Primitive forms of life may have originated around hydrothermal vents at the bottom of the ancient ocean. The Lost City hydrothermal vent field, fueled by just rock and water, provides an analog for not only primitive ecosystems but also potential extraterrestrial rock-powered ecosystems. The microscopic life covering the towering chimney structures at the Lost City has been previously documented, yet little is known about the carbon cycling in this ecosystem. These results provide a better understanding of how carbon from the deep subsurface can fuel rich microbial ecosystems on the seafloor.
KEYWORDS: Chloroflexi, hydrothermal vents, metagenomics, methanogens, serpentinization
ABSTRACT
The Lost City hydrothermal field on the Mid-Atlantic Ridge supports dense microbial life on the lofty calcium carbonate chimney structures. The vent field is fueled by chemical reactions between the ultramafic rock under the chimneys and ambient seawater. These serpentinization reactions provide reducing power (as hydrogen gas) and organic compounds that can serve as microbial food; the most abundant of these are methane and formate. Previous studies have characterized the interior of the chimneys as a single-species biofilm inhabited by the Lost City Methanosarcinales, but they also indicated that this methanogen is unable to metabolize formate. The new metagenomic results presented here indicate that carbon cycling in these Lost City chimney biofilms could depend on the metabolism of formate by Chloroflexi populations. Additionally, we present evidence for metabolically diverse, formate-utilizing Sulfurovum populations and new genomic and phylogenetic insights into the unique Lost City Methanosarcinales.
IMPORTANCE Primitive forms of life may have originated around hydrothermal vents at the bottom of the ancient ocean. The Lost City hydrothermal vent field, fueled by just rock and water, provides an analog for not only primitive ecosystems but also potential extraterrestrial rock-powered ecosystems. The microscopic life covering the towering chimney structures at the Lost City has been previously documented, yet little is known about the carbon cycling in this ecosystem. These results provide a better understanding of how carbon from the deep subsurface can fuel rich microbial ecosystems on the seafloor.
INTRODUCTION
The towering carbonate chimneys of the Lost City hydrothermal field protrude from the Atlantis Massif, a dome of ultramafic rock uplifted from the mantle. These chimneys differ from other deep-sea hydrothermal systems because they are driven primarily by rock-water reactions, known as serpentinization, rather than magmatic activity. The serpentinization reactions create high-pH fluids that mix with the surrounding cold seawater to form the calcium carbonate structures. Serpentinite-hosted systems are of astrobiological interest because they provide a source of energy for life that does not require sunlight or vigorous magmatic activity (1). These systems are thought to be present on icy worlds, such as Jupiter’s moon Europa and Saturn’s moon Enceladus (2, 3).
The dense microbial biofilms of Lost City chimneys are fueled by the carbon and energy released by serpentinization of the underlying ultramafic rock (4–8). The serpentinization reactions provide high concentrations of hydrogen gas, methane, and other simple organic compounds that serve as food and energy sources for microbes. The more extreme interiors of chimneys are anoxic and continually bathed in the warm serpentinizing fluids. The temperatures of venting fluids can reach >95°C, and the pH of the fluids can be as high as 11 (9). Previous studies have shown that these interiors are dominated by a single archaeal phylotype, the Lost City Methanosarcinales (5). In contrast, the chimney exteriors host a more complex microbial community, including organisms involved in the oxidation of sulfur and methane (e.g., Methylomonas, Thiomicrospira). These organisms likely thrive in the mixing zones, where they can take advantage of the cooling effect of the seawater and more efficient electron acceptors (e.g., oxygen) but still access the products of serpentinization supplied by venting fluids.
In general, little is known about the metabolic capabilities of Lost City organisms. Our previous research has shown that much of the microbial biomass at Lost City is derived from carbon that originated deep in the Earth’s subsurface (4, 10). In most ecosystems, inorganic carbon (CO2) serves as the starting carbon source for primary production. However, the Lost City fluids contain extremely low concentrations of dissolved inorganic carbon (DIC) due to its reduction to hydrocarbons and its rapid precipitation as calcium carbonate at a pH above ∼9 (11–13). The organic acid formate has been proposed to be an alternative primary carbon source; it is present in high concentrations in Lost City fluids (36 to 158 μM) and is expected to form abiotically in serpentinizing fluids (4, 14, 15). In support of this, our previous experiments have shown that the isotopic compositions of carbon (13C and 14C) in bacterial and archaeal lipids resemble those of formate from the vent fluids (4).
Formate is unable to enter carbon fixation pathways directly and needs to be converted to CO2 for autotrophic metabolism (16–19). The enzyme formate dehydrogenase catalyzes the reversible oxidation of formate to CO2. Obtaining formate from the environment requires active transport; therefore, any Lost City formate-utilizing species would carry genes encoding a formate transporter and formate dehydrogenase. This study identifies two formate-utilizing populations of the Lost City chimneys based on metagenomic evidence, including the presence of formate transporters and formate dehydrogenases. These formate-utilizing organisms may enable mantle-derived carbon to become available to the other microbial inhabitants of Lost City chimneys that are unable to use formate, such as the Methanosarcinales.
RESULTS AND DISCUSSION
Metagenomic assembly and binning.
We performed shotgun paired-end sequencing of environmental DNA extracted from a sample of chimney material collected at Marker 5 within the Lost City hydrothermal field. The metagenome consisted of 145,937,844 read pairs (after quality filtering), which were assembled into 730,351 contigs with an N50 of 2,518 bp and a maximum length of 250,900 bp. The assembled contigs represent 62.47% of all read pairs in the metagenome. Each contig of >1,000 bp was assigned a taxonomy by the PhyloPythiaS+ program (20), and the overall taxonomic composition of these contigs is shown in Fig. S1 in the supplemental material. These results are consistent with those of previous studies that have described the chimney biofilm communities as being dominated by Gammaproteobacteria, including Thiotrichales and Methylococcales (6, 7, 21).
Contigs were binned into metagenome-assembled genomes (MAGs), only seven of which initially contained <10% contamination and >18% completeness (Table S1) after automated binning. None of these seven MAGs contained strong evidence for formate utilization. Therefore, we manually explored the other MAGs with evidence of formate metabolism.
Two types of formate transporters have been characterized. In formate-utilizing methanogens, the gene fdhC is thought to be necessary for the transport of formate into the cell (22). A different formate/nitrite transporter (focA) from the same FNT protein family has been described in Escherichia coli (23). E. coli requires focA for removal of the formate produced during mixed-acid fermentations, but the protein is known to be bidirectional and can therefore bring formate or nitrite into the cell (24). In order to identify potential formate-using populations in our metagenomes, we examined all bins containing fdhC or focA. We identified genes for five formate transporters in the metagenome, three of which were found on contigs with evidence of nitrite metabolism, but no other genes involved in formate metabolism. The genes for the other two formate transporters were located on contigs with adjacent genes involved in formate metabolism. Therefore, we manually refined these two MAGs, as well as a third representing the Lost City Methanosarcinales phylotype (8, 12).
