Mining activities are accompanied by significant environmental and financial liabilities, including the release of acid mine drainage (AMD). AMD is caused by accelerated chemical and biological oxidation of sulfide minerals in mine wastes and is characterized by low pH and high concentrations of sulfate and metal(loid)s. Microorganisms assume important roles in the catalysis of redox reactions. Our research elucidates linkages among the biogeochemistry of mine wastes and remediation systems and microbial community and activity. This study assesses the performance and utility of geosynthetic-clay-liner cover systems for management of acid-generating mine wastes. Analyses of the microbial communities in tailings isolated beneath an engineered cover system provide a better understanding of the complex biogeochemical processes involved in the redox cycling of key elements, contribute to the remediation of mine wastes, and provide a valuable tool for assessment of the effectiveness of the remediation system.
KEYWORDS: mine waste, tailings, geosynthetic clay liner, biogeochemistry, sulfur oxidizers, iron oxidizers, acid mine drainage, biodiversity
ABSTRACT
The abandoned Kam Kotia Mine (Canada) is undergoing remediation. A geosynthetic-clay-liner (GCL) cover system was installed in the Northern Impounded Tailings (NIT) area in 2008 to isolate acid-generating tailings from water and oxygen and to mitigate sulfide oxidation. The cover system includes a vegetated uppermost soil layer underlain by a granular protective layer (sand), a clay moisture-retaining layer, a GCL, a granular capillary-break material (cushion sand), and a crushed waste rock-capillary break layer installed above the tailings. The goal of this study was to characterize the microbiology of the covered tailings to assess the performance of the cover system for mitigating sulfide bio-oxidation. Tailings beneath the GCL were characterized by high sulfur and low carbon content. The bulk pH of the tailings pore water was circumneutral (∼5.5 to 7.3). Total genomic DNA was extracted from 36 samples recovered from the constituent layers of the cover system and the underlying tailings and was analyzed in triplicates using high-throughput amplicon sequencing of 16S rRNA genes. Iron-oxidizing, sulfur-oxidizing, sulfate-reducing, and aerobic heterotrophic microorganisms were enumerated by use of most probable number enumeration, which identified heterotrophs as the most numerous group of culturable microorganisms throughout the depth profile. Low relative abundances and viable counts of microorganisms that catalyze transformations of iron and sulfur in the covered tailings, compared to previous studies on unreclaimed tailings, indicate that sulfide oxidation rates have decreased due to the presence of the GCL. Characterization of the microbial community can provide a sensitive indicator for assessing the performance of remediation systems.
IMPORTANCE Mining activities are accompanied by significant environmental and financial liabilities, including the release of acid mine drainage (AMD). AMD is caused by accelerated chemical and biological oxidation of sulfide minerals in mine wastes and is characterized by low pH and high concentrations of sulfate and metal(loid)s. Microorganisms assume important roles in the catalysis of redox reactions. Our research elucidates linkages among the biogeochemistry of mine wastes and remediation systems and microbial community and activity. This study assesses the performance and utility of geosynthetic-clay-liner cover systems for management of acid-generating mine wastes. Analyses of the microbial communities in tailings isolated beneath an engineered cover system provide a better understanding of the complex biogeochemical processes involved in the redox cycling of key elements, contribute to the remediation of mine wastes, and provide a valuable tool for assessment of the effectiveness of the remediation system.
INTRODUCTION
Sulfidic ore deposits are an important source of base and precious metals and other mined commodities. However, mining of sulfide ores generates sulfide-bearing mine wastes, including waste rock and mill tailings. Oxidation of sulfide minerals in mine wastes can occur via either direct or indirect mechanisms, with the primary oxidants being atmospheric O2 or Fe3+, respectively (1). Sulfide mineral oxidation can lead to the generation of acid mine drainage (AMD), characterized by low pH and elevated concentrations of sulfate, iron, and metal(loid)s (2). AMD generation can be catalyzed via the metabolic activity of autochthonous microorganisms that catalyze dissimilatory oxidoreduction of sulfur and iron (3, 4). The microbiology of AMD-impacted systems has been thoroughly reviewed in previous studies (3–5). Numerous previous reports focused on microbial community composition in mill tailings using culture-dependent techniques, such as most probable number (MPN) enumerations (6–9). Molecular methods have been used to identify cultured microorganisms in tailings samples (10, 11) and also as a culture-independent approach to describe microbial diversity (12–14). High-throughput amplicon sequencing of 16S rRNA genes has been used to describe bacterial and archaeal communities (BACs) present in mine waste environments (15–20). Culture-based approaches provide insights into physiological traits and metabolic potential present within environmental samples, whereas high-throughput, culture-independent microbial ecology techniques provide insights into the composition of the entire BAC. Relatively few studies (21) to date have utilized both culture-based and high-throughput molecular techniques to characterize mill tailings.
Bacteria that are widely distributed in AMD environments include acidophiles, characterized by pH optima < 3 and with optimal mesophilic growth temperatures ranging from 17 to 45°C, belonging to the phyla Proteobacteria, Nitrospirae, Actinobacteria, Firmicutes, and Acidobacteria (5). Members of these taxa include (among others) chemolithoautotrophs that obtain energy primarily from iron, sulfur, or hydrogen metabolism, as well as mixotrophs and heterotrophs. The Archaea observed in mine wastes generally belong to the order Thermoplasmatales and display exclusively organotrophic growth, except for members of Ferroplasmaceae, which also use iron (22). Microbial catalysis of redox reactions in mine waste environments and microbiology of mine wastes have been extensively studied (3, 23). However, most studies focus on AMD (3), and knowledge of the microbiology of solid mine wastes is relatively limited (23).
Sustainable mine waste management practices are an increasingly important aspect of metal production in Canada. However, Canada is home to more than 10,000 abandoned mine sites (24), the legacy of which continues to impact local environments and communities. AMD can impact local groundwater systems and/or discharge directly into receiving surface water bodies (25). In Ontario alone, there are approximately 5,600 abandoned mines; the liability associated with the management of abandoned mines represents a significant financial burden (26). Active treatment of impacted waters originating from mine wastes is costly and, in the context of abandoned mines, is not ideal for a long-term solution, while emplacement of (dry or wet) covers on tailings impoundments has been demonstrated to be a successful method of reducing AMD and metal leaching (25, 27–29). Cover systems and physical barriers are designed to mitigate the influx of water and atmospheric oxygen. Dry cover designs for tailings are numerous and site specific; they are constructed from locally available solid materials and can be single or multilayered. Dry covers range from the direct establishment of native vegetation over the top of the wastes to complex composite covers (30). Materials used for dry covers include geotextiles, low-sulfide waste rock, oxide wastes, organic wastes, clay, and soils (25). Low-permeability geosynthetic clay liners (GCLs) are increasingly incorporated into cover systems for a wide variety of hydraulic and gas-containment applications in mine waste containment facilities (31, 32).
Aqueous geochemistry of AMD is intrinsically linked to tailings mineralogy, geochemistry, microbiology, and hydrology. Rates of sulfide oxidation, and the mobility of associated metal(loid)s, are dependent on multiple (bio)geochemical processes. At present, relatively few studies have evaluated the impact of dry covers on mine waste biogeochemistry or the impact of microbiological diversity on the long-term geochemical stability of tailings. In addition, despite increasing recognition of the impact of soil microbes on both AMD generation and belowground metabolic recovery, little is known regarding the complexity and functions of BACs in soils impacted by mine waste disposal. The main aim of the present study was to provide a large-scale evaluation of the microbiology of tailings installed within the Northern Impounded Tailings (NIT) at the Kam Kotia Mine to elucidate the impact of GCL installation on sulfide oxidation and AMD generation. Diversity, taxonomic composition, and metabolic activities of BACs were determined, with a special focus on microorganisms anticipated to drive the geochemical evolution of mine waste environments, including iron-oxidizing microorganisms (IOM), sulfur-oxidizing microorganisms (SOM), and sulfate-reducing bacteria (SRB).
RESULTS
Aqueous and solid-phase geochemistry.
The distinct reddish-brown color of the solid phase, and the presence of residual sulfide minerals, indicated that the upper layer (1.5 to 2.7 m below ground surface [mbgs]) of the covered tailings at site 1 was oxidized or partially oxidized. Tailings at depth > 2.7 mbgs at site 1, and at the other three sampling locations, were dark gray, and sulfide minerals were visible. Selected physicochemical properties of the tailings samples used in this study are shown in Table 1. Within the tailings, pH values ranged from 5.5 to 7.3 (compared to 7.0 to 7.8 within the cover system), and Eh ranged from +208 to +339 mV (versus +286 to +453 mV in the cover). Concentrations of soluble iron and dissolved sulfate were greater in the tailings pore water samples compared to values in the cover system; maximum total Fe concentration detected in the tailings reached 1.76 g liter−1 but remained below the detection limit in the cover material; maximum SO42− concentration in the tailings was 8.45 g liter−1 compared to 1.21 g liter−1 in the cover. Total sulfur in solid samples was much lower in the cover system layers relative to the tailings, which were typically characterized by concentrations of the 20 to 40 wt% range (with the exception of site 1, where S concentrations were < 0.15 wt%). Total solid-phase carbon content was low in all tailings samples (maximum, 0.6 wt%) and present mainly as inorganic C; mineralogical analysis confirmed the presence of calcite and dolomite in the unoxidized tailings. In the cover system, the highest carbon concentrations (∼3.3 wt%) were detected in the clay samples. Differences between most parameter means (pH, Eh, dissolved SO42−, total solid-phase C, and total solid-phase S) in the tailings and cover system were statistically significant (P < 0.05).
TABLE 1.
