It is highly desirable to produce biologics for diagnoses and therapeutic interventions on demand that could be used in a variety of settings. Among the many biologics, Nbs have attracted attention due to their small size, thermal stability, and broad utility in diagnoses, therapies, and fundamental research. Nbs originate from antibodies found in camelids, and >10 companies have invested in Nbs as potential drugs. Here, we present a system using cells of the bacterium Bacillus subtilis as a versatile platform for production of Nbs and then antigen detection via customized affinity columns. Importantly, B. subtilis carrying engineered genes for Nbs can form spores, which survive for years in a desiccated state. However, upon rehydration and exposure to nutrients, spores rapidly transition to growing cells which secrete encoded Nbs, thus allowing their manufacture and purification.
KEYWORDS: Bacillus subtilis, spores, nanobody, protein secretion, monoclonal antibodies, recombinant-protein production, synthetic biology
ABSTRACT
There is a growing need for a highly stable system to allow the production of biologics for diagnoses and therapeutic interventions on demand that could be used in extreme environments. Among the variety of biologics, nanobodies (Nbs) derived from single-chain variable antibody fragments from camelids have attracted great attention in recent years due to their small size and great stability with translational potentials in whole-body imaging and the development of new drugs. Intracellular expression using the bacterium Escherichia coli has been the predominant system to produce Nbs, and this requires lengthy steps for releasing intracellular proteins for purification as well as removal of endotoxins. Lyophilized, translationally competent cell extracts have also been explored as offering portability and long shelf life, but such extracts may be difficult to scale up and suffer from batch-to-batch variability. To address these problems, we present a new system to do the following: (i) engineer the spore-forming bacterium Bacillus subtilis to secrete Nbs that can target small molecules or protein antigens on mammalian cells, (ii) immobilize Nbs containing a cellulose-binding domain on a cellulose matrix for long-term storage and small-molecule capturing, (iii) directly use Nb-containing bacterial supernatant fluid to perform protein detection on cell surfaces, and (iv) convert engineered B. subtilis to spores that are resistant to most environmental extremes. In summary, our work may open a new paradigm for using B. subtilis as an extremely stable microbial factory to produce Nbs with applications in extreme environments on demand.
IMPORTANCE It is highly desirable to produce biologics for diagnoses and therapeutic interventions on demand that could be used in a variety of settings. Among the many biologics, Nbs have attracted attention due to their small size, thermal stability, and broad utility in diagnoses, therapies, and fundamental research. Nbs originate from antibodies found in camelids, and >10 companies have invested in Nbs as potential drugs. Here, we present a system using cells of the bacterium Bacillus subtilis as a versatile platform for production of Nbs and then antigen detection via customized affinity columns. Importantly, B. subtilis carrying engineered genes for Nbs can form spores, which survive for years in a desiccated state. However, upon rehydration and exposure to nutrients, spores rapidly transition to growing cells which secrete encoded Nbs, thus allowing their manufacture and purification.
INTRODUCTION
Living cellular systems, such as bacteria, yeasts, plants, and mammalian cells, have become the workhorse for production of a myriad of biologics and compounds. More recently, lyophilized cell extracts (LCE) from living cells have enabled decentralized pharmaceutical production and room temperature storage, which offer opportunities for safe deployment of genetically encoded materials (1, 2). However, current LCE systems have not been proven to be stable for several years without loss of activity at room temperature, and there is no evidence that these systems are sufficiently resistant to various harsh conditions to allow them to perform their functions in extreme environments (1).
To address the issues noted above, in this work, we have proposed to engineer and insert DNA coding for desired biologics in cells of the bacterium Bacillus subtilis, which readily forms highly resistant spores, thus allowing long-term “gene” storage with excellent stability. Notably, these spores are highly resistant to desiccation and to radiation at doses that exceed the threshold for damage to humans, and spores have been studied for decades to elucidate the biological mechanisms underlying their extreme resistance (Fig. 1) (3–6). Despite their dormancy, spores can rapidly return to vegetative growth in 1 to 2 h via germination and outgrowth and then proliferate in the presence of nutrients (7). Importantly, in our work, we have utilized the protein expression and secretory systems of vegetative B. subtilis cells to design a pipeline (see Results) to produce nanobodies (Nbs), which are a distinct class of proteins derived from single-domain antibody fragments found in Camelidae (e.g., camels and llamas) (Fig. 1) (8). These Nbs will be produced with an affinity tag, allowing their purification for eventual antigen detection or purification as well. Nbs have small sizes (12 to 17 kDa), high solubility, and high thermal stability compared to conventional antibodies (8, 9). Due to their small sizes, Nbs are particularly suited for single-molecule imaging at nanoscale resolution, clinical diagnostics such as positron emission tomography-computed tomography imaging, and therapeutics for a variety of diseases (10–12). Currently, most, if not all, Nbs are produced intracellularly in microbial cells, predominantly Escherichia coli (10, 11, 13). However, Nb isolation from E. coli cells requires lysis of the bacteria, which may significantly prolong the downstream processing time. Additionally, E. coli contains endotoxins, which can cause multiple undesirable effects in humans, and thus E. coli extracts require complete removal of endotoxins in the development of Nbs produced in this organism as therapeutic biologics (14–16).
FIG 1.
A scheme to illustrate the structure and properties of B. subtilis spores as stable living systems to manufacture camelid-derived Nbs on demand, as these spores are extremely resistant to space vacuum, cosmic radiation, temperature fluctuations, UV (if shielded), or acidic pH.
In addition to those in E. coli, production of Nbs has been explored in yeast and Brevibacillus expression systems (17, 18) or through display of Nbs on the surface of B. subtilis spores for sensing (http://2016.igem.org/Team:Freiburg/Binding), but using B. subtilis to secrete Nbs into the medium has not previously been considered. One simple advantage in using this Gram-positive bacterium is that it contains minimal endotoxin levels (19), and thus extracellular production of Nbs with extremely low endotoxin levels in B. subtilis could simplify downstream purification (19). B. subtilis is well known to secrete many of the proteins it synthesizes (20, 21). However, many eukaryotic proteins (e.g., mammalian) are poorly produced by the Sec-dependent secretion pathway in B. subtilis, likely because this secretion system requires an unfolded state of target proteins during export of proteins from the cytoplasm to the environment (20), where abundant proteases are also secreted by B. subtilis (21). Although protease-deficient B. subtilis strains have been developed to mitigate the problem of proteolysis, secreting some intact mammalian proteins in high yields by B. subtilis remains problematic, possibly due to misfolding of target proteins (22). In planning this work, however, we speculated that the high thermostability and relatively small size of Nbs may render these proteins amenable to the B. subtilis secretory system. Indeed, we demonstrate that a panel of four different Nbs specific for small molecules (caffeine and methotrexate) or eukaryotic cell surface-associated protein antigens, cytotoxic-T-lymphocyte-associated protein 4 (CTLA-4), and programmed death-ligand 1 (PD-L1) can be readily secreted in an intact state and at high yields (up to 20 mg protein per liter in a shake flask mode) by B. subtilis. In addition, by fusing an anti-caffeine Nb with a cellulose-binding domain (CBD), the Nb can be immobilized on cellulose-based materials, including thin paper and columns, for long-term storage or small molecule capturing on a low-cost substrate. Importantly, direct secretion of Nbs in the B. subtilis culture supernatant fluid can potentially enable the rapid detection of antigen targets without purification. In turn, the latter advances may also pave the way for the development of low-cost, portable and extremely stable detection systems, and even biologics production systems, all in extreme environments.
RESULTS
Schematic pipeline for expression of Nbs, their purification, and use for antigen purification as well.
The overall scheme for ultimate purification of antigens is outlined in Fig. 2. The pipeline begins with transformation of B. subtilis with the gene for the Nb selected plus appropriate signals for transcription and translation of the gene in growing cells and excretion of the protein. The secreted Nb will contain either a His or a CBD tag, allowing easy purification of the protein on an appropriate affinity column. The column-bound protein can then be used to purify the Nb’s antigen, or the Nb can be eluted for use in antigen detection. The various individual steps in this pipeline are described and discussed below.
FIG 2.