Sulfurovum.
The Sulfurovum MAG was refined to be 95.9% complete and to have 2.19% contamination by examining the hierarchical clustering of contigs, as visualized in the anvi’o platform, and by inspecting the taxonomic assignment (by the PhyloPythiaS+ method) of each contig. The fragments mapped to this MAG comprised 0.41% of the total assembly coverage (Table S2). Of the three MAGs discussed here, the Sulfurovum MAG contains the lowest number of protein-encoding genes (2,036), but 90% of these were annotated with a functional prediction. This MAG also has the highest number of complete KEGG modules (25) (Data Set S1).
The Sulfurovum MAG includes formate dehydrogenase and formate transporter genes, in addition to genes for a complete KEGG pathway for selenocompound metabolism (ko00450), responsible for synthesizing l-selenocysteinyl-tRNA. The selenocysteine residue is a key feature of the catalytic subunit in formate dehydrogenase (FdhA) and is thought to be directly involved in proton transfer from formate (26).
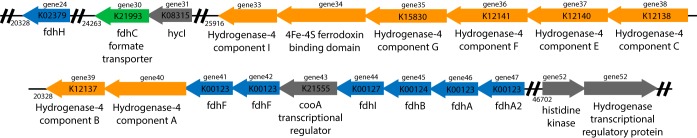
The contig containing the formate transporter gene (fdhC) contains multiple formate hydrogenlyase (FHL; hydrogenase-4 [Hyd-4] form) genes starting 1,653 bp away (Fig. 1). Interestingly, the formate hydrogenlyase on this contig is homologous to the Hyd-4 form, which is unaffected by alkaline pH (27). In E. coli, the bidirectional FHL complex links formate oxidation to proton reduction and is operational during mixed-acid fermentations (28). During these fermentations in E. coli, formate is formed by the pyruvate formate-lyase enzyme and transported outside of the cell (29). It is unlikely that the FHL complex is involved in mixed-acid fermentation by Sulfurovum because the MAG did not contain genes for a pyruvate formate-lyase. Therefore, FdhC and the FHL complex most likely bring in formate from the environment and carry out the membrane-bound conversion of formate to CO2, which can then enter a carbon fixation pathway.
FIG 1.
The formate transporter (fdhC) contig in the Lost City Sulfurovum MAG. Relevant genes are reported here in the order that they are found on the contig. Gene order is indicated above the arrows that indicate the forward or reverse direction of transcription. Double lines indicate where genes are omitted from this visual representation of the contig. Orange genes are involved in the formate hydrogenlyase (FHL) complex; blue genes (fdhH, fdhF, fdhI, fdhB, fdhA, fdhA2) indicate formate dehydrogenase genes that may interact with the FHL complex; the green gene (fdhC) is involved in formate transport. All other genes, including regulatory genes, are reported in gray. KEGG identification numbers are indicated on arrows where appropriate. hycI, hydrogenase 3 maturation protease; cooA, CRP/FNR family CO-sensing transcription factor.
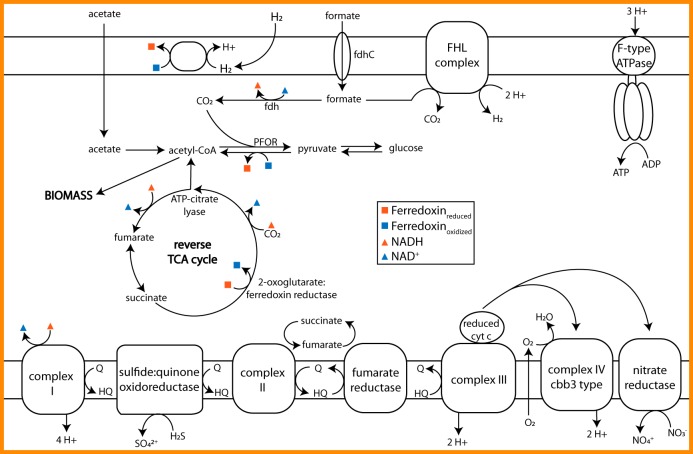
Key genes for carbon fixation via the reductive tricarboxylic acid (TCA) cycle were found in the Sulfurovum MAG: genes for pyruvate synthase (PFOR; heterodimer type), ATP citrate lyase, 2-oxoglutarate reductase, and fumarate reductase (Fig. 2). In addition, genes for gluconeogenesis are present, suggesting that fixed carbon can be stored as glucose. It is unclear if Sulfurovum can run the TCA cycle in the forward direction for heterotrophic growth. Succinate dehydrogenase genes are present in the MAG, but there was no evidence for citrate synthase. Desulfobacter hydrogenophilus is known to use ATP citrate lyase (instead of citrate synthase) in both the forward and the reverse directions, and this may be possible for this Sulfurovum population as well (30). Alternatively, instead of using a bidirectional TCA cycle to break down glucose reserves, Sulfurovum may ferment it into lactate; indeed, the lactate dehydrogenase gene is present in this MAG.
FIG 2.
Overview of the Lost City Sulfurovum central carbon metabolism pathways and electron transport chain. Abbreviations: fdh, formate dehydrogenase; fdhC, formate transporter; FHL, formate hydrogenlyase complex; HQ, hydroquinone; Q, quinone; cty c, cytochrome c; PFOR, pyruvate:ferredoxin oxidoreductase.
The Sulfurovum MAG contains genes for a complex electron transport chain, suggesting a metabolically diverse lifestyle (Fig. 2). Complex I (NADH dehydrogenase) likely serves as a versatile entry point for many catabolic reactions. We also found a sulfide:quinone oxidoreductase (SQOR), indicating that Sulfurovum might use sulfide as an electron donor. Evidence for an electron transport chain in this MAG includes the presence of genes for three terminal electron acceptors: fumarate reductase, cytochrome c oxidase (complex IV), and nitrate reductase.
This MAG also contains genes for a number of cofactor ABC transporters, including those for tungstate (required for formate dehydrogenase activity), molybdate, iron, thiamine, and zinc (26). We also found evidence of genes for transporters for macronutrients, such as l-amino acids, branched-chain amino acids, phospholipids, and phosphate.
In addition to these nutrient-acquiring transporters, the MAG contains the gene for a transporter responsible for excreting capsular polysaccharides. After intracellular construction, these molecules are exported to form a capsule around the cell which is involved in both biofilm formation and environmental stress protection (31). We also found a lipopolysaccharide export system, indicating that the Sulfurovum population at Lost City builds an outer membrane like other characterized Sulfurovum species (32).
Sulfurovum lithotrophicum, the type species for the genus, was first isolated from hydrothermal sediments off the coast of Okinawa, Japan (33). The genome for this species closely resembles our Lost City Sulfurovum MAG. It contains genes for a reverse TCA cycle and a sulfide:quinone oxidoreductase (SQOR) and can use O2 or NO3− as an electron acceptor (34). However, unlike our Sulfurovum MAG, this and most characterized Sulfurovum species are also able to oxidize sulfur (S0) or thiosulfate through the sulfur oxidation (SOX) system (35–37). Both Sulfurovum aggregans and Sulfurovum lithotrophicum have been shown to use hydrogen, but not formate, as an electron donor (36, 38).