Selected physicochemical characteristics of the Kam Kotia tailings samples, determined in pore watera
| Site | Sample type | Depth (mbgs) | pH | Eh (mV) | Fe (g liter−1) | SO42− (g liter−1) | δ18O (‰) | C (wt%)b | S (wt%)b |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Soil | 0.05 | NA | NA | NA | NA | NA | NA | NA |
| Sand | 0.30 | 7.2 | +326 | 0.00 | 0.00 | −11.0 | NA | NA | |
| Clay | 0.75 | NA | NA | NA | NA | NA | NA | NA | |
| Cushion sand | 1.10 | 7.5 | +453 | 0.00 | 1.15 | −10.6 | NA | NA | |
| Rock | 1.30 | NA | NA | NA | NA | NA | NA | NA | |
| Tailings | 2.00 | NA | NA | NA | NA | NA | 0.28 | 0.09 | |
| 2.67 | NA | NA | NA | NA | NA | 0.12 | 0.13 | ||
| 3.18 | 5.5 | +293 | 1.76 | 8.45 | −11.8 | 0.31 | 0.11 | ||
| 3.67 | 5.9 | +232 | 0.49 | 5.69 | −11.7 | 0.18 | 0.04 | ||
| 4.24 | 6.2 | +208 | 0.33 | 5.13 | −11.1 | NA | NA | ||
| 4.68 | NA | NA | NA | NA | NA | 0.27 | 0.05 | ||
| 3 | Soil | 0.05 | NA | NA | NA | NA | NA | NA | NA |
| Sand | 0.30 | 7.8 | +326 | 0.00 | 0.01 | −10.6 | 0.92 | 0.00 | |
| Clay | 0.65 | 7.4 | +363 | 0.00 | 0.00 | −11.2 | 3.26 | 0.00 | |
| Cushion sand | 0.95 | 7.3 | +286 | 0.00 | 0.56 | −10.0 | 1.47 | 0.01 | |
| Rock | 1.40 | NA | NA | NA | NA | NA | NA | NA | |
| Tailings | 2.16 | 6.8 | +244 | 0.02 | 1.19 | −10.8 | 0.08 | 20.81 | |
| 2.44 | 6.3 | +297 | 0.18 | 1.65 | −10.3 | 0.13 | 20.92 | ||
| 3.25 | NA | NA | NA | NA | NA | 0.61 | 5.20 | ||
| 3.43 | 5.9 | +258 | NA | NA | NA | NA | NA | ||
| 4 | Soil | 0.05 | NA | NA | NA | NA | NA | NA | NA |
| Sand | 0.30 | NA | NA | NA | NA | NA | 0.73 | 0.01 | |
| Clay | 0.70 | NA | NA | NA | NA | NA | 3.21 | 0.04 | |
| Cushion sand | 1.00 | 7.5 | +306 | 0.00 | 0.23 | −11.0 | NA | NA | |
| Rock | 1.37 | 7.0 | +406 | 0.00 | 1.21 | −11.9 | NA | NA | |
| Tailings | 1.80 | 6.8 | +293 | 0.01 | 1.46 | −10.7 | 0.06 | 26.06 | |
| 2.02 | 6.8 | +253 | 0.03 | 1.37 | −10.9 | 0.06 | 19.70 | ||
| 2.47 | 6.3 | +326 | 0.05 | 1.42 | −12.1 | 0.05 | 30.02 | ||
| 5 | Soil | 0.05 | NA | NA | NA | NA | NA | NA | NA |
| Sand | 0.40 | NA | NA | NA | NA | NA | NA | NA | |
| Clay | 0.95 | NA | NA | NA | NA | NA | NA | NA | |
| Cushion sand | 1.25 | NA | NA | NA | NA | NA | 0.13 | 0.00 | |
| Rock | 1.75 | NA | NA | NA | NA | NA | NA | NA | |
| Tailings | 1.97 | NA | NA | NA | NA | NA | 0.06 | 33.72 | |
| 2.18 | 7.3 | +339 | 0.00 | 1.78 | −11.0 | 0.07 | 35.85 | ||
| 2.56 | NA | NA | NA | NA | NA | 0.06 | 24.93 |
mbgs, meters below ground surface; NA, not available.
Determined in solid-phase samples.
Mean aqueous concentrations of transition metals other than Fe, including Ag, Cd, Co, Cr, Cu, and Ni, were generally < 110 μg liter−1 in tailings pore water samples, with the exception of Mn (mean concentration of 4.6 mg liter−1) and Zn (0.95 mg liter−1). Low Al concentrations were also observed (mean, ∼270 μg liter−1) in tailings pore water samples. Arsenic concentrations varied significantly across sites in the NIT; mean aqueous concentrations ranged from 0.01 mg liter−1 at site 1 up to 11 mg liter−1 at site 4. Arsenic concentrations as high as 18 mg liter−1 were measured in tailings pore water at a depth of 2.4 mbgs at site 4.
Enumeration of viable microorganisms.
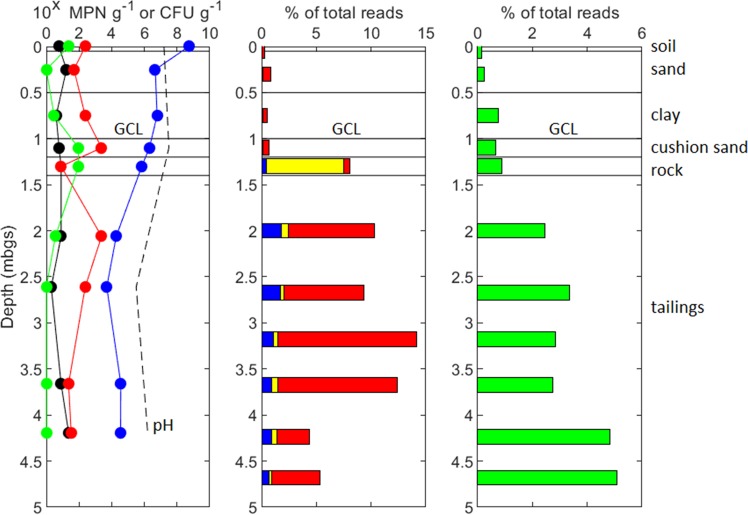
Figure 1 (left panel) shows numbers of culturable microbial populations within each tested layer through the NIT profile at site 1, determined by colony counting. Aerobic heterotrophs were the most abundant culturable microorganisms in both the cover and the underlying tailings. Heterotroph populations declined with depth, with the maximum value of 5.3 × 108 CFU g−1 observed at the surface, decreasing to relatively stable values of 104 CFU g−1 in the tailings. The numbers of acidophilic IOM (aIOM) were approximately 102 MPN g−1, with a local minimum of 8 MPN g−1 in the rock cover layer and two local maxima of 2.4 × 103 MPN g−1 in the cushion sand and top portion of the tailings. Viable populations of acidophilic SOM (aSOM) were lower than aIOM and were typically observed at abundances of approximately 10 MPN g−1. Acidithiobacillus ferrooxidans was identified by plating onto selective solid media (followed by PCR using 27F/1387R primers, Sanger sequencing, and BLAST search) as the most numerous aIOM/aSOM in both cover and tailings samples. SRB numbers were low, ranging from 0 to 93 MPN g−1, peaking in the cover layers below GCL. For comparison, relative abundances of aIOM/aSOM and SRB (determined by high-throughput sequencing) in samples collected at site 1 are shown in Fig. 1 (center and right panels, respectively).
FIG 1.
A vertical profile for heterotrophs (blue circles), sulfate reducers (SRB) (green circles), acidophilic iron oxidizers (aIOM) (red circles), and acidophilic sulfur-oxidizers (aSOM) (black circles) in solid samples taken from the NIT at the Kam Kotia Mine, determined by MPN. The data points represent means of triplicates for heterotrophs and SRB, and of five replicates for acidophiles. Dashed line indicates single measure of pH. MPN data are compared to percentages of total reads of SOM/IOM (blue bars), SOM (yellow bars), IOM (red bars), and SRB (green bars), as determined by high-throughput 16S rRNA amplicon sequencing of samples collected at site 1.
Overview of 16S rRNA gene sequence data statistics.
Triplicate solid-phase samples from each layer were sequenced in the same sequencing run. All mean values in this section refer to samples resulting from pooling the triplicate samples (which contained >10,000 sequences each). A total of 6,967,367 raw sequence reads were obtained, with a mean of 193,538 ± 10,595 reads per sample. About 8.4% of sequences were flagged as chimeric, and in total, 18.6% of reads were lost to quality trimming (e.g., removal of selected taxons). A total effective sequence number was 5,673,921 with 157,609 ± 13,192 reads per sample and a mean of 1,119 ± 145 OTUs (operational taxonomic units; 97% sequence similarity) per library (Table S1). The difference between mean OTU numbers in tailings (449 ± 76 OTUs per sample) and cover samples (1,655 ± 180 OTUs per sample) was highly significant (P < 0.01).
Taxonomy of entire BACs at Kam Kotia.
The prokaryotic taxonomy in the key components of the Kam Kotia NIT cover system was first established on the phylum level (Fig. 2). As expected, the layers of the cover system harbored a diverse range of bacterial and archaeal taxa and were dominated by those widely distributed in most soil environments, such as the phyla Proteobacteria (mean, ∼34% of total reads in the cover; the phylum contains, among other members, also sulfur- and/or iron-metabolizing Acidithiobacillus and Thiobacillus spp.), Actinobacteria (11.5%, including also iron-metabolizing acidophiles), and Bacteroidetes (8.1%). The BAC composition in the tailings beneath the cover system was characterized by community composition similar to the overlying cover system; however, the tailings samples typically contained higher relative abundances of sequences affiliated with the phyla Firmicutes (mean, 12.4% in tailings versus 2.7% in cover system, disregarding rock layer; including, for example, Sulfobacillus, Acidibacillus, and Alicyclobacillus) and Euryarchaeota (3.1% versus 0.3%; for example, Ferroplasma), both of which include members that obtain energy through dissimilatory sulfur and iron metabolism. A proportion of the sequences present in our samples could only be assigned at the domain level (especially unclassified bacteria; mean, ∼5.0% of total reads), indicating the presence of novel or unknown lineages.
FIG 2.
