Exopolysaccharides are key components of the surfaces of their bacterial producers, contributing to protection, microbial and host interactions, and even virulence. They also have significant applications in industry, and understanding their biosynthetic mechanisms may allow improved production of novel and valuable polymers. Four categories of bacterial exopolysaccharide biosynthesis have been described in detail, but novel enzymes and glycosylation mechanisms are still being described. Our findings that a putative bactoprenol glycosyltransferase and flippase are essential to homopolysaccharide biosynthesis in Lactobacillus johnsonii FI9785 indicate that there may be an alternative mechanism of glucan biosynthesis to the glucansucrase pathway. Disturbance of this synthesis leads to a slow-growth phenotype. Further elucidation of this biosynthesis may give insight into exopolysaccharide production and its impact on the bacterial cell.
KEYWORDS: exopolysaccharide, alpha glucan, Lactobacillus johnsonii, proteomics, glycosyltransferase, nuclear magnetic resonance
ABSTRACT
Lactobacillus johnsonii FI9785 makes two capsular exopolysaccharides—a heteropolysaccharide (EPS2) encoded by the eps operon and a branched glucan homopolysaccharide (EPS1). The homopolysaccharide is synthesized in the absence of sucrose, and there are no typical glucansucrase genes in the genome. Quantitative proteomics was used to compare the wild type to a mutant where EPS production was reduced to attempt to identify proteins associated with EPS1 biosynthesis. A putative bactoprenol glycosyltransferase, FI9785_242 (242), was less abundant in the Δeps_cluster mutant strain than in the wild type. Nuclear magnetic resonance (NMR) analysis of isolated EPS showed that deletion of the FI9785_242 gene (242) prevented the accumulation of EPS1, without affecting EPS2 synthesis, while plasmid complementation restored EPS1 production. The deletion of 242 also produced a slow-growth phenotype, which could be rescued by complementation. 242 shows amino acid homology to bactoprenol glycosyltransferase GtrB, involved in O-antigen glycosylation, while in silico analysis of the neighboring gene 241 suggested that it encodes a putative flippase with homology to the GtrA superfamily. Deletion of 241 also prevented production of EPS1 and again caused a slow-growth phenotype, while plasmid complementation reinstated EPS1 synthesis. Both genes are highly conserved in L. johnsonii strains isolated from different environments. These results suggest that there may be a novel mechanism for homopolysaccharide synthesis in the Gram-positive L. johnsonii.
IMPORTANCE Exopolysaccharides are key components of the surfaces of their bacterial producers, contributing to protection, microbial and host interactions, and even virulence. They also have significant applications in industry, and understanding their biosynthetic mechanisms may allow improved production of novel and valuable polymers. Four categories of bacterial exopolysaccharide biosynthesis have been described in detail, but novel enzymes and glycosylation mechanisms are still being described. Our findings that a putative bactoprenol glycosyltransferase and flippase are essential to homopolysaccharide biosynthesis in Lactobacillus johnsonii FI9785 indicate that there may be an alternative mechanism of glucan biosynthesis to the glucansucrase pathway. Disturbance of this synthesis leads to a slow-growth phenotype. Further elucidation of this biosynthesis may give insight into exopolysaccharide production and its impact on the bacterial cell.
INTRODUCTION
Production of exopolysaccharides (EPS) has a large impact on the nature of the bacterial surface and hence on interactions with the environment, hosts and host defense systems, and other microbes (1, 2). EPS can protect bacteria against environmental conditions, both outside and inside the host (1, 3, 4), and in the case of pathogens such as Streptococcus pneumoniae, they can have an important association with immune evasion and virulence (5). EPS can have immunomodulatory and protective properties in the host (6–9) and can affect the composition and function of the gut microbiota (10, 11). EPS can also play a crucial role in biofilm formation, adhesion to host cells, and colonization (3, 12–15). In addition to their biological importance, bacterial EPS have a range of technological applications in food, pharmaceutical, and other industries and may also have potential health benefits due to their activities in immune stimulation, antitumor activity, and lowering of blood cholesterol or as prebiotics (1, 2, 16, 17).
Lactobacillus johnsonii FI9785 is a poultry isolate which has shown promise as a competitive exclusion agent against Clostridium perfringens (18) and Campylobacter jejuni (19). This strain makes 2 capsular exopolysaccharides—EPS2, a heteropolysaccharide containing glucose and galactose encoded by a 14-gene eps operon of the Wzx/Wzy type, and EPS1, a branched dextran homopolysaccharide with an α-(1→6) backbone and α-(1→2) branches which are present on every unit of the backbone and consist of a single glucose (Glc) residue (20, 21). This is an unusual structure which has not been described in other bacteria, although a small percentage of α-(1→2) branches were seen in dextran produced by Leuconostoc citreum E497 (22). Glucansucrases have been shown to synthesize homopolysaccharides in lactic acid bacteria, using sucrose as a substrate (17). However, L. johnsonii FI9785 makes EPS1 in the absence of sucrose, and there is no glucansucrase gene present in the genome, suggesting a different mode of biosynthesis (20). In previous work, the 14-gene eps operon (loci FI9785_1170 to FI9785_1183 inclusive, now renamed FI9785_RS05260 to FI9785_RS05325) was removed by deletion mutagenesis to create the mutant strain Δeps_cluster (20), and a second mutant strain where just the transcriptional regulator epsA (FI9785_1183) was deleted was also constructed (23). Although these mutations were expected to just affect the synthesis of EPS2 and not EPS1, these strains did not show an EPS layer by transmission electron microscopy (TEM), and nuclear magnetic resonance (NMR) analysis of EPS extractions failed to identify either EPS1 or EPS2 (20, 23, 24). In this work, we compared the proteome of the wild-type L. johnsonii FI9785 EPS producer with the Δeps_cluster mutant strain to attempt to identify proteins involved in homopolysaccharide biosynthesis.