The lack of genes for a SOX system and the presence of formate-metabolizing genes in the Lost City Sulfurovum MAG are novel for the genus, although it is possible that SOX genes were missed during binning. The genomic capability to scavenge amino acids, form biofilms, and retain genetic flexibility for multiple electron acceptors supports the possibility that this organism has a mixotrophic lifestyle capable of adapting to a fluctuating environment with varying ratios of seawater and hydrothermal fluids. These results suggest that the Sulfurovum population is adapted to a transition zone between the interior and the exterior of the chimneys.
Chloroflexi.
After manual refining, the Chloroflexi MAG is estimated to be 70% complete with 4.73% contamination. The mapped fragments made up 0.62% of the metagenomic assembly (Table S2). Of the three MAGs discussed here, this MAG contains the highest number of protein-encoding genes (3,936), but only 84% of these were annotated with functional predictions. This bin also has the highest number of incomplete KEGG modules (39) (Data Set 1).
The Chloroflexi MAG contains a focA formate transporter adjacent to a formate dehydrogenase alpha subunit (fdhA) gene and three genes encoding the catalytic subunit of NAD(H) dehydrogenase (nuoG, hoxF, hoxE) (Fig. 3). The beta subunit (fdhB) was located together with fdhA on a different contig. As with the Sulfurovum MAG, the Chloroflexi MAG contains genes for a complete KEGG pathway for selenocompound metabolism, responsible for synthesizing l-selenocysteinyl-tRNA.
FIG 3.
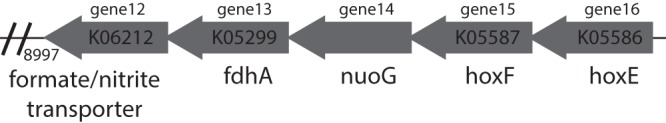
The formate transporter (focA) contig found in the Lost City Chloroflexi MAG. Note the adjacent gene encoding the alpha subunit of formate dehydrogenase (fdhA). Double lines indicate where genes are omitted from this visual representation of the contig. KEGG identification numbers are indicated on arrows where appropriate. nuoG, NADH-quinone oxidoreductase subunit G; hoxF and hoxE, bidirectional (NiFe) hydrogenase diaphorase subunit.
The Chloroflexi MAG contains genes for transporters for tungstate (required for formate dehydrogenase activity), iron, and thiamine. As with Sulfurovum, the Chloroflexi populations might scavenge macronutrients, as the MAG contains genes for the following transporters: l-amino acids, branched-chain amino acids, phospholipids, and phosphate. Because the MAG is estimated to be only 70% complete, it is likely that it contains genes for additional transporters not identified in this study.
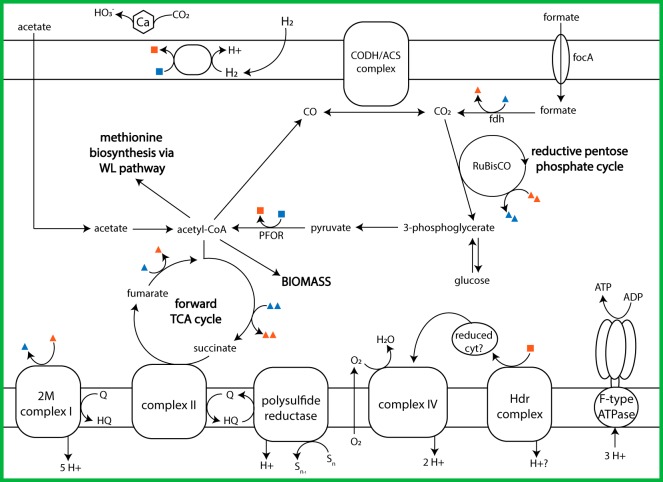
In addition to formate-utilizing genes, the Chloroflexi MAG contains genes for a nearly complete reductive pentose phosphate cycle (Fig. 4). The MAG also includes genes for a carboxysome-specific carbonic anhydrase, suggesting that this organism uses a carboxysome to concentrate CO2 around ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (40). Carboxysomes are used by many organisms when the concentration of CO2 outside the cell is lower than the Km of RuBisCO (39). If this Chloroflexi couples the carboxysome shell with the conversion of formate to CO2, it could be an effective adaptation to the lack of CO2 in the chimney environment.
FIG 4.
Overview of the Lost City Chloroflexi central carbon metabolism pathways and electron transport chain. The definitions of symbols are the same as those provided in the key to Fig. 2. Abbreviations: RuBisCO, ribulose-1,5-bisphosphate carboxylase/oxygenase; Hdr, heterodisulfide reductase; fdh, formate dehydrogenase; CODH/ACS, carbon monoxide dehydrogenase/acetyl-CoA synthase; focA, formate/nitrite transporter; WL pathway, Wood-Ljungdahl pathway; Ca, carbonic anhydrase; cyt, cytochrome; PFOR, pyruvate:ferredoxin oxidoreductase; HQ, hydroquinone; Q, quinone.
The one enzyme missing from the reductive pentose phosphate cycle in this MAG is glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which is also involved in glycolysis. Interestingly, thermophilic organisms use a distinct form of GAPDH to cope with the heat instability of glyceraldehyde-3-phosphate (30, 41), so it is possible that the Chloroflexi MAG contains the gene for an as yet unidentified variant of GAPDH. Another adaptation of thermophilic growth is in the structure of fructose-bisphosphate aldolase, an enzyme involved in gluconeogenesis (41). The gene for this enzyme is also included in the Chloroflexi MAG and is most closely related to that of the thermophile Caldilinea aerophila.
The Chloroflexi MAG also contains the genes for an incomplete Wood-Ljungdahl pathway. Typically, the presence of genes for this pathway in a bacterial genome indicates that it is involved in carbon fixation during acetogenesis, but the lack of genes for two key enzymes in this MAG casts doubt on that scenario. One of these missing enzymes (methylene-tetrahydrofolate reductase [MTHFR]) is essential for acetogenesis. The second missing enzyme is acetate kinase, which is involved in the last step of acetate formation. While the absence of genes in an incomplete MAG must be interpreted with caution, a similar partial Wood-Ljungdahl pathway has been described in the dehalogenating Chloroflexi Dehalococcoides mccartyi (42). Although D. mccartyi is missing MTHFR, the species is capable of de novo methionine biosynthesis through the partial Wood-Ljungdahl pathway via the cleavage of acetyl coenzyme A (CoA) synthetase (ACS). As the Lost City Chloroflexi MAG contains no genes for other pathways for methionine biosynthesis, this organism, like D. mccartyi, may use the incomplete Wood-Ljungdahl pathway for methionine biosynthesis rather than for carbon fixation.