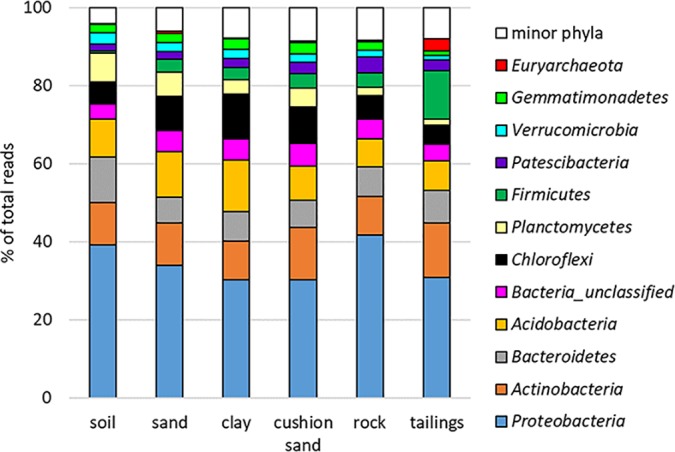
Percentages of total reads of dominant lineages (phylum level) in different layers of the NIT cover system and underlying tailings at the Kam Kotia Mine, as determined by high-throughput 16S rRNA amplicon sequencing. Minor phyla with relative abundance < 1% were grouped together.
In total, 1,673 different genera were identified in the samples collected in the cover system and underlying tailings. Major genera (or higher taxa when identification to the genus level was not possible) are listed in Table S2. The sum of minor genera (<0.5% of total amplicons) accounted for 70.7% of total reads. The detected prokaryotic genera use a wide range of metabolic strategies and have been observed in a variety of diverse environments. Next to genera including species catalyzing oxidoreductions of Fe and S (described in detail in the section “Iron- and sulfur-metabolizing prokaryotic genera”), common soil bacteria (e.g., Actinobacteria accounting for 0.6% of total reads), plant symbionts (Rhizobiales, 1.3%), animal pathogens (e.g., Clostridiales, 0.33%; Enterobacteriaceae, 0.13%), and others were among the most abundant taxa.
Diversity of BACs at Kam Kotia.
High Good’s coverage (calculated for an OTU definition of 0.03), ranging from 93.8 to 99.8% (Table S1), indicated that the samples represented the BACs within each layer of the cover system well. In order to assess how the entire BACs differed in one environment compared to another, BAC compositional heterogeneity (β-diversity) was examined. β-diversity provides insights into mechanisms that drive biodiversity changes, and its investigation is therefore especially important in ecological communities that are subjected to significant environmental disturbances. β-diversity between BACs at four sites at the NIT at the Kam Kotia Mine was investigated by two-dimensional nonmetric multidimensional scaling (2D-NMDS) (Fig. S1); good quality of the ordination is indicated by a low stress value (2D stress, 0.116). Point dispersions were tested for significance using the permutation test for homogeneity of multivariate dispersions (number of permutations, 1,000), indicating the same “multivariate spread” among the groups (P > 0.05). Pairwise comparisons between sites are shown in Table S3. The null hypothesis of permutational multivariate analysis of variance (PERMANOVA) was confirmed, and pairwise testing showed no significant differences among the four sites (P > 0.05; Table S4). Samples collected from each layer at the four different sites could thus be considered replicates for further processing.
The NIT at Kam Kotia Mine represents a very heterogeneous habitat, owing to the differing physicochemical conditions within the constituent layers of the cover system and the underlying tailings. Each layer of the cover system is characterized by different physicochemical properties, and, with the possible exception of the rock layer constructed from the waste rock, differs greatly from the tailings. Fig. 3 shows a 2D-NMDS plot comparing prokaryotic communities within each layer of the NIT Kam Kotia Mine profile. The Bray-Curtis statistic was used for comparing the BAC similarity; almost identical clustering was achieved with the Jaccard index (not shown). To test for statistical differences, PERMANOVA or ANOSIM (analysis of similarities) is often used. However, the assumption of similar dispersion in Fig. 3 was not met, as tested by the permutation test for homogeneity of multivariate dispersions (number of permutations, 1,000). Pairwise comparisons between layers are shown in Table S5, which demonstrated significantly greater dispersion in tailings (P < 0.05) compared to the layers of the cover system.
FIG 3.
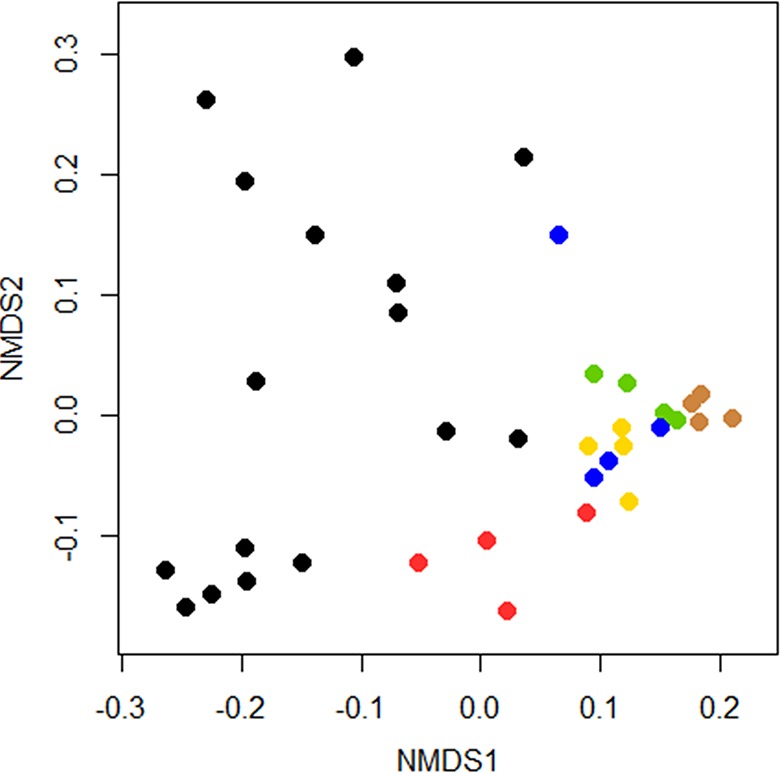
Two-dimensional nonmetric multidimensional scaling (2D-NMDS; stress, 0.116) plot of Bray-Curtis similarity matrices of microbial communities in soil (brown circles), sand (green circles), clay (blue circles), cushion sand (yellow circles), and rock layer (red circles) of the NIT cover system, and underlying tailings at the Kam Kotia Mine (black circles), determined by high-throughput amplicon sequencing. Points represent the composition of a community, and the distance between any two points represents the difference between those two communities.
Although α- and β-diversity did not seem to be affected by increased metal(loid) concentrations and low-pH regions in the tailings, significantly lower (P < 0.05) richness (number of OTUs; investigated by Chao’s index; data not shown) was determined in the tailings samples compared to the cover system samples. The results indicate that even though a reduced number of genera can be expected in mill tailings, their even distribution might compensate for this difference.
Relationships between BACs and geochemical variables.
The composition of prokaryotic populations in a specific environment is defined by many environmental parameters and variables. A number of relationships between BACs in the Kam Kotia tailings samples and selected physicochemical parameters were found and are shown as NMDS plots in Fig. 4. Little clustering was observed in the pH plot (Fig. 4A). Greater clustering of points representing community compositions was observed among samples containing higher concentrations (within the experimental ranges) of total iron (Fig. 4B), sulfate (Fig. 4C), and carbon (Fig. 4D), and also among samples with lower sulfur content (Fig. 4E).
FIG 4.
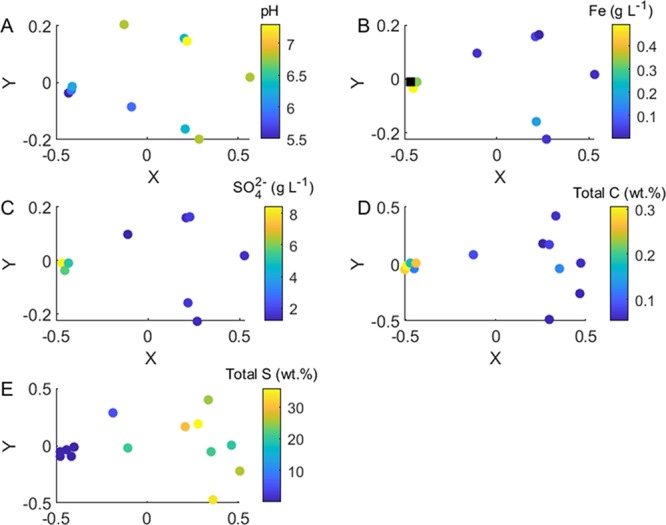
2D-NMDS plots comparing similarities of microbial communities in tailings (based on Bray-Curtis matrices) in dependence on pH (A), concentrations of Fe (g liter−1) (B), SO42− (g liter−1) (C), total C (wt%) (D), and total S (wt%) (E), determined by high-throughput amplicon sequencing. Total C and S were determined in solid phases, other parameters in pore water samples. For used metadata, see Table 1. For better visualization, this value (1.764 g liter−1) (■) was not included in the color range.
Iron- and sulfur-metabolizing prokaryotic genera.
Fig. 5 shows proportions of total reads of genera that catalyze dissimilatory oxidoreduction of iron and sulfur in each layer of the NIT profile. Physiological characteristics of the detected sulfur- and/or iron-metabolizing genera are summarized in Table 2. In the tailings, relative abundances of IOM/SOM (Fig. 5A) and SRB (Fig. 5B) accounted for 5.72 ± 1.65 and 2.32 ± 0.50% of total amplicons (mean ± standard deviation [SD]; 95% confidence interval), respectively. Both values were significantly higher (P < 0.05) than the proportions of IOM/SOM and SRB in the cover system (disregarding the rock layer), where they accounted for 0.97 ± 0.12 and 0.41 ± 0.13% of total amplicons (mean ± SD; 95% confidence interval), respectively. The rock layer of the cover system is comprised of waste rock with similar chemical composition to the tailings; elevated relative abundances of iron and sulfur oxidizers were found within the waste-rock layer (mean, ∼7.84% of total reads).
FIG 5.