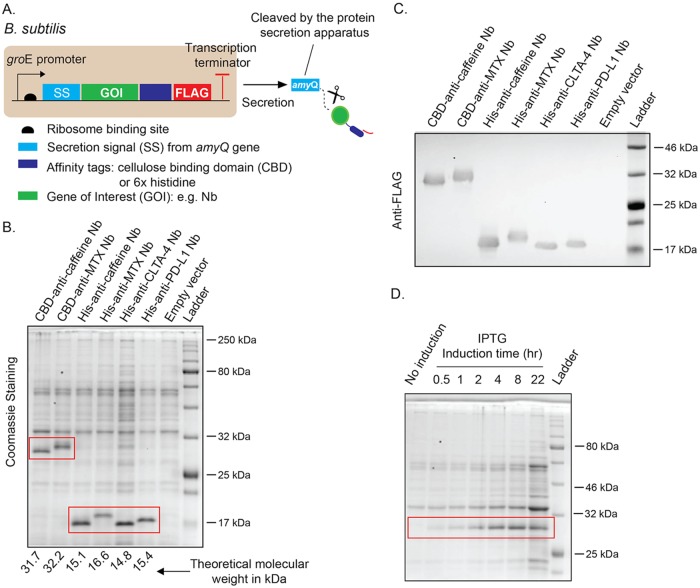
A pipeline for Nb production, including dry spore storage, inoculation of spores into medium to allow for spore germination and outgrowth, bacterial growth, and protein secretion and purification. Vegetative cells of B. subtilis are transformed with plasmids that encode genes of interest (GOI), such as a particular Nb, and the Nb is fused with a cellulose binding domain (CBD) to allow for cellulose-based immobilization or with a 6×Histidine tag (His-tag) to enable metal affinity purification on a cobalt nitriloacetic acid (NTA) resin. The column-bound protein can then be used to purify the Nb’s antigen (e.g., small molecules), or the Nb can be eluted for use in antigen detection via conjugation of fluorescent dyes or radioisotope labeling. For long-term storage, B. subtilis is induced to form spores, and when the Nb is needed, the spores can be inoculated in culture medium to enable spore germination, outgrowth, and subsequent vegetative growth and then synthesis and secretion of the Nb via the Sec-dependent protein secretion pathway. amyQ, protein secretion signal peptide; groE, a strong promoter from the B. subtilis groE operon, which has been converted into an IPTG-inducible promoter by addition of the lac operator from E. coli.
Extracellular secretion of Nbs in B. subtilis.
B. subtilis secretes proteases into the culture supernatant, which can dramatically lower the yields and quality of secreted proteins. To minimize this problem in our work, we used a modified B. subtilis strain, WB800N, which is deficient in eight extracellular proteases (23). The final fusion gene cassette was cloned into a vector named pHT43 (24), which contains a strong promoter that is derived from the B. subtilis groE operon and has been converted into an isopropyl-β-d-thiogalactoside (IPTG)-inducible promoter by addition of the lac operator from E. coli. Downstream of the promoter region are the ribosome binding site (RBS) and the amyQ secretion signal sequence (Fig. 3A). Because the amyQ secretion signal sequence requires the bacterial Sec pathway to export proteins extracellularly, exported proteins, such as Nbs, must first be unfolded to thread through the protein export machinery anchored on the bacterial membrane. As a result, many mammalian recombinant proteins have failed to be secreted at detectable levels due to degradation or misfolding. However, we hypothesized that recombinant proteins that are highly thermally stable and/or have relatively small molecular weights, such as Nbs, may not be subject to this problem in the Sec-dependent secretion pathway in B. subtilis. To confirm this hypothesis, we chose to produce four different Nbs as follows: (i) anti-caffeine, (ii) anti-methotrexate (MTX), (iii) anti-mouse CTLA-4, and (iv) anti-mouse PD-L1. The first two Nbs bind small molecules with potential applications in small-molecule purification, while the latter two detect surface antigens on immune cells and tumor cells with potential clinical diagnostic applications. Notably, these four Nbs, two with the CBD tag and two with the His tag, were readily detected in the supernatant of bacterial cultures after IPTG induction for 6 h in comparison to that of empty vector-transformed bacteria (Fig. 3B). The Nb yields were estimated to be 15 to 20 mg from one liter of bacterial culture in a shake flask mode, which is comparable to yields in shake-flask-based host systems for Nb production in E. coli (25). To confirm that the protein bands detected on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were truly the proteins of interest, we performed Western blotting using an antibody specific for the FLAG epitope cloned in the fusion proteins. As shown in Fig. 3C, the fusion proteins’ migration positions on SDS-PAGE were those expected based on their calculated molecular weights shown in Fig. 3B.
FIG 3.
(A) Scheme to use B. subtilis to produce camelid-derived Nbs. (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of B. subtilis strains engineered to secrete four different Nbs upon growth starting from dormant spores. The Nbs are as follows: (i) anti-caffeine, (ii) anti-methotrexate (MTX), (iii) anti-cytotoxic-T-lymphocyte-associated protein 4 (CTLA-4; mouse), and (iv) anti-programmed death-ligand 1 (PD-L1; mouse). For immobilization on cellulose paper with the purpose of filter paper-based tests or Nb-mediated small-molecule capturing, the Nbs are fused with a CBD. Alternatively, the Nbs are fused to a His tag to enable metal affinity purification. The image shown is an SDS-PAGE gel of total proteins secreted by B. subtilis and precipitated by trichloroacetic acid (TCA) after induction with 1 mM IPTG overnight as described in Materials and Methods. The red rectangles indicate the target proteins, which are absent in the noninduced culture. The theoretical molecular weight of each fusion protein is indicated at the bottom of the SDS-PAGE gel. (C) Western blotting to confirm the presence of target proteins. Recombinant proteins are fused with a FLAG epitope for detection by an anti-FLAG antibody. (D) Kinetics of B. subtilis secretion of an anti-caffeine Nb fused with a CBD after IPTG induction. Bacteria expressing the Nb fusion were induced by 1 mM IPTG. A volume of 0.5 ml culture supernatant from each indicated time point was harvested and precipitated by TCA for SDS-PAGE. The recombinant protein (∼31.7 kDa) was readily detectable 30 min after IPTG induction. The red rectangle indicates the target protein.
Extracellular expression bypasses the need for lysing bacteria to extract proteins but may have relatively slower production rates than those of intracellular expression due to the requirement for proteins to unfold and refold when using the Sec-dependent secretion pathway. To understand the kinetics of Nb secretion from B. subtilis, we collected bacterial culture supernatants at different times after addition of the inducer IPTG. Following trichloroacetic acid (TCA) precipitation of total proteins from the supernatant, Nbs with the His tag (PD-L1 and CTLA-4) (see Fig. S1 in the supplemental material) or the CBD tag (caffeine) were detectable by SDS-PAGE as early as 0.5 h after induction and levels increased up to 4 h (Fig. 3D). These results suggest that when an inducible protein secretion system is desirable in B. subtilis, protein secretion can occur within a 1-h time frame, which can enable the on-demand production of Nb-based biologics.
The fusion protein consisting of a CBD and an Nb is functionally active following secretion from B. subtilis.
Immobilizing thermostable Nbs on a substrate such as cellulose with low cost and a long shelf life may have implications for the detection of small molecules using hapten-specific Nbs under extreme conditions (26, 27). To this end, we turned our attention to the CBD from the cipA gene in the bacterium Clostridium thermocellum. This CBD has been shown to be resistant to denaturation at high temperatures (Tm = 70°C) due to the thermophilic nature of C. thermocellum (28). While this CBD has been used as an affinity tag to enable protein immobilization and purification, it has been predominantly expressed inside E. coli to make recombinant fusion proteins, which necessitates lysis of bacteria via enzymes (e.g., lysozyme). In contrast, the ease of producing recombinant proteins extracellularly by the Sec-dependent secretory pathway in B. subtilis motivated us to produce fusion proteins consisting of a CBD and an Nb. We speculated that the small size and high thermostability of the CBD (∼17 kDa) may allow for fast refolding and resistance to protease-mediated degradation, which has been a problem for heterologous protein expression using the Sec-dependent secretion system in B. subtilis.