The Chloroflexi MAG has genes for both glycolysis/gluconeogenesis pathways and a forward TCA cycle. This suggests that the organism could store carbon fixed through the reductive pentose phosphate pathway as glucose reserves and grow heterotrophically when carbon is limited in the environment. Additional evidence for a flexible mixotrophic lifestyle for this MAG includes genes for three carbohydrate transporters (multiple sugar, ribose, and d-xylose transport systems), suggesting that this Chloroflexi population is capable of metabolizing additional complex carbon sources.
Genomic evidence points to a modified structure of the NADH:quinone oxidoreductase (complex I) in this Chloroflexi population. An additional NuoM subunit is responsible for translocating an extra proton per reaction cycle in these modified complexes (43). The operon arrangement for these 2M complexes is unique to cultured Chloroflexi species (43). In particular, these species encode an additional nuoM2, located between the nuoL and nuoM1 genes (as in nuoLM1M2N), and nuoBCDI are separated from the operon with a fused nuoCD. These genomic features are consistent with two contigs in our Chloroflexi MAG. The nuoM sequence, which forms a clade with the modified nuoM2 gene of Anaerolinea thermophila (Fig. S2), is surrounded by three nuo genes: nuoN; the nuoM sequence, which forms a clade with the nuoM1 gene of Anaerolinea thermophila; and nuoL (as in nuoLM1M2N). The nuoBCDI genes are found on a separate contig in the MAG. The increased proton-pumping ability for the modified 2M complex I has been proposed to be beneficial for energy conservation in alkaliphilic environments or slow-growing organisms (43).
The other membrane-bound complexes encoded by the Chloroflexi MAG are also consistent with an anaerobic, mixotrophic lifestyle (Fig. 4). Additional energy conservation appears to be mediated by a modified NADH:quinone oxidoreductase (2M complex I), succinate dehydrogenase (complex II), polysulfide reductase, cytochrome c oxidase (complex IV), heterodisulfide reductase, and an F-type ATPase typical of bacteria. Electrons could be donated by formate or carbohydrates, but the terminal electron acceptor is unclear.
The MAG has the gene for cytochrome c oxidase (complex IV), which would indicate oxygen as the terminal electron acceptor, but the lack of the gene for cytochrome oxidoreductase, such as a cytochrome bc or b6f complex (complex III), is perplexing. The genome of anaerobic Sulfurimonas gotlandica strain GD1 contains the gene encoding cytochrome c oxidase, but the enzyme’s suspected function is to occasionally remove inhibitory oxygen rather than to serve as a terminal electron acceptor (44). Furthermore, the Chloroflexi genus Anaerolineae is described to be obligately anaerobic, yet many species contain genes for aerobic respiration in their genomes (45). Due to the presence of genes encoding numerous oxygen-sensitive enzymes (aldehyde ferredoxin oxidoreductase, the carbon monoxide dehydrogenase [CODH]/ACS complex, anaerobic forms of glycerol-3-phosphate dehydrogenase, and sulfatase maturase) in the Chloroflexi MAG, it is likely that the cytochrome c oxidase’s role is to remove oxygen rather than to serve in the last step of an aerobic respiratory chain. Alternatively, complex III genes could be missing simply due to the incomplete nature of the MAG.
If complex III is missing, heterodisulfide reductase (Hdr) may be involved in electron transfer from hydrogen (via ferredoxin reduced by hydrogenases) to cytochromes, as proposed for other species (46, 47). This complex could work in tandem with cytochrome c oxidase to remove intracellular oxygen. If oxygen is not the terminal electron acceptor, then the presence of polysulfide reductase (Psr) indicates that polysulfide compounds are the most likely terminal electron acceptors. The Chloroflexi polysulfide reductase sequence has 40% identity with the Psr sequence from Thermus thermophilus, which has been shown to use polysulfide as its terminal electron acceptor (48).
Considering the presence of multiple genes encoding oxygen-sensitive enzymes and similarities to the genomes of anaerobic organisms, we propose that the Chloroflexi population is an anaerobic population adapted to the interiors of Lost City chimneys, where it would have access to abundant formate. By building biomass from formate, Chloroflexi populations would convert mantle-derived carbon into organic matter that could subsequently be utilized by other members of the community.
Methanosarcinales.
The manually refined Methanosarcinales MAG was estimated to be 84.87% complete with 5.26% contamination. The mapped fragments made up 4.41% of the entire assembly (Table S2). The MAG contained 2,324 protein-encoding genes, only 77% of which could be assigned a predicted function. The Methanosarcinales MAG has 33 complete and 28 incomplete KEGG modules (Data Set S1).
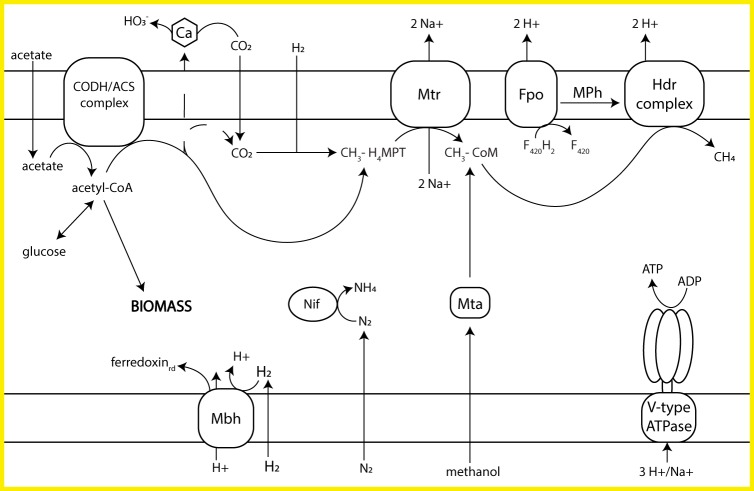
We identified this MAG as the previously described Lost City Methanosarcinales phylotype due to the taxonomic assignment of Methanosarcinales for all contigs and the presence of nitrogenase reductase (nifH, nitrogen fixation) and methyl coenzyme M reductase (mcrA, methanogenesis) gene sequences that matched those of previously sequenced genes (8, 12) (Tables S3 and S4). In agreement with our previous analysis with a smaller data set, we found no evidence that the Methanosarcinales population is able to utilize formate as a carbon source (4). Although novel, previously undiscovered formate metabolism genes may exist, no archaeal formate dehydrogenase gene sequences (fdhA, fdhB) or fdhC sequences affiliated with methanogens were detected in the metagenome. The MAG does contain a genomic inventory that would allow the Methanosarcinales to utilize CO2, acetate, and methanol for methanogenesis (Fig. 5). The genes for transporters for tungstate and molybdate were also identified; these metals are cofactors required for many of the enzymes in the methanogenic pathway.
FIG 5.