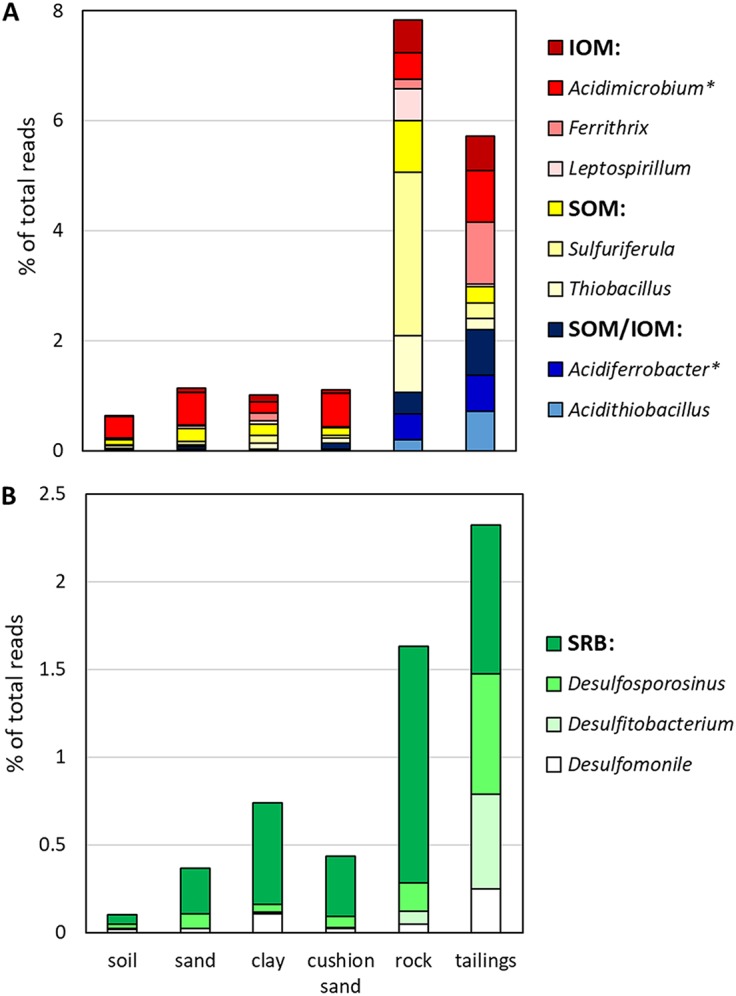
Percentages of total reads of microorganisms catalyzing dissimilatory oxidation (A) and reduction (B) of iron and sulfur in layers of the NIT cover system and underlying tailings at the Kam Kotia Mine, as determined by high-throughput amplicon sequencing. Most numerous bacterial genera within each group are shown separately. IOM, iron-oxidizing microorganisms; SOM, sulfur-oxidizing microorganisms; SOM/IOM, sulfur- and iron-oxidizing microorganisms; SRB, sulfate- and sulfur-reducing bacteria. Genera that were pooled with respective higher taxa are marked with asterisks. For sample depths, see Table 3. Physiological characteristics of the detected sulfur- and/or iron-metabolizing genera are summarized in Table 2.
TABLE 2.
Metabolic traits of genera detected in the Kam Kotia tailings samples that are known to catalyze the dissimilatory oxidoreduction of iron and/or sulfur
| Microorganism | Mean % of total reads in cover system | Mean % of total reads in tailings | pH responsea | Sulfur oxidation | Iron oxidation | Sulfate reduction | Iron reductionb |
|---|---|---|---|---|---|---|---|
| Acidimicrobiiac | 0.44 | 0.89 | EA | +d | + | ||
| Ferrithrix | 0.07 | 1.13 | EA | + | + | ||
| Sulfuriferula | 0.65 | 0.28 | MA & N | + | |||
| Geobacter | 0.44 | 0.41 | N | + | |||
| Desulfosporosinus | 0.08 | 0.68 | N & MA | + | + | ||
| Thiobacillus | 0.26 | 0.20 | N | + | |||
| Desulfitobacterium | 0.02 | 0.54 | N & MA | + | + | ||
| Acidithiobacillus | 0.05 | 0.71 | EA | + | + | + | |
| Desulfomonile | 0.05 | 0.25 | N & MA | + | + | ||
| Sulfurifustis | 0.14 | 0.07 | N | + | |||
| Desulfurivibrio | 0.14 | 0.10 | A | + | + | ||
| Acidithrix | 0.05 | 0.22 | EA & MA | + | + | ||
| Alicyclobacillus | 0.06 | 0.44 | EA & MA | + | + | + | |
| Desulfobulbaceaec | 0.08 | 0.05 | N | + | + | ||
| Leptospirillum | 0.14 | 0.06 | EA | + | |||
| Desulfuromonadalesc | 0.04 | 0.01 | N | + | + | ||
| Ferrovum | 0.02 | 0.13 | EA | + | + | ||
| Acidibacter | 0.14 | 0.10 | MA | + | |||
| Sulfobacillus | 0.02 | 0.20 | EA | + | + | + | |
| Acidiferrobacteraceaec | 0.05 | 0.50 | EA | + | + | + | |
| Desulfobacca | 0.04 | 0.09 | N | + | + | ||
| Acidibacillus | 0.04 | 0.19 | EA | + | + | + | |
| Acidiphilium | 0.04 | 0.11 | EA | + | |||
| Desulfovibrio | 0.03 | 0.03 | N & MA | + | + | ||
| Desulfurispora | 0.02 | 0.08 | N | + | + | ||
| Gallionellaceaec | 0.07 | 0.03 | N & MA | + | |||
| Acidimicrobiaceaec | 0.01 | 0.04 | EA | + | + | ||
| Acidiferrobacterc | 0.05 | 0.16 | EA | + | + | + | |
| Sulfuricellaceaec | 0.04 | <0.01 | N | + | |||
| Desulfobacteraceaec | 0.04 | <0.01 | N & MA | + | + | ||
| Ferroplasma | 0.01 | 0.15 | EA | + | + | ||
| Desulfobacteralesc | 0.01 | <0.01 | N & MA | + | + | ||
| Desulfobulbus | 0.02 | 0.15 | N | + | + | ||
| Desulforhopalus | 0.01 | 0.03 | N | + | + | ||
| Desulfocapsa | 0.03 | <0.01 | N | + | + | ||
| Sulfurimonas | 0.01 | 0.03 | N | + | |||
| Desulfovibrionalesc | 0.01 | 0.25 | N & MA | + | + | ||
| Thiomonas | 0.02 | 0.01 | MA | + | |||
| Gallionella | 0.02 | 0.01 | N | + | |||
| Ferribacterium | 0.01 | 0.01 | N | + | |||
| Sulfuritalea | 0.03 | <0.01 | N | + | |||
| Ferrimicrobium | 0.01 | 0.08 | EA | + | + | ||
| Acidimicrobialesc | <0.01 | 0.01 | EA | + | + | ||
| Desulfatiglans | 0.01 | <0.01 | N | + | + | ||
| Desulfomicrobium | 0.01 | 0.03 | N | + | + | ||
| Desulfuromonas | <0.01 | 0.03 | N | + | + | ||
| Desulfovirga | 0.01 | <0.01 | N | + | + | ||
| Sulfuricella | 0.03 | 0.06 | N | + | |||
| Sulfurirhabdus | 0.01 | <0.01 | N | + | |||
| Sulfurospirillum | <0.01 | <0.01 | N | (Sulfur) | + | ||
| Sulfuricurvum | 0.01 | <0.01 | N | + | |||
| Alicyclobacillaceaec | <0.01 | <0.01 | EA & MA | + | + | + | |
| Acidicaldus | 0.01 | <0.01 | EA | + | + | ||
| Ferrovibrio | <0.01 | <0.01 | N | + | |||
| Desulfatirhabdium | 0.01 | 0.01 | N | + | + | ||
| Desulfobacula | 0.01 | <0.01 | N | + | + | ||
| Desulfonema | <0.01 | <0.01 | N | + | + | ||
| Desulfoplanes | <0.01 | <0.01 | N | + | + | ||
| Desulfovibrionaceaec | <0.01 | <0.01 | N & MA | + | + | ||
| Sideroxydans | 0.01 | <0.01 | N | + | + | ||
| Ferritrophicum | <0.01 | <0.01 | N & MA | + | |||
| Thiovirga | <0.01 | 0.01 | N | + |
EA, extremely acidophilic; MA, moderately acidophilic; N, neutrophilic; A, alkaliphilic.
Both direct and indirect.
Higher taxa that could not be identified on the genus level.
Plus sign indicates at least one species of the genus has been reported to catalyze the dissimilatory reaction referred to.
DISCUSSION
Bacteria inhabiting acidic waters and sediments associated with AMD belong primarily to the phyla Proteobacteria, Nitrospirae, Actinobacteria, Firmicutes, and Acidobacteria, and also Bacteroidetes are often detected. Archaea populating mine sites generally belong to Euryarchaeota (5, 15–17, 33). Our taxonomic results at the phylum level corresponded well to the published findings. Cyanobacteria are a phylum of bacteria that obtain energy through photosynthesis. However, some Cyanobacteria can also live via organoheterotrophy (34). Also, chemolithotrophic metabolism has been reported in Cyanobacteria, although in the presence of light (35). These organisms accounted for 0.78% of total reads in the NIT samples, and their activity could contribute to overall biogeochemical cycling of C, S, and/or Fe in the system by competing for organic carbon with heterotrophic sulfur- and/or iron-metabolizing species, as well as consuming short-chain organic carbon compounds, which inhibit chemolithotrophs (36). Many studies investigating microbial populations in mill tailings have observed the predominance of typical leaching bacteria, such as Acidithiobacillus, Leptospirillum, Sulfobacillus, or Ferroplasma, at abundances reaching tens of percents in unremediated tailings (17, 18, 20, 33, 37). In our study, sulfur- and iron-oxidizing prokaryotes were detected with low abundances (their mean abundance in tailings reaching 5.72%, determined by 16S rRNA gene amplicon sequencing).