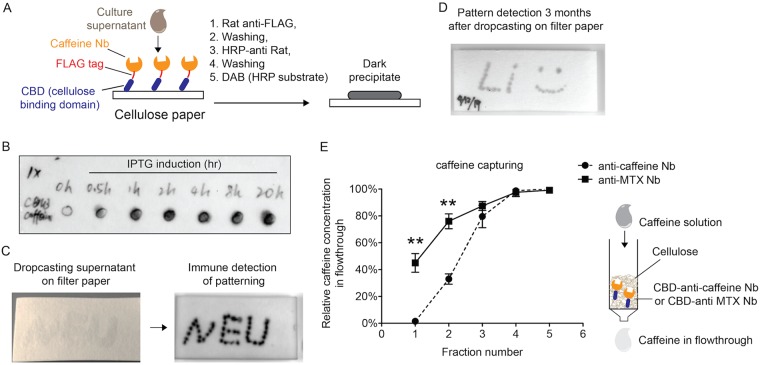
To determine whether CBD fusion proteins were bifunctional (i.e., cellulose binding and Nb-specific target recognition), we focused on the CBD–anti-caffeine Nb as a proof of concept. We first spotted supernatant fluid containing secreted fusion proteins on a piece of cellulose filter paper (Fig. 4B, circled areas); upon air drying, the paper was stained with a rat antibody against the FLAG epitope in the fusion protein, followed by an anti-rat secondary antibody conjugated with horseradish peroxidase (HRP) and HRP substrates, yielding a black precipitate (Fig. 4A). As shown in Fig. 4B, black staining inside the circles indicated binding of the fusion protein to the cellulose paper, and the protein abundance in the supernatant increased with induction time as evidenced by the increase in intensity of the black staining. As a negative control, no fusion protein was detected in the supernatant from uninduced (i.e., 0 h) bacteria. Alternatively, we generated a pattern of “NEU” (Northeastern University) on cellulose paper using the same supernatant and detected the pattern by immunoblotting (Fig. 4C). We also demonstrated the stability of the immobilized fusion proteins 3 months after application to and drying the fusion protein on filter paper using a different pattern, with the original sample still detectable by immunoblotting (Fig. 4D). Therefore, we have successfully utilized B. subtilis to secrete a CBD-based fusion protein that can be immobilized on cellulose, with potential applications in biosensing or biologic storage based on cellulose paper.
FIG 4.
(A) Scheme to detect recombinant fusion proteins after dropping bacterial supernatant on cellulose-based filter paper. (B) Confirmation of binding of fusion proteins to cellulose filter paper by adapting a Western blotting protocol. Bacterial supernatants containing the CBD-anti-caffeine Nb fusion proteins were harvested at different times after 1 mM IPTG induction, spotted into circles, and then detected with anti-FLAG serum as described in panel A. (C) Alternatively, the supernatant from a 4 h induction was patterned on filter paper as shown by the image on the left. Proteins were then detected using an anti-FLAG antibody (image on the right) as described in panel A. (D) CBD-anti-caffeine Nb fusion protein remained immobilized and stable 3 months after drying the protein-containing supernatant on cellulose-based filter paper after analysis as described in panel A. (E) Percentage of remaining caffeine in five continuous flowthrough fractions (100 μl per fraction) relative to the starting concentration (150 μg/ml in PBS). A customized syringe column was prepacked with ∼100 μl regenerated amorphous cellulose (RAC) and then loaded with bacterial supernatant containing the CBD-anti-caffeine Nb or the CBD-anti-MTX Nb (negative control) until it was saturated as determined by running flowthrough fractions in SDS-PAGE. After extensive washing of the RAC columns with PBS, 150 μg/ml caffeine in PBS was loaded, and caffeine concentrations in five flowthrough fractions were measured at 273 nm and results are given as the mean ± standard deviation (SD) (n = 3), with all values relative to the original caffeine concentration applied to the columns. **, P < 0.01.
To confirm that the Nb component in the CBD-Nb fusion protein retained its target recognition ability, we devised a chromatography column prototype by packing a 1-ml syringe with 0.1 ml regenerated amorphous cellulose (RAC) (∼50 mg dry weight) (Fig. 4E). RAC was prepared by hydrolyzing cellulose microcrystals with 85% phosphoric acid, such that the hydrolyzed cellulose has a larger surface area to maximize its binding to the CBD. Next, we loaded the column with bacterial supernatant containing the fusion protein and monitored the saturation of the column by detecting the presence of CBD-anti-caffeine Nb from the flowthrough via SDS-PAGE (see Fig. S2 in the supplemental material). By quantifying the amount of CBD-anti-caffeine Nb in the original supernatant and the flowthrough fractions, in which the fusion proteins were found to saturate the column, it was estimated that ∼0.5 mg of CBD-anti-caffeine Nb was bound to 0.1 ml RAC (∼50 mg dry weight). These procedures in principle converted a cellulose-based column to an affinity purification system, where the CBD-Nb fusion protein serves as a bridge between a low-cost, stable immobile phase (i.e., the cellulose substrate) and molecules of interest from the mobile phase (e.g., caffeine).
As a proof of concept for purification of Nb antigens, we applied 150 μg/ml caffeine solution prepared in phosphate-buffered saline (PBS) to an RAC column carrying immobilized CBD-anti-caffeine Nb or CBD-anti-MTX Nb (a negative control), collected five flowthrough fractions (100 μl per fraction), and analyzed the fractions with a UV spectrophotometer. The measurements showed that caffeine levels in the mobile phase were significantly reduced when passing through the RAC column that carried the CBD-anti-caffeine Nb in comparison to those of the CBD-anti-MTX Nb, thus demonstrating specific target recognition by the secreted CBD-anti-caffeine Nb (Fig. 4E) compared to nonspecific caffeine adsorption by the RAC column carrying the nonspecific Nb, CBD-anti-MTX Nb. Using data from only the first three flowthrough fractions from the RAC column with the CBD-anti-caffeine Nb and subtracting the nonspecific caffeine binding to the column indicated that there was ∼10 μg caffeine bound to the column that had ∼500 μg immobilized CBD-anti-caffeine Nb fusion protein; this amount of caffeine bound is close to one caffeine per one fusion protein. Consequently, these results demonstrate that the two components in the CBD-anti-caffeine Nb fusion protein preserved their respective functions upon secretion from B. subtilis. Moreover, directly applying the supernatant containing the desired CBD-Nb fusion proteins to a cellulose-based column conveniently generates a simple chromatography system to capture small molecules. This approach may be highly tunable for target binding and purification by swapping the Nb component in the fusion protein.
Detection of protein antigens using Nbs secreted from B. subtilis.
Nbs for immune checkpoint blockade (ICB) such as anti-PD-L1 and CTLA-4 have found utility in cancer immunotherapy as well as in stratifying tumor specimens to inform patient responses to FDA-approved ICB therapeutics. While the majority of anti-PD-L1 and anti-CTLA-4 Nbs are currently produced intracellularly in E. coli, the lengthy processes of lysing bacteria and removing endotoxins may dramatically increase costs and demand a centralized facility for production. In contrast, we were able to directly load bacterial culture supernatant containing His-tagged anti-PD-L1 or anti-CTLA-4 Nb to cobalt nitriloacetic acid (NTA) agarose resin in a column format as illustrated in Fig. 2B, and the proteins subsequently eluted from these columns exhibited high purity and homogeneity (see Fig. S3 in the supplemental material). Of note, in the shake flask expression system, anti-PD-L1 and anti-CTLA-4 Nbs can be secreted into the supernatant of B. subtilis culture at ∼15 mg/liter of culture. To further confirm that these Nbs remain functional following secretion and purification, we chose the mouse dendritic cell (DC) line DC 2.4 because it is known to constitutively express the immunoinhibitory receptor PD-L1 on the cell surface (Fig. 5A), and we also validated the expression of PD-L1 by a commercial antibody specific for mouse PD-L1 (see Fig. S4 in the supplemental material). DC 2.4 cells were immunostained with purified anti-PD-L1 Nb or anti-caffeine Nb, followed by a fluorophore-conjugated anti-FLAG antibody to detect the FLAG epitope that was fused to the Nbs. The stained samples were analyzed by flow cytometry and fluorescence microscopy, and the results of flow cytometry indicated the specific binding of purified anti-PD-L1 Nbs and no binding for the anti-caffeine Nb control in comparison to that of the unstained sample (Fig. 5B). In agreement with the flow cytometry data, imaging by fluorescence microscopy showed positive staining of PD-L1 on the surface of DC 2.4 cells and the absence of binding with the anti-caffeine Nb (Fig. 5C). To further test whether the purification step could be bypassed by directly using bacterial supernatant, we applied 200 μl of the original B. subtilis supernatant containing the anti-PD-L1 Nb to stain ∼1 million DC 2.4 cells for fluorescence microscopy (see Fig. S5 in the supplemental material). As shown in Fig. 5C, the bacterial supernatant with the PD-L1-specific Nb detected PD-L1 expression on DC 2.4 cells, but anti-caffeine Nb (negative control) did not.