Overview of the Lost City Methanosarcinales (LCMS) methanogenic pathway and membrane-bound complexes involved in energy conservation. Abbreviations: CODH/ACS, carbon monoxide dehydrogenase/acetyl-CoA synthase; Mbh, membrane-bound hydrogenase; Nif, nitrogenase; Mtr, methyl-H4MPT, coenzyme M methyltransferase; Mta, methanol:5-hydroxybenzimidazolylcobamide methyltransferase; Fpo, F420H2 dehydrogenase; MPh, methanophenazine; Hdr, heterodisulfide reductase; ferredoxinrd, reduced ferredoxin.
The Methanosarcinales MAG provides new information on the phylogenetic status of this species. Previous studies classified it within the order Methanosarcinales (5), but it has never been maintained as a cultivated isolate, despite many attempts, and a more specific phylogenetic classification has never been attempted. Its closest relatives have been previously reported to include members of the Methanosarcinaceae and Methanosaetaceae families (5, 7). These two families include the only species known to be capable of methane production from acetate (49). Each group of methanogens has distinct mechanisms for acetate activation and energy conservation. Acetate activation in Methanosarcina proceeds via two enzymes: acetate kinase and phosphotransacetylase. Methanosaeta species, in contrast, use acetyl-CoA synthetase (ACS) for acetate activation. The Lost City Methanosarcinales MAG includes genes for both ACS and acetate kinase (but not the gene for phosphotransacetylase, perhaps due to the incompleteness of the MAG), suggesting that it may be able to use both systems or a hybrid system.
The energy conservation strategy of Lost City Methanosarcinales appears to be more similar to that of Methanosaeta than to that of Methanosarcina. As in Methanosaeta, the MAG contains no genes for Ech hydrogenase, the Rnf complex, or the methanophenazine-reducing hydrogenase (Vho). Instead, the only gene for an energy-conserving complex external to the methanogenesis pathway present in the Methanosarcinales MAG is that for F420H2 dehydrogenase, which is also employed by Methanosaeta (49).
Phylogenetic analyses of methanogenesis genes from this MAG suggest that it is distinct from both Methanosaeta and Methanosarcina, perhaps forming a novel family within the order Methanosarcinales (Fig. S3). In two of the gene trees, those for formylmethanofuran-tetrahydromethanopterin formyltransferase and F420-dependent N5,N10-methylene- tetrahydromethanopterin reductase (ftr and mer), the Lost City gene was monophyletic with methylotrophic methanogens, such as Methanococcoides burtonii. A third phylogeny, F420-dependent N5,N10-methylene-tetrahydromethanopterin dehydrogenase (mtd), grouped the Lost City gene with hydrogenotrophic and formate-utilizing Methanosarcinales, such as Methanothermococcus okinawensis. In a fourth phylogeny, N5,N10-methenyl-tetrahydromethanopterin cyclohydrolase (mch), the Lost City gene was distinct from all other known species.
No genomes of Methanosaeta contain genes for membrane-bound hydrogenases that would allow H2 to serve as the electron donor (50–52). However, the Lost City MAG contains five genes annotated as ferredoxin-dependent membrane-bound hydrogenase (Mbh). This complex is known to translocate protons with the formation or cleavage of hydrogen gas, similar to the Ech hydrogenase found in many hydrogenotrophic methanogens (53). Therefore, the Lost City Methanosarcinales may be able to use this enzyme for hydrogenotrophic methanogenesis. For hydrogenotrophic growth, the Lost City Methanosarcinales would have to obtain CO2 from the chimney’s carbonate minerals (for which there is no known mechanism) or from another member of the community, such as the Sulfurovum or Chloroflexi populations described above.
The Methanosarcinales MAG contains the nifH, nifD, and nifK genes, encoding the nitrogenase complex involved in nitrogen fixation. Previous work found low δ15N values of Lost City chimneys, indicative of biological nitrogen fixation (25). The concentrations of biologically available nitrogen are relatively low (<6 μM) in Lost City fluids, but the concentrations of N2 resemble those in seawater (25). Therefore, the densely populated biofilm communities of Lost City chimneys must be reliant on nitrogen fixation, probably carried out at least in part by Methanosarcinales populations.
Conclusion.
The biofilms growing on Lost City chimneys are unique ecosystems where microbes must face the challenges of multiple extremes, including pHs of >10 and temperatures of up to at least 95°C. Our previous work demonstrated that the microbial communities inhabiting the chimneys are fueled by carbon venting from Earth’s mantle (4). The present study identifies the genomic potential of the Chloroflexi and Sulfurovum populations to utilize formate, which may be required to make mantle-derived carbon available to the rest of the chimney ecosystem.
The Lost City biofilms that inhabit the anoxic interiors of the chimneys have been described as containing a single species, the Lost City Methanosarcinales phylotype (4–8, 12). The single Methanosarcinales MAG reported here represents 4.41% of the chimney metagenome, >5 times more abundant than the other MAGs reported here. The Methanosarcinales phylotype has previously been shown to dominate the anoxic, interior zones of Lost City chimneys (5, 6, 8), yet it appears to be unable to use one of the most abundant carbon sources, formate. The Lost City Chloroflexi MAG, in contrast, contains the genes required for using formate and may be able to provide biologically available carbon to Methanosarcinales and other members of the biofilm community. This is an apparent conundrum, as the Chloroflexi MAG is seven times less abundant than the Methanosarcinales MAG in the chimney sample described here. One potential explanation is that Chloroflexi species are highly active and able to rapidly cycle carbon while maintaining a low abundance in the biofilm community. Alternatively, the Chloroflexi population may be more abundant in subsurface habitats underlying the chimneys, where formate is expected to be generated (4). Future research should test these hypotheses by experimentally investigating how microbial activity in subsurface environments can influence the food and energy available to the biofilm communities of the chimneys.
MATERIALS AND METHODS
Sample collection.
Sample H08_080105_Bio5slurpB1 from Marker 5 was collected in 2005 during a National Oceanic and Atmospheric Administration (NOAA) Ocean Explorer cruise with the ROV Hercules aboard the R/V Ronald H. Brown. The sample was immediately placed in a sterile Whirl-Pak sample bag upon arrival on deck and stored at −80°C until analysis. DNA was extracted from the samples according to a previously published protocol (6). A previous metagenomic analysis of this sample has been published (4), but the results presented here are from a different DNA extraction and a much deeper metagenomic sequencing effort. All laboratory and bioinformatic protocols are available at https://doi.org/10.5281/zenodo.3629892. Environmental DNA was extracted with a protocol modified from that described previously (5, 7, 54). The chimney sample was thawed and homogenized with a sterile mortar and pestle at room temperature, and 0.25-g subsamples were placed in a DNA extraction buffer containing 0.1 M Tris, 0.1 M Na-EDTA, 0.1 M KH2PO4, 1.5 M NaCl, 0.8 M guanidium HCl, and 0.5% Triton X-100. For lysis, samples were subjected to one freeze-thaw cycle, incubation at 65°C for 30 min, and beating with 0.1-mm glass beads in a Mini-BeadBeater 16 (Biospec Products). Purification was performed via extraction with phenol-chloroform-isoamyl alcohol, precipitation in 3 M sodium acetate and ethanol, washing in Amicon 30K Ultra centrifugal filters, and a final cleanup with 2× SPRI beads (55). The DNA concentration after purification was approximately 10 ng/μl, as measured with a Qubit fluorometer (Thermo Fisher), with an A260/A230 ratio of 1.6 and an A260/A280 ratio of 1.8, as measured with a NanoDrop spectrophotometer (Thermo Fisher).