Acidophiles that include species that oxidize both substrates accounted for 2.21% of total reads in the tailings samples, aIOM accounted for 2.56%, and aSOM for 0.39%. The extremely acidophilic iron oxidizers (and reducers) Ferrithrix and Acidimicrobiia were the most abundant IOM in the tailings samples; their mean proportions of the total amplicons reached 1.13 and 0.89% of total reads, respectively. Another extremely acidophilic SOM/IOM present in the tailings were Acidithiobacillus spp. (0.71%), Acidiferrobacteraceae (0.50%), and Alicyclobacillus (0.44%). The abovementioned iron- and/or sulfur-oxidizing bacteria are commonly found in acidic, sulfide mineral-bearing environments and have also been identified in more neutral and alkaline pH environments (3, 38, 39). Neutrophilic genera accounted for only 0.42% of total reads of sulfur- and iron-oxidizing genera, suggesting that the samples studied were highly dominated by acidophiles. Neutrophilic SOM were detected in the tailings samples, although at lower relative abundances (e.g., Thiobacillus 0.20%); mean relative abundance of neutrophilic IOM was low (<0.04%). The cover system (disregarding the rock layer) was host to similar sulfur- and/or iron-oxidizing genera as the underlying tailings, although in lower proportions (Sulfuriferula, 0.65%; Acidimicrobiia, 0.44%; Thiobacillus, 0.26%) (Table 2).
Viable populations of aIOM, aSOM, SRB, and neutrophilic heterotrophs were enumerated using MPN. Apart from heterotrophs, numbers of cultured prokaryotes did not vary between the cover system and the tailings, which is consistent with the above hypothesis of low rates of sulfide oxidation in the underlying tailings. The observed numbers of acidophilic sulfur and iron oxidizers were lower than determined in similar studies; Benner et al. (8) reported 103 aSOM g−1 and 105 acidophilic iron-oxidizing bacteria (aIOB) g−1 in tailings from a Ni/Cu mine; Southam and Beveridge (6) reported 108 aIOM g−1 in pH-neutral tailings from a Cu mine; and Mendez et al. (21) reported around 104 both aIOM g−1 and aSOM g−1 in extremely and moderately acidic Pb/Zn tailings. Our acidophile enumerations were very similar to those in the study by Blowes et al. (7) in Au tailings, which were, however, reported to be dominated by sulfur-oxidizing neutrophils. Lindsay et al. (9) observed dominance of neutrophilic SOM (nSOM) in a sulfide-rich tailings deposit characterized by neutral drainage. Our sequence data suggested low relative abundance of nSOM (0.38% of total reads) in comparison with aSOM (2.60%, including acidophiles that are capable of utilizing both iron and sulfur), indicating the dominance of acidophilic species in the Kam Kotia tailings. It has been postulated that acidophiles under bulk circumneutral-pH conditions can form acidic microenvironments at sulfide mineral surfaces through the formation of secondary Fe(III)-(oxy)hydroxide and Fe(III)-oxyhydroxysulfate phases that limit diffusive transport of oxidation products from sulfide mineral surfaces and therefore increase Fe(III) solubility within close proximity of these surfaces (38, 40).
The microbiology of Kam Kotia tailings and pore water chemistry prior to cover installation have been reported (41); Fortin et al. (41) enumerated aIOM and SRB in a 70-cm-deep profile composed of oxidized and acidic (pH 2 to 4) tailings. Iron oxidizers were found in greatest abundance (i.e., 105 cells g−1) at the surface and shallow subsurface (0 to 15 cm) of the uncovered tailings, with lower abundances detected at depth (≤103 aIOB g−1). Elevated concentrations of dissolved Fe and SO42− (up to ∼500 and ∼700 mM, respectively) in the surface pore waters, as well as a near depletion of the pyrite content in the tailings, coincided with the abundance of acidophilic IOM. Fortin et al. (41) also detected the presence of SRB (maximum, ∼104 SRB g−1) predominantly in the lower portion of the tailings profile. A comparison of our MPN results with those obtained prior to cover installation (41) clearly indicates a decrease in the abundance of iron-oxidizing prokaryotes in the Kam Kotia tailings after construction of the engineered cover system. We hypothesize that the reduced abundance of SRB (≤93 MPN g−1) was due to lowered sulfide oxidation rates (and thus sulfate availability to serve as a substrate). Improved pore water chemistry also indicated lower sulfide oxidation rates after the NIT was covered; maximum concentration of Fe decreased from ∼500 to 31.5 mM, and SO42− from ∼700 to 88.0 mM.
Many microorganisms capable of oxidizing organic carbon, such as Bacillus, Pseudomonas, Acidiphilium, Enterobacter, Alicyclobacillus, Acinetobacter, and Sulfobacillus, are typically observed in mill tailings and mine waste disposal areas (5, 23). Aerobic heterotrophs were abundant in the NIT cover system (especially in the surficial soil layer) and were also found in significant numbers in the underlying tailings despite the generally low organic carbon content. It has been reported that growth of both heterotrophs and SRB in tailings can be supported by dissolved organic carbon, such as exudates, lysates, and other compounds derived from autotrophic primary producers (42). The presence of viable aerobes beneath the cover system also suggests the existence of diverse microenvironments within the tailings. The counts of viable SRB, most of which are obligate anaerobes and highly sensitive to acidity, peaked in the anoxic environment immediately below the GCL. Gaseous O2 concentrations were determined above water table levels by use of gas chromatography. Due to a high water level at the site, oxygen data for most of the site 1 (where the MPN enumerations were performed) depth profile are not available. However, dissolved oxygen values, corresponding to measured gaseous O2 concentrations, dropped significantly below the GCL at sites 3 to 5 (from 8.74 mg liter−1 in the layers of the cover system above the GCL to 1.66 to 4.16 mg liter−1 in the layers below the GCL), indicating the potential for development of anaerobic microenvironments where SRB could thrive. Sequencing results indicated greatest relative abundance of SRB in the underlying tailings, with Desulfosporosinus being the most abundant sulfate-reducing genus (0.5% of total reads in the tailings). The culturable populations of SRB in the Kam Kotia tailings determined in this study were generally much less numerous than values for anoxic Cu/Zn tailings reported in literature, typically reaching abundances of 106 CFU g−1 dry weight (41, 43–45). Even higher maximum SRB numbers (109 CFU g−1 dry weight Cu/Zn tailings) were observed by Praharaj and Fortin (46). SRB populations in the uncovered Kam Kotia tailings determined by Fortin et al. (41) reached 104 cells g−1 of anoxic tailings, remaining lower than IOM. Both our MPN and sequence results suggest low potential for sulfate reduction in the Kam Kotia covered tailings, probably due to competition for limited organic carbon substrates between SRB and other heterotrophs.
Although elevated relative abundances of the SOM/IOM and SRB (determined by sequencing) were observed in the underlying tailings compared to the NIT cover system, the MPN values did not differ between the two sample groups. This suggests that even though DNA levels were elevated in the tailings, a portion of the iron- and sulfur-metabolizing prokaryotes might have been metabolically inactive. Bias in the culture or DNA-based methodology should also be considered a potential explanation for this finding. Interpretation of both sequence and MPN data requires caution due to limitations specific to each technique. In the case of MPN, growth and activity of the investigated groups of prokaryotes could be affected by different growth conditions in the laboratory compared to the field. Detection of DNA by sequencing, on the other hand, does not reflect cell viability.
Prokaryotic β-diversity within and among AMD sites changes in response to geochemical conditions, with pH being a primary parameter driving these changes (15, 17, 33, 47). Decreasing microbial diversity with decreasing pH has been previously observed in mill tailings (17, 21). Other environmental factors which control BACs in mine wastes are temperature, dissolved oxygen (DO) (48), concentrations of dissolved metal(loid)s (17, 18) and sulfate, and total organic carbon (37). All samples collected in this study had circumneutral pH, and the BACs were therefore defined predominantly by substrate concentrations.
Microorganisms play important roles in the catalysis of redox reactions and environmental geochemistry of mine wastes, and although a certain degree of caution with microbiological data interpretation is needed, the data obtained during this study can help elucidate linkages between BAC composition and the biogeochemistry of mine-impacted environments. Analyses of BACs in these environments provide a better understanding of the complex microbially catalyzed processes involved in the redox cycling of Fe, C, S, and other key elements.
Conclusion.
Nine years after a GCL cover was installed over acid-generating tailings in the North Impounded Tailings area at the Kam Kotia Mine, low relative abundances and low viable counts of iron and sulfur oxidizers were determined in the covered tailings. Although acidic drainage was reported prior to installation of the cover system (41), tailings pore water samples collected through the present study were characterized by bulk circumneutral pH and improved quality. Both our microbiological and geochemical results indicate that the remediation efforts have significantly lowered sulfide oxidation rates in the acid-generating tailings. Our results also show that the sulfidic mill tailings are characterized by diverse bacterial communities, and geochemical parameters other than pH can impact controls on the composition of the microbial communities inhabiting acid-generating mine wastes.
MATERIALS AND METHODS
Site description.
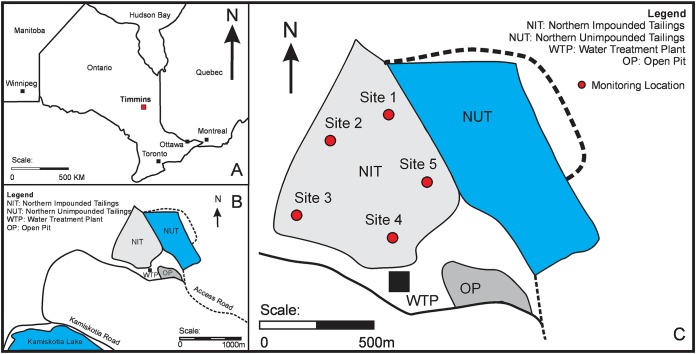
The Kam Kotia Mine is located 24 km northwest of Timmins, ON, Canada (48°36′ N, 81037′ W, Fig. 6A), and was the site of Cu (chalcopyrite, CuFeS2), Zn (sphalerite, [Zn, Fe]S), and secondary Ag and Au extraction from the early 1940s until 1972. Six million tons of sulfide-rich tailings were generated and were deposited without containment on 500 ha of land, producing AMD that severely impacted adjacent water bodies. The dominant acid-generating minerals are pyrite and minor amounts of chalcopyrite (49, 50). Near-surface tailings contained elevated concentrations of As, Cu, and Zn and had a pH of ∼2.5, which increased to pH 5 at depth (51). A five-phase plan of rehabilitation of the mine site was initiated in 2000 and includes construction and operation of a lime treatment plant, tailings and waste rock relocation and neutralization, construction of engineered covers over the AMD-generating materials, and revegetation of the impacted lands. The tailings were relocated into two main containment cells (Fig. 6B and C), where distinct cover strategies—a monolayer water cover in the Northern Unimpounded Tailings (NUT) area and a GCL cover in the NIT area—have been implemented to minimize AMD (49, 52). The GCL, consisting of bentonite clay between two layers of geotextile, was installed in 2008. The cover system design is a layer configuration consisting from the tailings upward of crushed waste rock-capillary break layer (thickness of 0.3 m; referred to as rock in this document), granular capillary break material (0.3 m; cushion sand), GCL, clay moisture-retaining layer (0.3 m; clay), granular protective layer (0.5 m; sand), and an organically amended vegetated layer (0.1 m; soil). Relocation, lime amendment, and submergence of acid-generating mine wastes to minimize further sulfide oxidation have previously been shown to be effective for neutralizing the acidity and sequestering metals in oxidized mill tailings (53, 54).