FIG 5.
(A) Scheme to use two different cell surface antigens (PD-L1 and CTLA-4) to validate Nbs secreted from B. subtilis. (B) Recombinant Nb anti-PD-L1 purified from the supernatant of B. subtilis cells expressing this protein can readily detect PD-L1 on the surface of an immortalized murine dendritic cell line, DC 2.4. In contrast, recombinant Nbs against caffeine do not. The histograms from flow cytometry analysis are shown. Nbs are fused with a FLAG epitope and detected by an allophycocyanin (APC)-conjugated anti-FLAG antibody, all as described in Materials and Methods. (C) Direct detection of the cell surface antigen PD-L1 by bacterial supernatant. B. subtilis cell supernatant containing anti-PD-L1 or anti-caffeine Nb was incubated with DC 2.4 cells followed by APC-conjugated anti-FLAG antibody and analysis of cells by flow cytometry. (D) Immunofluorescence microscopy shows that recombinant Nb anti-PD-L1, but not the anti-caffeine Nb, can detect PD-L1 expression on the surface of DC 2.4 dendritic cells as purple fluorescence. (E) Detection of CTLA-4-positive cells in unstimulated primary cells harvested from lymph nodes in C57BL/6 mice. Cells were stained with a commercial antibody specific for mouse CTLA-4 or the corresponding control to validate the percentage of CTLA-4-positive cells. Meanwhile, cells were first incubated with purified anti-CTLA-4 Nb or anti-caffeine Nb (negative control), which contain a FLAG epitope, and then stained with APC-anti-FLAG. FSC denotes cell size via forward size scattering. Only live cells were analyzed by flow cytometry, and unstained cells were used to gate for negative cell population, all as described in Materials and Methods.
In addition to detecting abundant antigens on a homogeneous population of cells such as the immortalized dendritic cells DC 2.4, we also explored identification of targets present in only a low percentage of cells (e.g., ∼1%) (Fig. 5D), which may have implications in the development of sensitive diagnostic reagents using B. subtilis-secreted Nbs. According to the literature, CTLA-4 has a very low level of expression in unstimulated (naive) T lymphocyte cells, and only a small fraction of cells is positive for CTLA-4 (29, 30). As shown in Fig. 5E, ∼1% of CTLA-4-positive cells in unstimulated primary cells, which were harvested from lymph nodes in C57BL/6 mice, were detected by both a commercially available antibody specific for mouse CTLA-4 and the anti-CTLA-4 Nb purified from the supernatant of B. subtilis. Consequently, we conclude that Nbs secreted by B. subtilis are functional, and this new Nb production platform has potential applications in generation of Nb-based diagnostics and drug development.
Development of a highly resistant B. subtilis spore-based biologic production platform.
Upon nutrient starvation, B. subtilis can undergo sporulation to generate spores, and these spores have been studied for decades to understand the biological mechanisms underlying their extreme resistance (4, 6). Despite their dormancy and resistance, spores can rapidly return to vegetative growth within 2 h in the presence of nutrients. In light of these findings, we asked can we use spores to store genetic information that encodes and programs the generation of Nbs under extreme environments? To this end, we induced vegetative cells of B. subtilis carrying genes for Nbs to form spores and then followed established protocols to obtain highly purified spores which are essential for studies on spore resistance, killing, and germination (Fig. 6A) (31, 32). Afterwards, we exposed purified spores to four environmental extremes that are relevant to different applications as follows: (i) desiccation, (ii) wet heat (80°C), (iii) UV irradiation (254-nm UV radiation [UV254] and 365-nm UV radiation [UV365]), and (iv) acidic pH.
FIG 6.
(A) Phase contrast imaging of spores encoding CTLA-4 after the HistoDenz-based density gradient purification as described in Materials and Methods (the scale bar is 5 μm). Arrowheads indicate individual spores. (B) B. subtilis spores encoding anti-CTLA-4 Nb are resistant to desiccation air drying on filter paper for 24 and 48 h at 37°C, while vegetative cells are not. Colony growth was induced after wetting filter paper and streaking it on LB agar plates. (C) Spores with the above-mentioned desiccation treatment for 24 and 48 h maintain the ability to produce the recombinant Nbs anti-CTLA-4 and anti-PD-L1 without noticeable losses in yield in comparison to that of the untreated group (i.e., “0 h”) (red box). Randomly selected individual colonies at all time points from spores expressing the recombinant Nbs anti-CTLA-4 or anti-PD-L1 were grown, extracted, and extracts analyzed by SDS-PAGE as described in Materials and Methods. (D) Spores expressing anti-CTLA-4 Nb are tolerant to wet heat (80°C) for up to 3 h, while vegetative cells are killed within 1 min. LB plates show growth of B. subtilis spores or vegetative cells after incubating bacterial suspensions (an OD600 of 0.01) in 1× PBS at 80°C for the indicated times. The sections labeled “0 hr” indicate no heat treatment. (E) Spores with the above-mentioned heat treatment for 3 min and 3 h maintain the ability to produce the recombinant Nbs anti-CTLA-4 and anti-PD-L1 without noticeable losses in yield in comparison to that of the untreated group (red box). Randomly selected individual colonies per time point were grown, extracted, and extracts analyzed by SDS-PAGE as described in Materials and Methods. (F) Spores are resistant to acidic pH, while vegetative cells are not. LB plates show growth of B. subtilis spores or vegetative cells after acid challenges at pH 1.1 and 2.9 for the indicated times. (G) B. subtilis spores are more resistant to UV irradiation at 254 nm than vegetative cells. In contrast, spores and vegetative cells survived exposure to UV light at 365 nm for up to 40 min, without noticeable differences in viability. B. subtilis spores and vegetative cells, which were engineered to express anti-CTLA-4 Nb, all in 1× PBS, were irradiated with 254 nm and 365 nm UV irradiation for indicated times and then spread on a LB plate for enumeration of colonies. The sections labeled “0 min” indicate no UV irradiation. Arrow heads indicate colony growth after recovery from LB plates.
First, to enable long-term storage and great portability, it is desirable to store the engineered bacteria in a dry state, and when needed, they can outgrow and produce Nbs of interest. However, growing B. subtilis cells are vulnerable to desiccation. To demonstrate that spores that carry the Nb-encoding plasmid construct can serve as a stable platform against desiccation, we applied spores and vegetative cells (at an optical density at 600 nm [OD600] of 1.0) to a piece of filter paper followed by air drying and stored them at 37°C for 24 and 48 h. At the 24- and 48-h time points, filter paper carrying spores or vegetative cells was placed on agar growth plates and streaked to allow for recovery and colony formation. The result of the desiccation resistance test shows that spores were able to successfully germinate and proliferate, but the vegetative cells did not (Fig. 6B). Moreover, when the same desiccation-treated spores were inoculated into growth medium, they were able to produce Nbs (anti-CTLA-4 and anti-PD-L1) at a level comparable to that of freshly inoculated spores (Fig. 6C).
Next, engineered spores and vegetative cell controls, both of which express the anti-CTLA-4 Nb, were suspended at an OD of 0.01 and incubated at 80°C for 1 min, 3 min, 1 h, and 3 h. After each time point, spore and vegetative cell suspensions were plated for recovery and colony formation. The result of the heat resistance test shows that spores were able to germinate and proliferate after heat exposure for up to 3 h, but the vegetative cells did not survive even 1 min of exposure (Fig. 6D). Moreover, when the same heat-treated spores were inoculated into growth medium, they were able to produce Nbs (anti-CTLA-4 and anti-PD-L1) at a level comparable to that of the untreated strain (Fig. 6E).