Metagenome sequencing.
A Qsonica Q800R sonicator was used to fragment the metagenomic DNA to ∼500 to 700 bp. A metagenome library was constructed with 500 ng of fragmented DNA using a NEBNext Ultra DNA library preparation kit for Illumina according to the manufacturer’s instructions. Quality control and sequencing of the metagenomic libraries were conducted at the University of Utah High-Throughput Genomics Core Facility. Libraries were evaluated for quality on a Bioanalyzer DNA 1000 chip (Agilent Technologies), and then paired-end sequencing (2 × 125 bp) was performed on an Illumina HiSeq2500 platform with HiSeq (v4) chemistry. The library was multiplexed with one other library (from a second Lost City chimney sample, results from which are not reported here) on one Illumina lane, yielding 180 million read pairs (45 billion bases). Demultiplexing and conversion of the raw sequencing base-call data were performed through the CASAVA (v1.8) pipeline.
Metagenomic analysis.
Raw sequence data were processed by the W. J. Brazelton lab to trim adapter sequences with BBDuk (part of the BBTools suite, v35.85 [56]), to remove artificial replicates, and to trim the reads based on quality. Removal of replicates and quality trimming were performed with our seq-qc package (https://github.com/Brazelton-Lab/seq-qc). Paired-end reads were assembled with the MegaHit (v1.1.1) program, using kmers of 27 to 141. The Prodigal (v2.6.3) program was run in the anonymous gene prediction mode to identify open reading frames. Functional annotation was performed using the blastp function of the Diamond (v0.9.14) program with both the prokaryotes and T10000 (addendum annotations) databases from KEGG (release 83.2) with an E value of 1e−6. Annotations were selected by the highest-quality alignment, as determined by the bit score. Binning was performed with the ABAWACA (v1.00) program (https://github.com/CK7/abawaca). Contig taxonomy was assigned with PhyloPythiaS+ (v1.4) software (20). Curation of bins was performed in the anvi’o (v4) platform, using the default visualization of bins informed by tetranucleotide frequency as well as manual inspection of the PhyloPythiaS+ taxonomic assignment. The CheckM (v1.0.5) program was used to assess bin quality (57). Completion of the KEGG modules and pathways was determined using the KEGG Mapper online tool (https://www.genome.jp/kegg/mapper.html, accessed June 2019). Coverage was determined through read mapping with the bowtie2 (v2.3.2) and bedtools (v2.25.0) genomecov programs (58, 59). Reference proteins for phylogenetic trees were downloaded from NCBI GenBank in September 2019, and descriptions of these proteins can be found in Table S5 in the supplemental material. The multiple-sequence alignments were built with the MUSCLE (v3.8.31) program (60), and the phylogenies were inferred with the RaxML (v8.2.0) program and the -f a option with 100 bootstrap replicates.
Data availability.
All unassembled sequences related to this study are available at the NCBI Sequence Read Archive (BioSample accession number SAMN13035994), and MAG assemblies have been submitted to NCBI GenBank (BioSample accession numbers SAMN13172856 to SAMN13172858). All NCBI data may be found under BioProject accession number PRJNA577730. All SRA metadata, supplementary materials, and protocols are archived at https://doi.org/10.5281/zenodo.3629892. All custom software and scripts are available at https://github.com/Brazelton-Lab.
Supplementary Material
ACKNOWLEDGMENTS
We thank Emily Dart for DNA extractions and metagenomic library preparation, Alex Hyer and Christopher Thornton for computational assistance, and David Blair for assistance with metabolic pathways.
Funding was provided by the National Science Foundation (NSF projects 1536702 and 1536406), the Center for Dark Energy Biosphere Investigations, and the NASA Astrobiology Institute Rock-Powered Life Team.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lang SQ, Brazelton WJ. 2020. Habitability of the marine serpentinite subsurface: a case study of the Lost City hydrothermal field. Philos Trans A Math Phys Eng Sci 378:20180429. doi: 10.1098/rsta.2018.0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kargel JS, Kaye JZ, Head JW, Marion GM, Sassen R, Crowley JK, Ballesteros OP, Grant SA, Hogenboom DL. 2000. Europa’s crust and ocean: origin, composition, and the prospects for life. Icarus 148:226–265. doi: 10.1006/icar.2000.6471. [DOI] [Google Scholar]
- 3.Holm NG, Oze C, Mousis O, Waite JH, Guilbert-Lepoutre A. 2015. Serpentinization and the formation of H2 and CH4 on celestial bodies (planets, moons, comets). Astrobiology 15:587–600. doi: 10.1089/ast.2014.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lang SQ, Früh-Green GL, Bernasconi SM, Brazelton WJ, Schrenk MO, McGonigle JM. 2018. Deeply-sourced formate fuels sulfate reducers but not methanogens at Lost City hydrothermal field. Sci Rep 8:755. doi: 10.1038/s41598-017-19002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schrenk MO, Kelley DS, Bolton SA, Baross JA. 2004. Low archaeal diversity linked to subseafloor geochemical processes at the Lost City Hydrothermal Field, Mid-Atlantic Ridge. Environ Microbiol 6:1086–1095. doi: 10.1111/j.1462-2920.2004.00650.x. [DOI] [PubMed] [Google Scholar]
- 6.Brazelton WJ, Ludwig KA, Sogin ML, Andreishcheva EN, Kelley DS, Shen C-C, Edwards RL, Baross JA. 2010. Archaea and bacteria with surprising microdiversity show shifts in dominance over 1,000-year time scales in hydrothermal chimneys. Proc Natl Acad Sci U S A 107:1612–1617. doi: 10.1073/pnas.0905369107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. 2006. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl Environ Microbiol 72:6257–6270. doi: 10.1128/AEM.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brazelton WJ, Mehta MP, Kelley DS, Baross JA. 2011. Physiological differentiation within a single-species biofilm fueled by serpentinization. mBio 2:e00127-11. doi: 10.1128/mBio.00127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelley DS, Karson JA, Blackman DK, Früh-Green GL, Butterfield DA, Lilley MD, Olson EJ, Schrenk MO, Roe KK, Lebon GT, Rivizzigno P, AT3-60 Shipboard Party. 2001. An off-axis hydrothermal vent field near the Mid-Atlantic Ridge at 30°N. Nature 412:145–149. doi: 10.1038/35084000. [DOI] [PubMed] [Google Scholar]