FIG 6.
(A) Location of Timmins, Ontario, where the Kam Kotia Mine is located. (B) Site schematic of the Kam Kotia property. (C) Location of five sampling sites (1 to 5) within the Northern Impounded Tailings (NIT) area.
Excavation and core collection.
Five locations (sites 1 to 5) in the NIT area (Fig. 6C) were excavated layer by layer through the engineered cover system. Core samples were collected in September/October 2017, using a Pionjar hammer drill as described by Starr and Ingleton (55). Monitoring equipment for long-term geochemical observations was installed, after which each excavated pit was infilled with the appropriate cover material. The GCL was then repaired using overlapping sections of new GCL material and bentonite clay. Aqueous samples for chemical analyses (described in “Aqueous and solid-phase geochemistry”) were collected at the time of solid sample collection.
Bulk samples collected through the cover system (total thickness of which is ∼1.5 m) and cored tailings samples were subjected to microbiological analyses.
Enumeration of viable microorganisms.
Samples for MPN enumerations were taken at site 1 (Fig. 6C). Bulk samples of each layer of the cover system were collected in triplicates (≥1 m apart), which were subsequently pooled into sterile 50-ml centrifuge tubes. Tailings were cored to a depth of 4.7 mbgs. The cores were cut into sections, and four 10-cm-long sections (representing 2.0 to 2.12, 2.56 to 2.67, 3.61 to 3.71, and 4.14 to 4.24 mbgs) were capped. Bulk and core subsamples were stored at 4°C until they were processed in the laboratory within 1 week of collection. The MPN technique (56, 57), a method providing quantitative data based on incidence after serial logarithmic dilutions (in this study, 10 to 1010 dilutions), was used to demonstrate metabolic potential via enumeration of different groups of culturable microorganisms. Cultivations in liquid media were performed in total volumes of 10 ml for each of five replicates for each sample. Plating onto solid medium was executed in duplicates. All cultivations were conducted at a laboratory temperature (∼23°C) without agitation.
To enumerate aIOM and aSOM, 1 g of sample was added to each of 5 replicate sterile test tubes, each containing 9.0 ml of autoclave-sterilized pH 2.0 (for aIOM) or 3.0 (for aSOM) basal salt medium, supplemented with trace elements (58). The IOM medium was supplemented with 20 mM ferrous iron (from 1 M 0.2-μm filter-sterilized stock solution of Fe2SO4·7H2O, pH of 2.0) and aSOM medium with approximately 2% (wt/vol) elemental sulfur (powder, sterilized at 105°C for 60 min). Ferrous iron oxidation by aIOM was monitored using the ferrozine colorimetric assay (59). Positive growth of aSOM was indicated by a 0.5-unit decrease in pH, determined using a pH electrode (Thermo Scientific Orion Star A321 pH portable meter; Thermo Fisher Scientific, USA). Samples of aSOM and aIOM enrichments in the highest positive MPN dilutions were plated onto selective solid overlay media described by Johnson and Hallberg (60), FeSo and iFeo, respectively. Plates were incubated at room temperature for 10 days, and selected sulfur- and iron-oxidizing isolates were identified by Sanger sequencing of their 16S rRNA genes after PCR amplification with the 27F/1387R primer pair. To enumerate SRB, a similar MPN protocol to that used by Gould et al. (61) to monitor iron-reducing bacteria was applied. A modified Postgate C medium (pH ∼7.5) (62), containing 2.92 g liter−1 Na lactate (60%) and 1.28 g liter−1 Na acetate and supplemented with resazurin as an anaerobic indicator, was used. Serum bottles (20 ml) were incubated in an anaerobic chamber for 6 weeks and regularly monitored for precipitation, indicating biogenic H2S production by sulfate reduction. To enumerate heterotrophs, samples (cover materials and tailings) were serially diluted in sterile deionized water and plated in duplicate on R2A agar (Sigma-Aldrich, USA; pH ∼7.2). Colonies were counted after a 5-day incubation period under aerobic conditions.
High-throughput amplicon sequencing of 16S rRNA genes.
To analyze BAC diversity using high-throughput amplicon sequencing of 16S rRNA genes, a second set of samples was collected at sites 1 to 5. Both bulk samples of the cover system materials and core samples of tailings were collected as described for MPN and stored at −20°C until they were processed in the laboratory. Cores were opened, subsampled into sterile 50-ml centrifuge tubes, and refrozen. Cores collected at site 2, which contained only sand overlying peat/organic material, were not subjected to high-throughput sequencing. Table 3 summarizes the layer types and depths at which the microbial diversity was analyzed at each location.
TABLE 3.
Summary of types and depths of environmental samples collected from the NIT at the Kam Kotia Mine, in which the overall microbial diversity was assessed using high-throughput amplicon sequencing of 16S rRNA genesa
| Sample type | Depth (m) at: |
|||
|---|---|---|---|---|
| Site 1b | Site 3 | Site 4 | Site 5 | |
| Soil | 0.05 | 0.05 | 0.05 | 0.05 |
| Sand | 0.3 | 0.3 | 0.3 | 0.4 |
| Clay | 0.75 | 0.65 | 0.7 | 0.95 |
| Cushion sand | 1.1 | 0.95 | 1 | 1.25 |
| Rock | 1.3 | 1.4 | 1.37 | 1.75 |
| Tailings | 2 | 2.16 | 1.8 | 1.97 |
| 2.67 | 2.44 | 2.02 | 2.18 | |
| 3.18 | 3.25 | 2.47 | 2.56 | |
| 3.67 | 3.43 | NA | NA | |
| 4.24 | NA | NA | NA | |
| 4.68 | NA | NA | NA | |
NA, not available.
Selected groups of viable microorganisms were also enumerated by the most probable number technique.
Genomic DNA was extracted in triplicates from bulk samples and core subsamples using the DNeasy PowerSoil kit (Qiagen Inc., Germany), following the manufacturer’s protocol. DNA concentration and quality were determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, USA) and Qubit dsDNA HS assay kit (Thermo Fisher Scientific, USA). Quality DNA was selected and stored at −20°C prior to submission for sequencing by Metagenom Bio Inc. (Canada) using the modified universal primers 515R/806R to amplify a 291-bp region targeting the V4 region of 16S rRNA genes in a broad range of archaeal and bacterial phylotypes (63). After DNA amplification, an Illumina MiSeq sequencing of the amplicons was performed.
The sequence data were analyzed using the mothur program v.1.39.5 updated 20 March 2017 (64), and the mothur MiSeq Standard Operating Procedure (64; https://www.mothur.org/wiki/MiSeq_SOP) from 7 March 2018. Three out of 108 total samples that contained fewer than 10,000 sequences were removed, after which the triplicate samples were merged. Chimeric sequences were discarded based on predictions by vsearch using the Silva database for 16S rRNA gene sequences (release 132 for mothur) as a reference. After splitting the sequences into bins and clustering within each bin, the sequences were clustered into OTUs at a 97% similarity level by a de novo picking method. Taxonomic annotation of individual OTUs was based on a mothur-formatted version of the Silva database (release 132). Several taxa (unknown, mitochondria, and eukaryotes) were not considered for further data analyses. Cyanobacteria, which are in the MiSeq SOP recommended to be removed, were included in further processing. To control variation resulting from an unequal number of sequences across samples, subsampling was performed for each sample after OTU generation at a rarefaction level based on the sample with the fewest number of sequences (24,652 sequences were detected in the tailings sample collected of 1.8 mbgs at site 4).
To assess how well the samples represented the larger environments, Good’s coverages (65) were generated using mothur (66). A distance matrix of the sequence data generated in mothur was used to calculate Bray-Curtis dissimilarity matrices (67), which were further visualized using two-dimensional nonmetric multidimensional scaling (2D-NMDS) in the software R. Unlike the Jaccard coefficient (68), which is a presence-absence index, the Bray-Curtis dissimilarity is used to quantify the compositional dissimilarity based on relative abundance of each OTU. Multivariate homogeneity of groups’ dispersions (R vegan function betadisper) was used to statistically assess dispersions of the BAC within each tested layer along the NIT profile. When the assumption of same “multivariate spread” among groups was met, the pairwise PERMANOVA (permutational multivariate analysis of variance; R vegan function adonis) was used to test for significant differences between groups.
A taxonomy file generated in mothur provided taxonomy for each layer of the cover system and several depths of tailings at each of the four investigated locations. The sequences were classified to the phylum level, and minor phyla with mean relative abundance < 1% were grouped and plotted together with major phyla (mean relative abundance > 1%) for each layer of the cover system and underlying tailings. Proportions of SRB, IOM, SOM, and IOM/SOM were obtained by screening the taxonomy file for prokaryotic genera (or, in a few instances, higher taxa when identification to the genus level was not possible) containing at least one species with the investigated metabolic trait. Again, relative abundances were averaged for each layer of the NIT system before plotting.
Aqueous and solid-phase geochemistry.