Thirdly, some B. subtilis strains have been commercialized as probiotics to promote health (33, 34). Although the exact mechanisms underlying how probiotics mediate health benefits remain elusive and subject to debate, one potential application is to explore B. subtilis as a chassis to deliver Nbs to address diseases in the gastrointestinal tract (GIT). For example, studies have shown that B. subtilis strains can persist in the mouse GIT for up to 27 days (35). However, with oral delivery, probiotics must first survive the deleterious environment in the stomach, which has an extremely acidic pH. To examine the resistance of spores to acidic pH, engineered B. subtilis spores and vegetative cells encoding the Nb specific for CTLA-4 were incubated at pH 1.1 or 2.9 with 0.9% NaCl to maintain an isotonic condition. The viabilities of the spores and control group were measured at 0.5 to 2 h in the range of estimated durations for spores to transit through the stomach. Our results showed that spores survive these acidic conditions and can both germinate and grow vegetatively after nutrients are provided. Conversely, the vegetative cells incubated in acidic pH were unable to grow after nutrients were provided (Fig. 6F). Importantly, after a 2-h challenge at acidic pH, the engineered spores remained able to germinate, grow, and produce recombinant Nb upon IPTG induction, with expression levels comparable to those of neutral pH-treated spores (see Fig. S7 in the supplemental material).
Finally, we explored the resistance of engineered spores to UV light of two different wavelengths (254 nm and 365 nm) by holding a UV lamp with dual wavelength at a distance of 6 cm perpendicular to spores suspended in PBS, and with this setting, the intensities of UV radiation are 760 (254 nm) and 720 (365 nm) μW/cm2. As expected, under UV at 254 nm, both spores and vegetative cells failed to survive when the exposure time passed 5 min, but spores showed higher viability than vegetative cells (Fig. 6G). Of note, colonies from spores which were recovered after UV254 exposure times of 1 min, 3 min, and 5 min remained able to produce the anti-CTLA-4 Nb after inoculation in medium and induction with IPTG (see Fig. S8 in the supplemental material). In contrast to UV254 treatment, both spores and vegetative cells survived exposure to UV light at 365 nm even after irradiation for 40 min and with no noticeable differences in viability (Fig. 6G). These results suggest that spores will need to be shielded against short wavelength UV such as 254 nm if engineered spores are to be utilized as a biologics production platform in space exploration.
DISCUSSION
In this work, we have presented a new approach for engineering B. subtilis to secrete Nbs directly into culture medium. As a proof of concept, we have shown that four different Nbs targeting either small molecules (caffeine and methotrexate) or cell surface-associated protein antigens (PD-L1 and CTLA-4) can be readily produced in and secreted from B. subtilis giving 15 to 20 mg of Nbs per liter of culture supernatant in the shake flask mode, which can enable facile purification and streamline many downstream applications. Notably, this yield is comparable to that reported for E. coli-based systems for extracellular secretion of Nbs (25, 36), although intracellular expression of Nbs in E. coli has been shown to reach ∼100 mg/liter through extensive optimization of growth conditions as suggested by Zarschler et al. (37). However, intracellular expression does not have the convenience of harvesting proteins directly from the supernatant without the need to lyse bacteria, and the latter may be particularly cumbersome for production of Nbs in a large scale (37). Future work may further improve the yield by using a fed-batch mode to scale up Nb production and excretion in B. subtilis. Additionally, we have shown that when genetically fusing the caffeine-specific Nb to a cellulose-binding domain, bacterial supernatant containing the fusion protein can be directly applied to a cellulose-based substrate for long-term storage or be integrated into an affinity-based chromatography system for caffeine capture. Importantly, this strategy could be adapted to bind and purify other small molecules by swapping the caffeine-specific Nb component in the fusion protein with other hapten-specific Nbs.
The ability to produce Nbs that detect immune checkpoint ligands such as PD-L1 or receptors such as CTLA-4 may also be useful in the field of cancer immunotherapy and could also be extended to generate Nbs that block inflammatory cytokines such as TNF-α to diagnose or treat inflammation (38). In this work, we have used the FLAG epitope cloned at the C terminus of Nbs as a molecular handle for Nb detection via indirect immunostaining. To reduce the indirect staining step, future work can replace the FLAG epitope with a sortase tag to covalently conjugate Nbs with fluorescent dyes or other peptide sequences via sortase-mediated protein ligation (39, 40). Additionally, this secretory system may be integrated with other advanced technologies, such as nonnatural amino acids, to allow for side-specific conjugations and labeling (40–42). Finally, as expected, the engineered B. subtilis strains can be induced to form spores via nutrient starvation, and these engineered spores can survive desiccation, extreme ambient heat, and acidic pH very well as well as UV254 exposure to a certain extent. Importantly, spores surviving these treatments were still able to secrete their encoded Nb after spore germination, outgrowth, and growth. These resistance properties may then allow spores to serve as a highly stable storage form for bacteria that might be needed to function in an environment such as deep space (5). While spores can survive for many years in a desiccated state, upon rehydration and exposure to nutrients, spores rapidly transition back to growing cells, thus providing the capacity to manufacture and purify desired pharmaceuticals.
MATERIALS AND METHODS
Chemicals.
All chemicals and cell culture media were purchased from Fisher Scientific International Inc. (Cambridge, MA, USA) unless otherwise noted and were of the highest purity or analytical grade commercially available. Gibson Assembly master mix kits and cloning enzymes were obtained from New England Biolabs, Inc. (NEB) (Ipswich, MA, USA). DNA oligonucleotides were customized and ordered from Sigma-Aldrich (St. Louis, MO, USA). Plasmid extraction, agarose gel DNA purification, and PCR cleanup kits were purchased from CoWin Biosciences (Cambridge, MA, USA). The DC protein assay reagent package was purchased from Bio-Rad Laboratories, Inc. (Bio-Rad, Hercules, CA, USA).
Plasmids and bacterial strains.
Plasmid pHT43 and the B. subtilis WB800N bacterial strain were generous gifts from Hoang Duc Nguyen (University of Science, Vietnam National University, Vietnam). The pHT43 vector is a high-level expression vector for recombinant protein production and secretion into the B. subtilis culture medium (24). The expression is based on the strong σA-dependent promoter preceding the groE operon of B. subtilis, which has been converted into an efficiently controllable (IPTG-inducible) promoter by addition of the lac operator. The amyQ signal sequence encoding the signal peptide of α-amylase is located downstream of the Shine-Dalgarno sequence (ribosome binding site), followed by restriction sites for cloning the gene of interest. pHT43 is an E. coli/B. subtilis shuttle vector that provides ampicillin resistance to E. coli and chloramphenicol resistance to B. subtilis. WB800N (nprE aprE epr bpr mpr::ble nprB::bsrΔvpr wprA::hyg cm::neo; neoR ) is a variant of B. subtilis 168 deficient for eight extracellular proteases and carries a neomycin resistance gene (43). B. subtilis WB800N was grown in Luria-Bertani (LB) (1% tryptone, 1% yeast extract, and 0.5% NaCl) medium. Chloramphenicol and neomycin, at final concentrations of 5 and 10 μg/ml, respectively, were used as selection antibiotics. E. coli strain DH5α from NEB was used for the construction and propagation of all plasmids.
Recombinant plasmid construction.
Primers used in this work are listed in Table 1. Sequences for anti-mouse PD-L1 (PDB accession number 5DXW) and anti-mouse CTLA-4 (PDB accession number 5E5M) Nbs are from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) and were synthesized as gene fragments by Twist Bioscience, Inc. (San Francisco, CA, USA). E. coli expression pET21 vectors encoding anti-MTX and anti-caffeine Nbs were gifts from James Horn (Northern Illinois University, IL, USA). All Nb-encoding DNA fragments were codon optimized for B. subtilis expression by the software provided by Twist Bioscience. To construct recombinant plasmids containing His- or Flag-tagged Nbs or their fusion with a CBD, DNA fragments were amplified via PCR with Q5 high-fidelity polymerase (NEB), and the gel-purified PCR product was ligated into pHT43 predigested by BamHI and XbaI through Gibson assembly. After transformation into DH5α, bacteria were selected on LB agar plates containing 100 μg/ml ampicillin following incubation at 37°C for 14 h. Single colonies were inoculated in 2 ml LB medium and grown for 12 to 18 h at 37°C, plasmid isolated as described (24), and sent for sequence verification by Sanger sequencing. Correct pHT43-based plasmids were transferred into chemically prepared competent WB800N cells according to a previous protocol (24). In brief, competent cells were prepared in competent cell preparation medium consisting of 1.07% (wt/vol) dibasic potassium phosphate (Sigma-Aldrich), 0.52% (wt/vol) monobasic potassium phosphate (Sigma-Aldrich), 2% (wt/vol) glucose, 0.088% (wt/wt) sodium citrate dihydrate, 0.1% (wt/vol) 2.2% 1,000× ferric ammonium citrate, 0.25% (wt/vol) aspartic acid potassium salt, 0.05% (wt/vol) yeast extract, and 10 mM magnesium sulfate. B. subtilis strain WB800N was streaked on LB plates the day before use, and a single colony was inoculated into 1 ml competent cell preparation medium and shaken at 37°C until the transition from log to stationary phase (OD600 of ∼1.0). A 100-ng plasmid was added to the cells, incubation was continued for 2.5 h, 100 μl of the culture was spread on LB plates containing chloramphenicol and neomycin, and plates were incubated at 37°C overnight. At least two colonies were separately examined to verify their production of the appropriate recombinant protein.