- 10.Lang SQ, Fru GL, Bernasconi SM, Lilley MD, Proskurowski G, Mehay S, Butterfield DA. 2012. Microbial utilization of abiogenic carbon and hydrogen in a serpentinite-hosted system. Geochim Cosmochim Acta 92:82–99. doi: 10.1016/j.gca.2012.06.006. [DOI] [Google Scholar]
- 11.Proskurowski G, Lilley MD, Seewald JS, Früh-Green GL, Olson EJ, Lupton JE, Sylva SP, Kelley DS. 2008. Abiogenic hydrocarbon production at Lost City hydrothermal field. Science 319:604–607. doi: 10.1126/science.1151194. [DOI] [PubMed] [Google Scholar]
- 12.Kelley DS, Karson JA, Früh-Green GL, Yoerger DR, Shank TM, Butterfield DA, Hayes JM, Schrenk MO, Olson EJ, Proskurowski G, Jakuba M, Bradley A, Larson B, Ludwig K, Glickson D, Buckman K, Bradley AS, Brazelton WJ, Roe K, Elend MJ, Delacour A, Bernasconi SM, Lilley MD, Baross JA, Summons RE, Sylva SP. 2005. A serpentinite-hosted ecosystem: the Lost City hydrothermal field. Science 307:1428–1434. doi: 10.1126/science.1102556. [DOI] [PubMed] [Google Scholar]
- 13.Früh-Green GL, Kelley DS, Bernasconi SM, Karson JA, Ludwig KA, Butterfield DA, Boschi C, Proskurowski G. 2003. 30,000 years of hydrothermal vent field. Science 301:495–498. doi: 10.1126/science.1085582. [DOI] [PubMed] [Google Scholar]
- 14.McDermott JM, Seewald JS, German CR, Sylva SP. 2015. Pathways for abiotic organic synthesis at submarine hydrothermal fields. Proc Natl Acad Sci U S A 112:7668–7672. doi: 10.1073/pnas.1506295112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang SQ, Butterfield DA, Schulte M, Kelley DS, Lilley MD. 2010. Elevated concentrations of formate, acetate and dissolved organic carbon found at the Lost City hydrothermal field. Geochim Cosmochim Acta 74:941–952. doi: 10.1016/j.gca.2009.10.045. [DOI] [Google Scholar]
- 16.Blackmore MA, Quayle JR. 1968. Choice between autotrophy and heterotrophy in Pseudomonas oxalaticus. Biochem J 107:705–713. doi: 10.1042/bj1070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickering BS, Oresnik IJ. 2008. Formate-dependent autotrophic growth in Sinorhizobium meliloti. J Bacteriol 190:6409–6418. doi: 10.1128/JB.00757-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bar-Even A. 2016. Formate assimilation: the metabolic architecture of natural and synthetic pathways. Biochemistry 55:3851–3863. doi: 10.1021/acs.biochem.6b00495. [DOI] [PubMed] [Google Scholar]
- 19.Yishai O, Lindner SN, Gonzalez de la Cruz J, Tenenboim H, Bar-Even A. 2016. The formate bio-economy. Curr Opin Chem Biol 35:1–9. doi: 10.1016/j.cbpa.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Gregor I, Dröge J, Schirmer M, Quince C, McHardy AC. 2016. PhyloPythiaS+: a self-training method for the rapid reconstruction of low-ranking taxonomic bins from metagenomes. PeerJ 4:e1603. doi: 10.7717/peerj.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brazelton WJ, Baross JA. 2009. Abundant transposases encoded by the metagenome of a hydrothermal chimney biofilm. ISME J 3:1420–1424. doi: 10.1038/ismej.2009.79. [DOI] [PubMed] [Google Scholar]
- 22.Kaster A, Goenrich M, Seedorf H, Liesegang H, Wollherr A, Gottschalk G, Thauer RK. 2011. More than 200 genes required for methane formation from H2 and CO2 and energy conservation are present in Methanothermobacter marburgensis and Methanothermobacter thermautotrophicus. Archaea 2011:973848. doi: 10.1155/2011/973848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suppmann B, Sawers G. 1994. Isolation and characterization of hypophosphite‐resistant mutants of Escherichia coli: identification of the FocA protein, encoded by the pfl operon, as a putative formate transporter. Mol Microbiol 11:965–982. doi: 10.1111/j.1365-2958.1994.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 24.Falke D, Schulz K, Doberenz C, Beyer L, Lilie H, Thiemer B, Sawers RG. 2010. Unexpected oligomeric structure of the FocA formate channel of Escherichia coli: a paradigm for the formate-nitrite transporter family of integral membrane proteins. FEMS Microbiol Lett 303:69–75. doi: 10.1111/j.1574-6968.2009.01862.x. [DOI] [PubMed] [Google Scholar]
- 25.Lang SQ, Früh-Green GL, Bernasconi SM, Butterfield DA. 2013. Sources of organic nitrogen at the serpentinite-hosted Lost City hydrothermal field. Geobiology 11:154–169. doi: 10.1111/gbi.12026. [DOI] [PubMed] [Google Scholar]
- 26.Jormakka M, Byrne B, Iwata S. 2003. Formate dehydrogenase—a versatile enzyme in changing environments. Curr Opin Struct Biol 13:418–423. doi: 10.1016/s0959-440x(03)00098-8. [DOI] [PubMed] [Google Scholar]
- 27.Mnatsakanyan N, Vassilian A, Navasardyan L, Bagramyan K, Trchounian A. 2002. Regulation of Escherichia coli formate hydrogenlyase activity by formate at alkaline pH. Curr Microbiol 45:281–286. doi: 10.1007/s00284-002-3764-z. [DOI] [PubMed] [Google Scholar]
- 28.McDowall JS, Murphy BJ, Haumann M, Palmer T, Armstrong FA, Sargent F. 2014. Bacterial formate hydrogenlyase complex. Proc Natl Acad Sci U S A 111:E3948–E3956. doi: 10.1073/pnas.1407927111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knappe J, Sawers G. 1990. A radical-chemical route to acetyl-CoA: the anaerobically induced pyruvate formate-lyase system of Escherichia coli. FEMS Microbiol Rev 75:383–398. doi: 10.1111/j.1574-6968.1990.tb04108.x. [DOI] [PubMed] [Google Scholar]
- 30.Berg IA. 2011. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Wilksch JJ, Strugnell RA, Gee ML. 2015. Role of capsular polysaccharides in biofilm formation: an AFM nanomechanics study. ACS Appl Mater Interfaces 7:13007–13013. doi: 10.1021/acsami.5b03041. [DOI] [PubMed] [Google Scholar]
- 32.Hug I, Feldman MF. 2011. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology 21:138–151. doi: 10.1093/glycob/cwq148. [DOI] [PubMed] [Google Scholar]
- 33.Inagaki F, Takai K, Nealson KH, Horikoshi K. 2004. Novel sulfur-oxidizing chemolithoautotroph within the ε-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int J Syst Evol Microbiol 54:1477–1482. doi: 10.1099/ijs.0.03042-0. [DOI] [PubMed] [Google Scholar]
- 34.Jeon W, Priscilla L, Park G, Lee H, Lee N, Lee D, Kwon H, Ahn I, Lee C, Lee H, Ahn J. 2017. Complete genome sequence of the sulfur-oxidizing chemolithoautotrophic Sulfurovum lithotrophicum 42BKT. Stand Genomic Sci 12:54. doi: 10.1186/s40793-017-0265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori K, Yamaguchi K, Hanada S. 2018. Sulfurovum denitrificans sp. nov., an obligately chemolithoautotrophic sulfur-oxidizing epsilonproteobacterium isolated from a hydrothermal field. Int J Syst Evol Microbiol 68:2183–2187. doi: 10.1099/ijsem.0.002803. [DOI] [PubMed] [Google Scholar]
