of different hydrolases in different types of Daqu—a Chinese industrial fermentation starter. J Sci Food Agric 98:113–121. doi: 10.1002/jsfa.8445. [DOI] [PubMed] [Google Scholar]
- 46.Kum S, Yang S, Lee SM, Chang P, Choi YH, Lee JJ, Hurh BS, Kim Y. 2015. Effects of Aspergillus species inoculation and their enzymatic activities on the formation of volatile components in fermented soybean paste (doenjang). J Agric Food Chem 63:1401–1418. doi: 10.1021/jf5056002. [DOI] [PubMed] [Google Scholar]
- 47.Li P, Liang H, Lin W, Feng F, Luo L. 2015. Microbiota dynamics associated with environmental conditions and potential roles of cellulolytic communities in traditional Chinese cereal starter solid-state fermentation. Appl Environ Microbiol 81:5144–5156. doi: 10.1128/AEM.01325-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peyer LC, Zannini E, Arendt EK. 2016. Lactic acid bacteria as sensory biomodulators for fermented cereal-based beverages. Trends Food Sci Technol 54:17–25. doi: 10.1016/j.tifs.2016.05.009. [DOI] [Google Scholar]
- 49.Hammes WP, Stolz P, Gaenzle M. 1996. Metabolism of lactobacilli in traditional sourdoughs. Adv Food Sci 18:176–184. [Google Scholar]
- 50.Rathore RS, Garg N, Garg S, Kumar A. 2009. Starch phosphorylase: role in starch metabolism and biotechnological applications. Crit Rev Biotechnol 29:214–224. doi: 10.1080/07388550902926063. [DOI] [PubMed] [Google Scholar]
- 51.Stolz P, Hammes WP, Vogel RF. 1996. Maltose-phosphorylase and hexokinase activity in lactobacilli from traditionally prepared sourdoughs. Adv Food Sci 18:1–6. [Google Scholar]
- 52.Wu Q, Kong Y, Xu Y. 2016. Flavor profile of Chinese liquor is altered by interactions of intrinsic and extrinsic microbes. Appl Environ Microbiol 82:422–430. doi: 10.1128/AEM.02518-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Bai Z, Wang D, Zhang W, Zhang M, Lin F, Gao L, Hui B, Zhang H. 2014. Cultivable bacterial diversity and amylase production in three typical Daqus of Chinese spirits. Int J Food Sci Technol 49:776–786. doi: 10.1111/ijfs.12365. [DOI] [Google Scholar]
- 54.Li H, Lian B, Ding Y, Nie C, Zhang Q. 2014. Bacterial diversity in the central black component of Maotai Daqu and its flavor analysis. Ann Microbiol 64:1659–1669. doi: 10.1007/s13213-014-0809-z. [DOI] [Google Scholar]
- 55.Zhao M, Zhang D, Su X, Duan S, Wan J, Yuan W, Liu B, Ma Y, Pan Y. 2015. An integrated metagenomics/metaproteomics investigation of the microbial communities and enzymes in solid-state fermentation of Pu-erh tea. Sci Rep 5:10117. doi: 10.1038/srep10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tyanova S, Temu T, Cox J. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 57.Cheow ESH, Cheng WC, Lee CN, Kleijn D, Sorokin V, Sze SK. 2016. Plasma-derived extracellular vesicles contain predictive biomarkers and potential therapeutic targets for myocardial ischemic (MI) injury. Mol Cell Proteomics 15:2628–2640. doi: 10.1074/mcp.M115.055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bergauer K, Fernandez-Guerra A, Garcia JAL, Sprenger RR, Stepanauskas R, Pachiadaki MG, Jensen ON, Herndl GJ. 2018. Organic matter processing by microbial communities throughout the Atlantic water column as revealed by metaproteomics. Proc Natl Acad Sci U S A 115:E400–E408. doi: 10.1073/pnas.1708779115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soergel DAW, Dey N, Knight R, Brenner SE. 2012. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J 6:1440–1444. doi: 10.1038/ismej.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buee M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F. 2009. 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol 184:449–456. doi: 10.1111/j.1469-8137.2009.03003.x. [DOI] [PubMed] [Google Scholar]
- 61.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 64.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Hoiland K, Kjoller R, Larsson E, Pennanen T, Sen R, Taylor AF, Tedersoo L, Vralstad T, Ursing BM. 2005. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol 166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- 66.Tao Y, Li J, Rui J, Xu Z, Zhou Y, Hu X, Wang X, Liu M, Li D, Li X. 2014. Prokaryotic communities in pit mud from different-aged cellars used for the production of Chinese strong-flavored liquor. Appl Environ Microbiol 80:2254–2260. doi: 10.1128/AEM.04070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu Q, Chen L, Xu Y. 2013. Yeast community associated with the solid state fermentation of traditional Chinese Maotai-flavor liquor. Int J Food Microbiol 166:323–330. doi: 10.1016/j.ijfoodmicro.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 68.Liu J, Wu Q, Wang P, Lin J, Huang L, Xu Y. 2017. Synergistic effect in core microbiota associated with sulfur metabolism in spontaneous Chinese liquor fermentation. Appl Environ Microbiol 24:e01475-17. doi: 10.1128/AEM.01475-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All generated protein data were submitted to the iProX database (http://www.iprox.org) under the project number IPX0001641000/PXD014221. All sequences generated were submitted to the DDBJ database under the accession numbers DRA008485 and DRA008486. The genomic sequences of the isolated Rhizopus microsporus JJ01, Saccharomyces cerevisiae JJ01, Lactobacillus fermentum JJ01, and Lactobacillus helveticus JJ02 strains were deposited in GenBank under the project numbers MH782032, MK994013, MN841929, and MN841930.