TABLE 1.
Primers used for genes of interest
| Target gene | Primer | Sequencea |
|---|---|---|
| FLAG-CBM3-TEV | B83 | tacaaaaacatcagccgtaggatccCATCATCATCATCATCACAGC |
| B84 | gtacttgggaTCCCTGGAAGTACAGGTTTTC | |
| hPTH | B136 | cttgaagtcctctttcagggacccAGCGTTTCGGAAATTCAATTG |
| B137 | ctgccccggggacgtcgactctagaCTATTAGAAATTGTGCACATCC | |
| Caffeine/MTX | B85 | cttccagggaTCCCAAGTACAACTGGTAG |
| B86 | ctgccccggggacgtcgactctagaCTATTAGCTGCTTACGGTTAC | |
| hG-CSF | B138 | cttgaagtcctctttcagggacccACGCCGTTGGGCCCCGCC |
| B139 | ctgccccggggacgtcgactctagaCTATTACGGCTGTGCAAGATGACGCAA | |
| PD-L1 and CTLA-4 | B145 | ACAAAAACATCAGCCGTAGGA |
| B146 | CCGGGGACGTCGACTCTAG |
Capital letters are complementary to the gene of interest. Lowercase letters are sequences that overlap with the plasmid backbone.
Regenerated amorphous cellulose preparation.
RAC was prepared by adapting an established protocol for RAC-based affinity protein purification (1). Briefly, 1 g of cellulose microcrystalline powder with an average particle size of 50 μm according to the manufacturer (Fisher Scientific, Waltham, MA) was added to 3 ml double-distilled water to make a slurry followed by slow addition of 12.5 ml of 85% ice cold phosphoric acid. After incubation for 2 h on ice, 12.5 g of sodium hydroxide was added to adjust the pH to ∼7. The material was then dialyzed at room temperature in 3,500 molecular weight cutoff dialysis tubing for 48 h against 4 liters double-distilled water, with the water changed every 6 h. The final dialyzed RAC sample was lyophilized for future use.
Protein expression and purification.
Purified proteins were obtained by growing B. subtilis WB800N containing the pHT43-based plasmid encoding the desired protein in 200 ml LB medium at 37°C in a 2-liter shake flask at 250 rpm. Protein expression was induced with 1 mM IPTG when the culture reached an OD600 of 0.6 to 0.8, and growth was continued for 18 h at 28°C. The culture supernatant was harvested by centrifugation at 6,000 × g for 30 min at room temperature, and a small fraction of supernatant was subjected to TCA precipitation. Briefly, 1 volume (0.1 ml) of 100% TCA was added to 4 volumes (0.4 ml) of the supernatant sample followed by incubation at 4°C for 10 min and centrifugation at 13,000 × g for 15 min. The supernatant was discarded, and the pellet was washed twice with 250 μl 100% ice-cold acetone. Samples were dried at 95°C for 5 min before suspension in 100 μl 1× SDS-PAGE reducing buffer (62.5 mM Tris-HCl, pH 6.8, 2.5% SDS, 0.002% bromophenol blue, 0.7135 M β-mercaptoethanol, and 10% glycerol) and analyzed by SDS-PAGE and Western blotting.
Fifty milliliters of fresh culture supernatant containing His-tagged proteins was purified at room temperature by gravity flow using a 10-ml gravity-based column (Bio-Rad) with 100-μl cobalt agarose beads (GoldBio, St. Louis, MO, USA). The supernatant was passed through the chromatography column, after which the column was washed with 10 column volumes of wash buffer (33 mM phosphate, 500 mM NaCl, and 10 mM imidazole, pH 7.4). Protein was eluted with 5 to 6 fractions (100 μl per fraction) of elution buffer (33 mM phosphate, 150 mM NaCl, and 150 mM imidazole, pH 7.4). The OD280 of eluted fractions was measured, and fractions with detectable protein levels (>0.1 mg/ml) were combined. For buffer exchange to protein storage buffer (1 mM dithiothreitol [DTT], 20 mM HEPES, 500 mM NaCl, and 10% glycerol, pH 7.4), pooled supernatants were loaded on a 15-ml Amicon ultrafiltration column with a molecular weight cut off 3,000 Da. The loaded columns were centrifuged at 4°C and 2,880 × g to remove the elution buffer, diluted with storage buffer, and this process was repeated three times. The concentrated protein solution (∼250 μl) was transferred to a microcentrifuge tube and stored at −80°C for future use.
Cellulose-binding protein or its fusion protein were immobilized by gravity flow through the 0.1-ml RAC column in a 1-ml syringe as described above. The RAC column was first equilibrated with 1× phosphate-buffered saline (PBS; 137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, pH 7.4), and then supernatant containing desired proteins was added to the column until the column was saturated by quantifying the amount of fusion proteins in flowthrough fractions and comparing it to the original supernatant via SDS-PAGE and an enzyme-linked immunosorbent assay. The saturated RAC was washed with 10 column volumes of PBS followed by loading with 100 μl 150 μg/ml caffeine solution via gravity flow. Flowthrough fractions of 100 μl were collected for OD273 measurement using a UV spectrometer (Fisher Scientific), with a total of five fractions collected. Caffeine has a distinct UV absorbance at 273 nm, and a linear standard curve was plotted for caffeine concentrations ranging from 0.1 to 500 μg/ml and was used to measure the concentration of caffeine in flowthrough fractions.
DC protein assay.
For the DC assay of recombinant proteins, fresh A′ reagent was prepared by mixing 20 μl of reagent S with 1 ml reagent A and 25 μl placed in each well in a 96-well plate. Five dilutions of a protein standard were prepared containing 0.16, 0.32, 0.64, 1.28, and 2.56 mg/ml bovine serum albumin (BSA), and 3 μl of standards and samples was pipetted into reagent A′ followed by addition of 200 μl of reagent B to each well and incubation for 15 min. Protein concentration was determined by measuring OD750 in samples and protein standard. A technical repeat was also performed to derive a standard curve, and the linearity was more than 0.99.
Filter paper function assay and Western blotting.
Culture supernatant was applied in various patterns on filter paper (Fisher Scientific) and dried at room temperature. Samples were incubated with 5 ml of 5% nonfat milk for 30 min to block nonspecific binding sites. Anti-FLAG epitope (DYKDDDDK; BioLegend, San Diego, CA; catalog number 637301) was diluted 1:2,000 in 3 ml Tris-buffered saline (TBS) plus 5% nonfat milk and incubated overnight with the filter paper at 4°C. The filter paper was washed three times with 5 ml 1× TBS containing 0.05% Tween 20 (15 min per wash cycle). Afterward, the secondary antibody anti-rat IgG HRP (Cell Signaling Technology, Danvers, MA; catalog number 7077) was diluted at 1:2,000 in 3 ml TBS plus 5% nonfat milk and incubated on a rocking platform at room temperature for 1 h. After washing with 1× TBS with 5 ml 0.05% Tween 20, premixed Pierce 3,3′-diaminobenzidine (DAB) substrate (Fisher Scientific) was directly added on the cellulose filter paper. The reaction was terminated with water after dark spots appeared. For detection of specific proteins by Western blotting, briefly, proteins were first separated by 12% SDS-PAGE and then transferred to a nitrocellulose membrane (Fisher Scientific). The membranes were incubated with the antibodies and the DAB substrate as described above.
Flow cytometry with commercial antibodies, B. subtilis supernatant, and purified Nbs.