- 36.Mino S, Kudo H, Arai T, Sawabe T, Takai K, Nakagawa S. 2014. Sulfurovum aggregans sp. nov., a hydrogen-oxidizing, thiosulfate-reducing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent chimney, and an emended description of the genus Sulfurovum. Int J Syst Evol Microbiol 64:3195–3201. doi: 10.1099/ijs.0.065094-0. [DOI] [PubMed] [Google Scholar]
- 37.Park S-J, Ghai R, Martín-Cuadrado A-B, Rodríguez-Valera F, Jung M-Y, Kim J-G, Rheea S-K. 2012. Draft genome sequence of the sulfur-oxidizing bacterium “Candidatus Sulfurovum sediminum” AR, which belongs to the Epsilonproteobacteria. J Bacteriol 194:4128–4129. doi: 10.1128/JB.00741-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horikoshi M, Nakagawa Y, Shimamura S, Koki S, Takai K. 2010. Molecular characterization of inorganic sulfur‐compound metabolism in the deep‐sea epsilonproteobacterium Sulfurovum sp. NBC37‐1. Environ Microbiol 12:1144–1153. doi: 10.1111/j.1462-2920.2010.02155.x. [DOI] [PubMed] [Google Scholar]
- 39.Cannon GC, Heinhorst S, Kerfeld CA. 2010. Carboxysomal carbonic anhydrases: structure and role in microbial CO2 fixation. Biochim Biophys Acta 1804:382–392. doi: 10.1016/j.bbapap.2009.09.026. [DOI] [PubMed] [Google Scholar]
- 40.Van Lun M, Hub JS, Van Der Spoel D, Andersson I. 2014. CO2 and O2 distribution in Rubisco suggests the small subunit functions as a CO2 reservoir. J Am Chem Soc 136:3165–3171. doi: 10.1021/ja411579b. [DOI] [PubMed] [Google Scholar]
- 41.Verhees CH, Kengen SW, Tuininga JE, Schut GJ, Adams MWW, De Vos WM, Van Der Oost J. 2003. The unique features of glycolytic pathways. Biochem J 375:231–246. doi: 10.1042/BJ20021472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhuang WQ, Yi S, Bill M, Brisson VL, Feng X, Men Y, Conrad ME, Tang YJ, Alvarez-Cohen L. 2014. Incomplete Wood-Ljungdahl pathway facilitates one-carbon metabolism in organohalide-respiring Dehalococcoides mccartyi. Proc Natl Acad Sci U S A 111:6419–6424. doi: 10.1073/pnas.1321542111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chadwick GL, Hemp J, Fischer WW, Orphan VJ. 2018. Convergent evolution of unusual complex I homologs with increased proton pumping capacity: energetic and ecological implications. ISME J 12:2668–2680. doi: 10.1038/s41396-018-0210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grote J, Schott T, Bruckner CG, Glöckner FO, Jost G, Teeling H, Labrenz M, Jürgens K. 2012. Genome and physiology of a model Epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. Proc Natl Acad Sci U S A 109:506–510. doi: 10.1073/pnas.1111262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward LM, Hemp J, Shih PM, Mcglynn SE, Fischer WW. 2018. Evolution of phototrophy in the Chloroflexi phylum driven by horizontal gene transfer. Front Microbiol 9:260. doi: 10.3389/fmicb.2018.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mander GJ, Duin EC, Linder D, Stetter KO, Hedderich R. 2002. Purification and characterization of a membrane-bound enzyme complex from the sulfate-reducing archaeon Archaeoglobus fulgidus related to heterodisulfide reductase from methanogenic archaea. Eur J Biochem 269:1895–1904. doi: 10.1046/j.1432-1033.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 47.Heiden S, Hedderich R, Setzke E, Thauer RK. 1993. Purification of a cytochrome b containing H2:heterodisulfide oxidoreductase complex from membranes of Methanosarcina barkeri. Eur J Biochem 213:529–535. doi: 10.1111/j.1432-1033.1993.tb17791.x. [DOI] [PubMed] [Google Scholar]
- 48.Jormakka M, Yokoyama K, Yano T, Tamakoshi M, Akimoto S, Shimamura T, Curmi P, Iwata S. 2008. Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol 15:730–737. doi: 10.1038/nsmb.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welte C, Deppenmeier U. 2014. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim Biophys Acta 1837:1130–1147. doi: 10.1016/j.bbabio.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Barber RD, Zhang L, Harnack M, Olson MV, Kaul R, Ingram-Smith C, Smith KS. 2011. Complete genome sequence of Methanosaeta concilii, a specialist in aceticlastic methanogenesis. J Bacteriol 193:3668–3669. doi: 10.1128/JB.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu J, Zheng H, Ai G, Zhang G, Liu D, Liu X, Dong X. 2012. The genome characteristics and predicted function of methyl-group oxidation pathway in the obligate aceticlastic methanogens, Methanosaeta spp. PLoS One 7:e36756. doi: 10.1371/journal.pone.0036756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith KS, Ingram-Smith C. 2007. Methanosaeta, the forgotten methanogen? Trends Microbiol 15:150–155. doi: 10.1016/j.tim.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Hensley SA, Moreira E, Holden JF. 2016. Hydrogen production and enzyme activities in the hyperthermophile Thermococcus paralvinellae grown on maltose, tryptone, and agricultural waste. Front Microbiol 7:167. doi: 10.3389/fmicb.2016.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lever MA, Torti A, Eickenbusch P, Michaud AB, Šantl-Temkiv T, Jørgensen BB. 2015. A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Front Microbiol 6:476. doi: 10.3389/fmicb.2015.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohland N, Reich D. 2012. Cost-effective, high-throughput DNA sequencing. Genome Res 22:939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bushnell B. BBMap. sourceforge.net/projects/bbmap/.
- 57.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Wilks C, Antonescu V, Charles R. 2019. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 35:421–432. doi: 10.1093/bioinformatics/bty648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All unassembled sequences related to this study are available at the NCBI Sequence Read Archive (BioSample accession number SAMN13035994), and MAG assemblies have been submitted to NCBI GenBank (BioSample accession numbers SAMN13172856 to SAMN13172858). All NCBI data may be found under BioProject accession number PRJNA577730. All SRA metadata, supplementary materials, and protocols are archived at https://doi.org/10.5281/zenodo.3629892. All custom software and scripts are available at https://github.com/Brazelton-Lab.