Chemical analyses of tailings pore water and solid phase were performed. Field measurements were completed on unfiltered aqueous samples for pH, using an Orion Ross Ultra combination pH electrode, and redox potential (adjusted to be relative to a standard hydrogen electrode [Eh values]) was measured using an Orion 9678 BN redox electrode, both coupled with an Orion pH/mV meter. Water samples were filtered (0.45 μm) and stored at 4°C before chemical analyses; major cations were analyzed using inductively coupled plasma-optical emission spectrometry (iCAP 6000 ICP-OES, Thermo Scientific; EPA Method 6010C, 2000 [69]) for preserved samples (acidified with HNO3) and inductively coupled plasma-mass spectrometry (ICP-MS X Series II, Thermo Scientific; EPA Method 6020A, 1998 [69]). Major anions were analyzed using ion chromatography (Dionex IC-CO3 system; EPA Method 300.0, 1993 [69]). Solid samples recovered from the NIT at the Kam Kotia Mine were analyzed for total carbon and total sulfur content using an Eltra CS-2000 carbon/sulfur analyzer coupled with an induction furnace (CS-800).
Data availability.
Illumina sequence data used in this study have been deposited in the European Nucleotide Archive (ENA) (accession no. PRJEB33459). Sequence reads for 16S rRNA amplicons of isolated bacteria have been deposited in GenBank (accession numbers MN982233 and MN982234).
Supplementary Material
ACKNOWLEDGMENTS
Funding for this research was provided by the Natural Sciences and Engineering Research Council of Canada (NSERC)’s Toward Environmentally Responsible Resource Extraction Network (TERRE-NET) program (grant NETGP 479708-15). We gratefully acknowledge financial and logistical support provided by the Ontario Ministry of Energy, Northern Development, and Mines.
In addition, we thank R. Purdon, S. Reitzel, and K. Westhaver with the Ministry for their assistance on this project. J. Bain (University of Waterloo [UW]), L. Groza (UW), and J. McBeth (University of Saskatchewan) provided technical assistance and advice. G. Schudel (Université du Québec en Abitibi-Témiscamingue) and F. Budimir (UW) provided assistance with chart generation.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Nordstrom DK, Alpers CN. 1999. Geochemistry of acid mine waters, p 133–160. In Plumlee GS, Logsdon MJ (ed), The environmental chemistry of mineral deposits, part A: processes, techniques, and health issues. Society of Economic Geologists, Littleton, CO. [Google Scholar]
- 2.Schippers A. 2004. Biogeochemistry of metal sulfide oxidation in mining environments, sediments, and soils. Geol Soc Am Spec Pap 379:49–62. [Google Scholar]
- 3.Baker BJ, Banfield JF. 2003. Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44:139–152. doi: 10.1016/S0168-6496(03)00028-X. [DOI] [PubMed] [Google Scholar]
- 4.Johnson DB, Hallberg KB. 2003. The microbiology of acidic mine waters. Res Microbiol 154:466–473. doi: 10.1016/S0923-2508(03)00114-1. [DOI] [PubMed] [Google Scholar]
- 5.Mendez-Garcia C, Pelaez AI, Mesa V, Sanchez J, Golyshina OV, Ferrer M. 2015. Microbial diversity and metabolic networks in acid mine drainage habitats. Front Microbiol 6:475. doi: 10.3389/fmicb.2015.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Southam G, Beveridge TJ. 1992. Enumeration of thiobacilli within pH-neutral and acidic mine tailings and their role in the development of secondary mineral soil. Appl Environ Microbiol 58:1904–1912. doi: 10.1128/AEM.58.6.1904-1912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blowes DW, Jambor JL, Hanton-Fong CJ, Lortie L, Gould WD. 1998. Geochemical, mineralogical and microbiological characterization of a sulphide-bearing carbonate-rich gold-mine tailings impoundment, Joutel, Quebec. Appl Geochem 13:687–705. doi: 10.1016/S0883-2927(98)00009-2. [DOI] [Google Scholar]
- 8.Benner SG, Gould WD, Blowes DW. 2000. Microbial populations associated with the generation and treatment of acid mine drainage. Chem Geol 169:435–448. doi: 10.1016/S0009-2541(00)00219-9. [DOI] [Google Scholar]
- 9.Lindsay MBJ, Condon PD, Jambor JL, Lear KG, Blowes DW, Ptacek CJ. 2009. Mineralogical, geochemical, and microbial investigation of a sulfide-rich tailings deposit characterized by neutral drainage. Appl Geochem 24:2212–2221. doi: 10.1016/j.apgeochem.2009.09.012. [DOI] [Google Scholar]
- 10.Diaby N, Dold B, Pfeifer H-R, Holliger C, Johnson DB, Hallberg KB. 2007. Microbial communities in a porphyry copper tailings impoundment and their impact on the geochemical dynamics of the mine waste. Environ Microbiol 9:298–307. doi: 10.1111/j.1462-2920.2006.01138.x. [DOI] [PubMed] [Google Scholar]
- 11.Tan GL, Shu WS, Hallberg KB, Li F, Lan CY, Zhou WH, Huang LN. 2008. Culturable and molecular phylogenetic diversity of microorganisms in an open-dumped, extremely acidic Pb/Zn mine tailings. Extremophiles 12:657–664. doi: 10.1007/s00792-008-0171-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhang HB, Shi W, Yang MX, Sha T, Zhao ZW. 2007. Bacterial diversity at different depths in lead-zinc mine tailings as revealed by 16S rRNA gene libraries. J Microbiol 45:479–484. [PubMed] [Google Scholar]
- 13.Huang LN, Zhou WH, Hallberg KB, Wan CY, Li J, Shu WS. 2011. Spatial and temporal analysis of the microbial community in the tailings of a Pb-Zn mine generating acidic drainage. Appl Environ Microbiol 77:5540–5544. doi: 10.1128/AEM.02458-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Touceda-González M, Álvarez-López V, Prieto-Fernández Á, Rodríguez-Garrido B, Trasar-Cepeda C, Mench M, Puschenreiter M, Quintela-Sabarís C, Macías-García F, Kidd PS. 2017. Aided phytostabilisation reduces metal toxicity, improves soil fertility and enhances microbial activity in Cu-rich mine tailings. J Environ Manage 186:301–313. doi: 10.1016/j.jenvman.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 15.Chen LX, Li JT, Chen YT, Huang LN, Hua ZS, Hu M, Shu WS. 2013. Shifts in microbial community composition and function in the acidification of a lead/zinc mine tailings. Environ Microbiol 15:2431–2444. doi: 10.1111/1462-2920.12114. [DOI] [PubMed] [Google Scholar]
- 16.Korehi H, Blöthe M, Schippers A. 2014. Microbial diversity at the moderate acidic stage in three different sulfidic mine tailings dumps generating acid mine drainage. Res Microbiol 165:713–718. doi: 10.1016/j.resmic.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Hua Z-S, Chen L-X, Kuang J-L, Li S-J, Shu W-S, Huang L-N. 2014. Correlating microbial diversity patterns with geochemistry in an extreme and heterogeneous environment of mine tailings. Appl Environ Microbiol 80:3677–3686. doi: 10.1128/AEM.00294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon MJ, Yang J-S, Lee S, Lee G, Ham B, Boyanov MI, Kemner KM, O'Loughlin EJ. 2015. Geochemical characteristics and microbial community composition in toxic metal-rich sediments contaminated with Au–Ag mine tailings. J Hazard Mater 296:147–157. doi: 10.1016/j.jhazmat.2015.04.035. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Bond PL, Van Nostrand JD, Zhou J, Huang L. 2015. From lithotroph- to organotroph-dominant: directional shift of microbial community in sulphidic tailings during phytostabilization. Sci Rep 5:12978. doi: 10.1038/srep12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruneel O, Mghazli N, Hakkou R, Dahmani I, Maltouf AF, Sbabou L. 2017. In‑depth characterization of bacterial and archaeal communities present in the abandoned Kettara pyrrhotite mine tailings (Morocco). Extremophiles 21:671–685. doi: 10.1007/s00792-017-0933-3. [DOI] [PubMed] [Google Scholar]
- 21.Mendez M, Neilson J, Maier R. 2008. Characterization of a bacterial community in an abandoned semiarid lead-zinc mine tailing site. Appl Environ Microbiol 74:3899–3907. doi: 10.1128/AEM.02883-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golyshina OV. 2011. Environmental, biogeographic, and biochemical patterns of archaea of the family Ferroplasmaceae. Appl Environ Microbiol 77:5071–5078. doi: 10.1128/AEM.00726-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schippers A, Breuker A, Blazejak A, Bosecker K, Kock D, Wright TL. 2010. The biogeochemistry and microbiology of sulfidic mine waste and bioleaching dumps and heaps, and novel Fe(II)-oxidizing bacteria. Hydrometallurgy 104:342–350. doi: 10.1016/j.hydromet.2010.01.012. [DOI] [Google Scholar]
- 24.NAOMI (National Orphaned/Abandoned Mines Initiative). 2009. Performance report 2002-2008. National Orphaned/Abandoned Mines Initiative, Ottawa, Ontario, Canada. [Google Scholar]
- 25.Blowes DW, Ptacek CJ, Jambor JL, Weisener CG, Paktunc D, Gould WD, Johnson DB. 2014. The geochemistry of acid mine drainage, p 131–190. In Holland HD, Turekian KK (ed) Treatise on geochemistry, 2nd ed Elsevier, San Diego, CA, USA. [Google Scholar]
- 26.Cowan WR, Mackasey WO. 2006. Rehabilitating abandoned mines in Canada: a toolkit of funding options. Prepared for National Orphaned/Abandoned Mines Initiative. Cowan Minerals Ltd., Sudbury, Ontario, Canada. [Google Scholar]
- 27.Mbonimpa M, Aubertin M, Aachib M, Bussiere B. 2003. Diffusion and consumption of oxygen in unsaturated cover materials. Can Geotech J 40:916–932. doi: 10.1139/t03-040. [DOI] [Google Scholar]