The DC 2.4 cell line, obtained from Kenneth Rock (University of Massachusetts, Worcester, MA), was propagated in a 6-cm dish in RPMI 1640 medium with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin before use. To prepare cells for surface staining, cells were washed with PBS twice and incubated with 2 ml 5 mM EDTA PBS solution at 37°C for 30 min. Two milliliters fluorescence-activated cell sorter (FACS) buffer (1× PBS, 5% FBS, 2 mM EDTA) was directly added to the dish to mechanically dissociate the cells. Cells were spun down at 500 × g for 5 min and resuspended in FACS buffer to obtain 0.5 million cells for each staining reaction. Working concentrations of 0.5 μg/ml phycoerythrin (PE) anti-mouse PD-L1 (BioLegend; catalog number 124307), PE rat IgG2B, κ isotype control antibody (BioLegend; catalog number 400607), and 0.2 ml B. subtilis culture supernatant plus 1% BSA or 0.5 μg/ml purified Nbs with the FLAG epitope were incubated with cells for 1 h, washed with PBS once, and allophycocyanin (APC) anti-FLAG (BioLegend; catalog number 637307) was diluted 200-fold in 1× PBS containing 1% BSA. After 1 h incubation, cells were washed with PBS twice and resuspended in PBS for flow cytometry (Invitrogen Attune; Fisher Scientific). Blue and red lasers along with 574-nm/26-nm and 670-nm/14-nm filters in the flow cytometer were used for detection of PE and APC dyes, respectively.
Immunocytochemistry.
Staining for immunocytochemistry was the same as in the steps prior to flow cytometry. After the last washing step, cells were spun down at 500 × g for 3 min and mixed with 4% formaldehyde in PBS for 10 min followed by washing with PBS twice. An aqueous mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; 100 nM) was used to resuspend the cell pellet, and cells were immobilized on a SuperFrost Plus adhesion slide (Electron Microscopy Sciences, Hatfield, PA, USA). Images were taken by a Nikon epifluorescence microscope.
Spore preparation.
B. subtilis spores of the engineered strains were prepared on 2× Schaeffer’s glucose medium (2× SG) agar plates as described previously (31, 32). Plates spread with log-phase cells were incubated for 2 days at 37°C, and then spores were scraped from plates and suspended in cold sterile water. The suspended spores were then purified over 5 to 7 days by repeated rounds of centrifugation and suspension by sonication ∼3 times/day. Aliquots of the final purified spores were further purified by centrifugation through a high-density medium of 50% HistoDenz (Sigma Chemical Co., St. Louis, MO, USA). The pelleted spores were washed 4 to 5 times with sterile water, and spores were suspended in cold water at ∼109 spores/ml. These final spores were >98% free of growth of sporulating cells, germinated spores, or debris as seen by phase-contrast microscopy (see Fig. 6A).
Examining the resistance of engineered spores.
For desiccation studies, spore and vegetative cell samples at an OD600 of 1.0 in 100 μl were applied to a piece of Whatman filter paper. After air drying on filter paper for 24 and 48 h at 37°C, the papers were hydrated with fresh LB medium and aliquots spread on an LB plate containing the desired antibiotics to examine colony formation. For analyses of wet heat, acid, and UV resistance, spores and vegetative cells were diluted to an OD600 of 0.01 (∼106 spores/cells/ml) in either 0.9% (wt/wt) sodium chloride containing hydrochloride acid at pH 1.1 or 2.9 (acid resistance) or in PBS at 80°C (wet heat resistance). After various incubation times, 5-μl samples were spread on LB plates with appropriate antibiotics, plates were incubated at 37°C overnight, and colony formation was examined. For UV challenge experiments, spores and vegetative cells were first diluted to an OD600 of 0.01 in PBS in a 96-well optically transparent plate. A handheld UV lamp (UVP 95-0007-06 model UVGL-58, 6 W, dual 254 nm and 365 nm lamps, 760/720 μW/cm2 at 3 in. between the lamp and bacteria) was used to expose spores and vegetative cells at a distance of 6 cm between the lamp source and the culture at 254 nm for 0, 1, 3, and 5 min, and at 365 nm, spores and vegetative cells were exposed for 0, 20, and 40 min. Samples were then spread on LB plates with desired antibiotics, plates were incubated at 37°C overnight, and cell and spore viability was examined.
Statistical analysis.
Statistical analyses were performed using GraphPad Prism 5.0a for Mac OS X (San Diego, CA, USA). The Student's t test was used to determine statistical significance in the caffeine-capturing study.
Supplementary Material
ACKNOWLEDGMENTS
Our work was supported by Northeastern University Faculty startup funding (J.L.) and the Peer Reviewed Medical Research Program from the Department of Defense’s Congressionally Directed Medical Research Programs (J.L.).
We are grateful to Guoxin Rong at the Institute for Chemical Imaging of Living Systems at Northeastern University for acquiring images. We would like to express our gratitude to Sara Rouhanifard at Northeastern University’s Department of Bioengineering for generously sharing her lab’s fluorescent microscope with us, Ke Zhang at Northeastern University’s Department of Chemistry for sharing his lab’s equipment, and James Horn from Northern Illinois University for providing vectors encoding anti-MTX and anti-caffeine Nbs.
J.L. and P.S. designed the study. M.Y., G.Z., G.K., X.S. performed the experiments. J.L., P.S. M.Y., G.Z., G.K., and X.S. analyzed the data. J.L. and P.S. wrote the manuscript.
Northeastern University has filed a provisional patent on the technology described in this paper (serial no. 62/914,130).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Pardee K, Green AA, Ferrante T, Cameron DE, DaleyKeyser A, Yin P, Collins JJ. 2014. Paper-based synthetic gene networks. Cell 159:940–954. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardee K, Slomovic S, Nguyen PQ, Lee JW, Donghia N, Burrill D, Ferrante T, McSorley FR, Furuta Y, Vernet A, Lewandowski M, Boddy CN, Joshi NS, Collins JJ. 2016. Portable, on-demand biomolecular manufacturing. Cell 167:248–259. doi: 10.1016/j.cell.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Granger AC, Gaidamakova EK, Matrosova VY, Daly MJ, Setlow P. 2011. Effects of Mn and Fe levels on Bacillus subtilis spore resistance and effects of Mn2+, other divalent cations, orthophosphate, and dipicolinic acid on protein resistance to ionizing radiation. Appl Environ Microbiol 77:32–40. doi: 10.1128/AEM.01965-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L, Wang S, Cortesao M, Wu M, Moeller R, Setlow P, Li YQ. 2018. Single-cell analysis reveals individual spore responses to simulated space vacuum. NPJ Microgravity 4:26. doi: 10.1038/s41526-018-0059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulrich N, Nagler K, Laue M, Cockell CS, Setlow P, Moeller R. 2018. Experimental studies addressing the longevity of spores—the first data from a 500-year experiment. PLoS One 13:e0208425. doi: 10.1371/journal.pone.0208425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Setlow P. 2014. Spore resistance properties. Microbiol Spectr 2:TBS-0003-2012. doi: 10.1128/microbiolspec.TBS-0003-2012. [DOI] [PubMed] [Google Scholar]
- 7.Setlow P. 2014. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol 196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harmsen MM, De Haard HJ. 2007. Properties, production, and applications of camelid single-domain antibody fragments. Appl Microbiol Biotechnol 77:13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Sapienza G, Rossotti MA, Tabares-da Rosa S. 2017. Single-domain antibodies as versatile affinity reagents for analytical and diagnostic applications. Front Immunol 8:977. doi: 10.3389/fimmu.2017.00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jailkhani N, Ingram JR, Rashidian M, Rickelt S, Tian C, Mak H, Jiang Z, Ploegh HL, Hynes RO. 2019. Noninvasive imaging of tumor progression, metastasis, and fibrosis using a nanobody targeting the extracellular matrix. Proc Natl Acad Sci U S A 116:14181–14190. doi: 10.1073/pnas.1817442116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie YJ, Dougan M, Jailkhani N, Ingram J, Fang T, Kummer L, Momin N, Pishesha N, Rickelt S, Hynes RO, Ploegh H. 2019. Nanobody-based CAR T cells that target the tumor microenvironment inhibit the growth of solid tumors in immunocompetent mice. Proc Natl Acad Sci U S A 116:7624–7631. doi: 10.1073/pnas.1817147116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steeland S, Vandenbroucke RE, Libert C. 2016. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov Today 21:1076–1113. doi: 10.1016/j.drudis.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt FI, Lu A, Chen JW, Ruan JB, Tang C, Wu H, Ploegh HL. 2016. A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly. J Exp Med 213:771–790. doi: 10.1084/jem.20151790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petsch D, Anspach FB. 2000. Endotoxin removal from protein solutions. J Biotechnol 76:97–119. doi: 10.1016/s0168-1656(99)00185-6. [DOI] [PubMed] [Google Scholar]