- 28.INAP (International Network for Acid Prevention). 2014. The global acid rock drainage guide. International Network for Acid Prevention; http://www.gardguide.com. Accessed 9 March 2018. [Google Scholar]
- 29.Anawar HM. 2015. Sustainable rehabilitation of mining waste and acid mine drainage using geochemistry, mine type, mineralogy, texture, ore extraction and climate knowledge. J Environ Manage 158:111–121. doi: 10.1016/j.jenvman.2015.04.045. [DOI] [PubMed] [Google Scholar]
- 30.Patterson BM, Robertson BS, Woodbury RJ, Talbot B, Davis GB. 2006. Long-term evaluation of a composite cover overlaying a sulfidic tailings facility. Mine Water Environ 25:137–145. doi: 10.1007/s10230-006-0125-3. [DOI] [Google Scholar]
- 31.Bouazza A. 2002. Geosynthetic clay liners. Geotex Geomembranes 20:3–17. doi: 10.1016/S0266-1144(01)00025-5. [DOI] [Google Scholar]
- 32.Shackelford CD, Sevick GW, Eykholt GR. 2010. Hydraulic conductivity of geosynthetic clay liners to tailings impoundment solutions. Geotex Geomembranes 28:149–162. doi: 10.1016/j.geotexmem.2009.10.005. [DOI] [Google Scholar]
- 33.Diaby N, Dold B, Rohrbach E, Holliger C, Rossi P. 2015. Temporal evolution of bacterial communities associated with the in situ wetland-based remediation of a marine shore porphyry copper tailings deposit. Sci Total Environ 533:110–121. doi: 10.1016/j.scitotenv.2015.06.076. [DOI] [PubMed] [Google Scholar]
- 34.Lopo M, Montagud A, Navarro E, Cunha I, Zille A, de Cordoba PF, Moradas-Ferreira P, Tamagnini P, Urchueguía JF. 2012. Experimental and modeling analysis of Synechocystis sp. PCC 6803 growth. J Mol Microbiol Biotechnol 22:71–82. doi: 10.1159/000336850. [DOI] [PubMed] [Google Scholar]
- 35.Olsson-Francis K, Cockell CH. 2010. Use of cyanobacteria for in-situ resource use in space applications. Planet Space Sci 58:1279–1285. doi: 10.1016/j.pss.2010.05.005. [DOI] [Google Scholar]
- 36.dos Santos AM, Vieira KR, Sartori RB, dos Santos AM, Queiroz MI, Zepka LQ, Jacob-Lopes E. 2017. Heterotrophic cultivation of cyanobacteria: study of effect of exogenous sources of organic carbon, absolute amount of nutrients, and stirring speed on biomass and lipid productivity. Front Bioeng Biotechnol 5:1–7. doi: 10.3389/fbioe.2017.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao E, Krumins V, Dong Y, Xiao T, Ning Z, Xiao Q, Sun W. 2016. Microbial diversity and community structure in an antimony-rich tailings dump. Appl Microbiol Biotechnol 100:7751–7763. doi: 10.1007/s00253-016-7598-1. [DOI] [PubMed] [Google Scholar]
- 38.Dockrey JW, Lindsay MBJ, Mayer KU, Beckie RD, Norlund KLI, Warren LA, Southam G. 2014. Acidic microenvironments in waste rock characterized by neutral drainage: bacteria–mineral interactions at sulfide surfaces. Minerals 4:170–190. doi: 10.3390/min4010170. [DOI] [Google Scholar]
- 39.Nordstrom DK, Blowes DW, Ptacek CJ. 2015. Hydrogeochemistry and microbiology of mine drainage: an update. Appl Geochem 57:3–16. doi: 10.1016/j.apgeochem.2015.02.008. [DOI] [Google Scholar]
- 40.Mielke RE, Pace DL, Porter T, Southam G. 2003. A critical stage in the formation of acid mine drainage: colonization of pyrite by Acidithiobacillus ferrooxidans under pH-neutral conditions. Geobiology 1:81–90. doi: 10.1046/j.1472-4669.2003.00005.x. [DOI] [Google Scholar]
- 41.Fortin D, Davis B, Beveridge TJ. 1996. Role of Thiobacillus and sulfate-reducing bacteria in iron biocycling in oxic and acidic mine tailings. FEMS Microbiol Ecol 21:11–24. doi: 10.1111/j.1574-6941.1996.tb00329.x. [DOI] [Google Scholar]
- 42.Johnson DB. 1998. Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol Ecol 27:307–317. doi: 10.1111/j.1574-6941.1998.tb00547.x. [DOI] [Google Scholar]
- 43.Fortin D, Beveridge TJ. 1997. Microbial sulfate reduction within sulfidic mine tailings: formation of diagenetic Fe sulfides. Geomicrobiol J 14:1–21. doi: 10.1080/01490459709378030. [DOI] [Google Scholar]
- 44.Fortin D, Roy M, Rioux J-P, Thibault PJ. 2000. Occurrence of sulfate-reducing bacteria under a wide range of physico-chemical conditions in Au and Cu-Zn mine tailings. FEMS Microbiol Ecol 33:197–208. doi: 10.1111/j.1574-6941.2000.tb00742.x. [DOI] [PubMed] [Google Scholar]
- 45.Fortin D, Rioux J-P, Roy M. 2002. Geochemistry of iron and sulfur in the zone of microbial sulfate reduction in mine tailings. Water Air Soil Pollut Focus 2:37–56. doi: 10.1023/A:1019983024680. [DOI] [Google Scholar]
- 46.Praharaj T, Fortin D. 2004. Indicators of microbial sulfate reduction in acidic sulfide-rich mine tailings. Geomicrobiol J 21:457–467. doi: 10.1080/01490450490505428. [DOI] [Google Scholar]
- 47.Kuang J-L, Huang L-N, Chen L-X, Hua Z-S, Li S-J, Hu M, Li J-T, Shu W-S. 2013. Contemporary environmental variation determines microbial diversity patterns in acid mine drainage. ISME J 7:1038–1050. doi: 10.1038/ismej.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Méndez-García C, Mesa V, Sprenger RR, Richter M, Diez MS, Solano J, Bargiela R, Golyshina OV, Manteca Á, Ramos JL, Gallego JR, Llorente I, Martins dos Santos VAP, Jensen ON, Peláez AI, Sánchez J, Ferrer M. 2014. Microbial stratification in low pH oxic and suboxic macroscopic growths along an acid mine drainage. ISME J 8:1259–1274. doi: 10.1038/ismej.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.SENES Consultants Limited, Lakefield Research Limited, ESG International Inc., Denison Environmental Services. 2000. Final report, Kam Kotia mine property rehabilitation study – phase 1. Company report prepared for Ontario Ministry of Energy, Northern Development and Mines. SENES, Richmond Hill, Ontario, Canada. [Google Scholar]
- 50.Van Huyssteen E. 1995. Tailings sampling results referenced in Kam Kotia E.L.O. Environmental summary report prepared by Klohn-Crippen. CANMET, Natural Resources Canada, Ottawa, Ontario, Canada. [Google Scholar]
- 51.Winterhalder K. 1992. The experimental use of various covers and native transplants for the revegetation of the Kam-Kotia tailings site, Timmins, Ontario. In Achieving Land Use Potential through Reclamation: Proceedings of the 9th Annual National Meeting. American Society for Surface Mining and Reclamation, Duluth, Minnesota. [Google Scholar]
- 52.Wardrop Engineering Inc. 2005. Design brief Kam Kotia mine – north impounded tailings composite cover. Ontario Ministry of Energy, Northern Development and Mines, Thunder Bay, Ontario, Canada. [Google Scholar]
- 53.Davis A, Eary LE, Helgen S. 1999. Assessing the efficacy of lime amendment to geochemically stabilize mine tailings. Environ Sci Technol 33:2626–2632. doi: 10.1021/es9810078. [DOI] [Google Scholar]
- 54.Morin KA, Hutt NM. 2001. Relocation of net-acid-generating waste to improve post-mining water chemistry. Waste Manag 21:185–190. doi: 10.1016/s0956-053x(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 55.Starr RC, Ingleton RA. 1992. A new method for collecting core samples without a drilling rig. Ground Water Monit Remediat 12:91–95. doi: 10.1111/j.1745-6592.1992.tb00413.x. [DOI] [Google Scholar]
- 56.Cochran WG. 1950. Estimation of bacterial densities by means of the “most probable number.” Biometrics 6:105–116. doi: 10.2307/3001491. [DOI] [PubMed] [Google Scholar]
- 57.Garthright WE, Blodgett RJ. 2003. FDA’s preferred MPN methods for standard, large or unusual tests, with a spreadsheet. Food Microbiol 20:439–445. doi: 10.1016/S0740-0020(02)00144-2. [DOI] [Google Scholar]
- 58.Ňancucheo I, Rowe OF, Hedrich S, Johnson DB. 2016. Solid and liquid media for isolating and cultivating acidophilic and acid-tolerant sulfate-reducing bacteria. FEMS Microbiol Lett 363:1–6. doi: 10.1093/femsle/fnw083. [DOI] [PubMed] [Google Scholar]
- 59.Stookey LL. 1970. Ferrozine–a new spectrophotometric reagent for iron. Anal Chem 42:779–781. doi: 10.1021/ac60289a016. [DOI] [Google Scholar]
- 60.Johnson DB, Hallberg KB. 2007. Techniques for detecting and identifying acidophilic mineral-oxidising microorganisms, p 237–261. In Rawlings DE, Johnson DB (ed), Biomining. Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- 61.Gould WD, Stitchbury M, Francis M, Lortie L, Blowes DW. 2003. An MPN method for the enumeration of iron-reducing bacteria In Mining and the Environment III Laurentian University, Sudbury, Ontario, Canada. [Google Scholar]
- 62.Postgate JR. 1984. The sulfate reducing bacteria. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 63.Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R. 2015. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–364. doi: 10.2307/2333344. [DOI] [Google Scholar]
- 66.Schloss PD, Westcott SL, Ryabin R, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bray J, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 68.Jaccard P. 1901. Étude comparative de la distribution florale dans une portion des Alpes et des Jura. (In French.) Bull Soc Vaud Sci Nat 37:547–579. doi: 10.5169/seals-266450. [DOI] [Google Scholar]
- 69.US EPA 2012. Selected analytical methods for environmental remediation and recovery (SAM) 2012. EPA/600/R-12/555, 2012. US Environmental Protection Agency, Washington, DC, USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Illumina sequence data used in this study have been deposited in the European Nucleotide Archive (ENA) (accession no. PRJEB33459). Sequence reads for 16S rRNA amplicons of isolated bacteria have been deposited in GenBank (accession numbers MN982233 and MN982234).