- 15.Hirayama C, Sakata M. 2002. Chromatographic removal of endotoxin from protein solutions by polymer particles. J Chromatogr B Analyt Technol Biomed Life Sci 781:419–432. doi: 10.1016/s1570-0232(02)00430-0. [DOI] [PubMed] [Google Scholar]
- 16.Aida Y, Pabst MJ. 1990. Removal of endotoxin from protein solutions by phase separation using Triton X-114. J Immunol Methods 132:191–195. doi: 10.1016/0022-1759(90)90029-u. [DOI] [PubMed] [Google Scholar]
- 17.McMahon C, Baier AS, Pascolutti R, Wegrecki M, Zheng S, Ong JX, Erlandson SC, Hilger D, Rasmussen SGF, Ring AM, Manglik A, Kruse AC. 2018. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat Struct Mol Biol 25:289–296. doi: 10.1038/s41594-018-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onishi H, Mizukami M, Hanagata H, Tokunaga M, Arakawa T, Miyauchi A. 2013. Efficient production of anti-fluorescein and anti-lysozyme as single-chain anti-body fragments (scFv) by Brevibacillus expression system. Protein Expr Purif 91:184–191. doi: 10.1016/j.pep.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Westers L, Westers H, Quax WJ. 2004. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim Biophys Acta 1694:299–310. doi: 10.1016/j.bbamcr.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 20.Harwood CR, Cranenburgh R. 2008. Bacillus protein secretion: an unfolding story. Trends Microbiol 16:73–79. doi: 10.1016/j.tim.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Cai D, Rao Y, Zhan Y, Wang Q, Chen S. 2019. Engineering Bacillus for efficient production of heterologous protein: current progress, challenge and prospect. J Appl Microbiol 126:1632–1642. doi: 10.1111/jam.14192. [DOI] [PubMed] [Google Scholar]
- 22.Schumann W. 2007. Production of recombinant proteins in Bacillus subtilis. Adv Appl Microbiol 62:137–189. doi: 10.1016/S0065-2164(07)62006-1. [DOI] [PubMed] [Google Scholar]
- 23.Wu SC, Yeung JC, Duan Y, Ye R, Szarka SJ, Habibi HR, Wong SL. 2002. Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall-bound protease on antibody fragment production. Appl Environ Microbiol 68:3261–3269. doi: 10.1128/aem.68.7.3261-3269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen HD, Phan TT, Schumann W. 2007. Expression vectors for the rapid purification of recombinant proteins in Bacillus subtilis. Curr Microbiol 55:89–93. doi: 10.1007/s00284-006-0419-5. [DOI] [PubMed] [Google Scholar]
- 25.Salema V, Fernandez LA. 2013. High yield purification of nanobodies from the periplasm of E. coli as fusions with the maltose binding protein. Protein Expr Purif 91:42–48. doi: 10.1016/j.pep.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Ding LL, Wang ZY, Zhong PY, Jiang H, Zhao ZX, Zhang YR, Ren Z, Ding Y. 2019. Structural insights into the mechanism of single domain VHH antibody binding to cortisol. FEBS Lett 593:1248–1256. doi: 10.1002/1873-3468.13398. [DOI] [PubMed] [Google Scholar]
- 27.Wagner HJ, Wehrle S, Weiss E, Cavallari M, Weber W. 2018. A two-step approach for the design and generation of nanobodies. Int J Mol Sci 19:3444. doi: 10.3390/ijms19113444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voutilainen SP, Nurmi-Rantala S, Penttilä M, Koivula A. 2014. Engineering chimeric thermostable GH7 cellobiohydrolases in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 98:2991–3001. doi: 10.1007/s00253-013-5177-2. [DOI] [PubMed] [Google Scholar]
- 29.Lindsten T, Lee KP, Harris ES, Petryniak B, Craighead N, Reynolds PJ, Lombard DB, Freeman GJ, Nadler LM, Gray GS. 1993. Characterization of CTLA-4 structure and expression on human T cells. J Immunol 151:3489–3499. [PubMed] [Google Scholar]
- 30.Wang W, Hou X, Yang X, Liu A, Tang Z, Mo F, Yin S, Lu X. 2019. Highly sensitive detection of CTLA-4-positive T-cell subgroups based on nanobody and fluorescent carbon quantum dots. Oncol Lett 18:109–116. doi: 10.3892/ol.2019.10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paidhungat M, Setlow B, Driks A, Setlow P. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J Bacteriol 182:5505–5512. doi: 10.1128/jb.182.19.5505-5512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setlow P. 2019. Observations on research with spores of Bacillales and Clostridiales species. J Appl Microbiol 126:348–358. doi: 10.1111/jam.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujiya M, Musch MW, Nakagawa Y, Hu S, Alverdy J, Kohgo Y, Schneewind O, Jabri B, Chang EB. 2007. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 1:299–308. doi: 10.1016/j.chom.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Rhayat L, Maresca M, Nicoletti C, Perrier J, Brinch KS, Christian S, Devillard E, Eckhardt E. 2019. Effect of Bacillus subtilis strains on intestinal barrier function and inflammatory response. Front Immunol 10:564. doi: 10.3389/fimmu.2019.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tam NK, Uyen NQ, Hong HA, Duc Le H, Hoa TT, Serra CR, Henriques AO, Cutting SM. 2006. The intestinal life cycle of Bacillus subtilis and close relatives. J Bacteriol 188:2692–2700. doi: 10.1128/JB.188.7.2692-2700.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruano-Gallego D, Fraile S, Gutierrez C, Fernández LÁ. 2019. Screening and purification of nanobodies from E. coli culture supernatants using the hemolysin secretion system. Microb Cell Fact 18:47. doi: 10.1186/s12934-019-1094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zarschler K, Witecy S, Kapplusch F, Foerster C, Stephan H. 2013. High-yield production of functional soluble single-domain antibodies in the cytoplasm of Escherichia coli. Microb Cell Fact 12:97. doi: 10.1186/1475-2859-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandenbroucke K, de Haard H, Beirnaert E, Dreier T, Lauwereys M, Huyck L, Van Huysse J, Demetter P, Steidler L, Remaut E, Cuvelier C, Rottiers P. 2010. Orally administered L. lactis secreting an anti-TNF nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol 3:49–56. doi: 10.1038/mi.2009.116. [DOI] [PubMed] [Google Scholar]
- 39.Virant D, Traenkle B, Maier J, Kaiser PD, Bodenhofer M, Schmees C, Vojnovic I, Pisak-Lukats B, Endesfelder U, Rothbauer U. 2018. A peptide tag-specific nanobody enables high-quality labeling for dSTORM imaging. Nat Commun 9:930. doi: 10.1038/s41467-018-03191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schumacher D, Helma J, Schneider AFL, Leonhardt H, Hackenberger C. 2018. Nanobodies: chemical functionalization strategies and intracellular applications. Angew Chem Int Ed Engl 57:2314–2333. doi: 10.1002/anie.201708459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hallam TJ, Wold E, Wahl A, Smider VV. 2015. Antibody conjugates with unnatural amino acids. Mol Pharm 12:1848–1862. doi: 10.1021/acs.molpharmaceut.5b00082. [DOI] [PubMed] [Google Scholar]
- 42.Axup JY, Bajjuri KM, Ritland M, Hutchins BM, Kim CH, Kazane SA, Halder R, Forsyth JS, Santidrian AF, Stafin K, Lu Y, Tran H, Seller AJ, Biroc SL, Szydlik A, Pinkstaff JK, Tian F, Sinha SC, Felding-Habermann B, Smider VV, Schultz PG. 2012. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc Natl Acad Sci U S A 109:16101–16106. doi: 10.1073/pnas.1211023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeong H, Jeong DE, Park SH, Kim SJ, Choi SK. 2018. Complete genome sequence of Bacillus subtilis strain WB800N, an extracellular protease-deficient derivative of strain 168. Microbiol Resour Announc 7:e01380-18. doi: 10.1128/MRA.01380-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.