RESULTS
Comparative quantitative proteomic analyses identified proteins affected by deletion of the eps cluster.
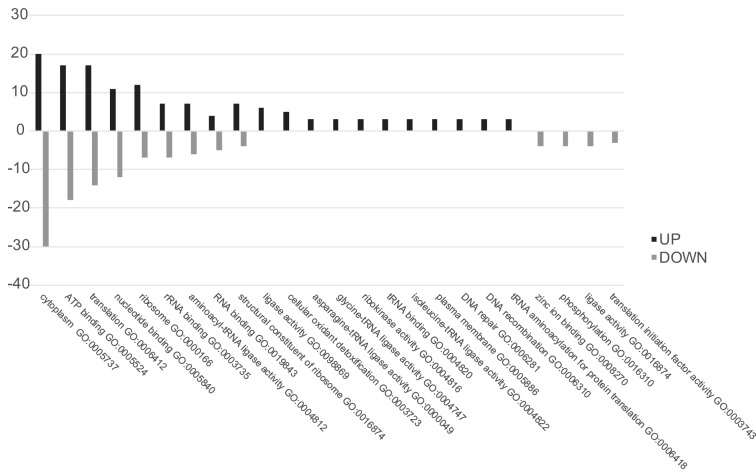
In order to identify proteins involved in EPS biosynthesis, the proteome of the wild type was compared to that of a mutant with a reduced EPS capsule to highlight proteins which were missing or downregulated in the mutant. Proteomic analysis of the soluble fractions of L. johnsonii FI9785 and the Δeps_cluster strain identified several proteins which were differently expressed between the two strains. The protein samples were trypsin digested and labeled by iTRAQ (isobaric tag for relative and absolute quantitation) reagents, mixed and analyzed using nano-liquid chromatography tandem mass spectroscopy (nLC MS/MS) or directly analyzed without labeling for the label-free experiment. Andromeda analyses resulted in the identification of 699 soluble proteins (see Data Set S1 in the supplemental material), 49 of which were differentially expressed in the Δeps_cluster strain versus the wild type (WT; Table 1). The volcano plots in Fig. 1 show the proteins which changed in abundance, obtained in iTRAQ (Fig. 1A) and label-free (Fig. 1B) experiments. The two different quantitative approaches allowed the quantitation of identical proteins with a similar ratio in the mutant and the control, e.g., D0R1R2, supporting the accuracy of the analyses, but also identified different proteins, allowing an in-depth characterization of proteins altered in the Δeps_cluster strain. A total of 20 proteins were found at a higher level in the Δeps_cluster strain, 4 identified by iTRAQ and 17 by the label-free approach, with only one found by both methods; the remaining 29 proteins were at higher levels in the WT, 17 found by iTRAQ and 14 by the label-free method, with 2 proteins identified by both methods (Table 1). In Fig. 2, enriched Gene Ontology (GO) terms of proteins found at different levels in the L. johnsonii FI9785 and Δeps_cluster strains are described. Soluble proteins, mainly present in the cytoplasm, are involved in ATP binding (GO:0005524), translation (GO:0006412), nucleotide binding (GO:0000166), and transferase activity (GO:0016740) in the mutant strain. Almost half of the proteins with altered abundance were associated with ribosomal structure, translation, and protein biosynthesis, but some were more and some less abundant in the Δeps_cluster strain, with no discernible pattern. No other biological processes seemed to be strongly impacted in the Δeps_cluster strain. Although EPS is known to protect the cells from stress, there were no notable changes in stress response except a higher level of thiol peroxidase, commonly involved in cell redox homeostasis (Table 1).
TABLE 1.
Quantified Lactobacillus johnsonii proteins using MaxQuant software in iTRAQ and label-free experimentsa
| Protein accession no. | Protein name | Gene name | No. of razor and unique peptides | Mol wt (kDa) | Score | log2 (D/WT) | D/WT ratio | WT/D ratio | iTRAQb | Label freeb | GO biological process |
|---|---|---|---|---|---|---|---|---|---|---|---|
| D0R1P2 | Uncharacterized protein | FI9785_401 | 4 | 22 | 234 | 3.71 | 13.12 | 0.08 | X | − | |
| D0R498 | Thiol peroxidase | tpx | 3 | 18 | 29 | 2.41 | 5.30 | 0.19 | X | Cell redox homeostasis; oxidation/reduction process; cellular oxidant detoxification | |
| D0R5C5 | Ribosomal silencing factor RsfS | rsfS | 3 | 14 | 111 | 2.22 | 4.67 | 0.21 | X | Mature ribosome assembly; negative regulation of ribosome biogenesis; negative regulation of translation; regulation of translation | |
| D0R3E4 | 50S ribosomal protein L28 | rpmB | 3 | 7 | 26 | 2.17 | 4.49 | 0.22 | X | Translation | |
| D0R3T4 | Aspartate-tRNA ligase | aspS | 26 | 71 | 155 | 1.38 | 2.60 | 0.39 | X | Translation; tRNA aminoacylation for protein translation; aspartyl-tRNA aminoacylation | |
| D0R3V1 | Glycine-tRNA ligase beta subunit | glyS | 16 | 78 | 150 | 1.23 | 2.35 | 0.43 | X | Translation; arginyl tRNA aminoacylation; glycyl tRNA aminoacylation | |
| D0R277 | Uncharacterized protein | FI9785_219 | 7 | 17 | 87 | 1.16 | 2.24 | 0.45 | X | − | |
| D0R5K0 | Aspartyl/glutamyl-tRNA (Asn/Gln) amidotransferase subunit C | gatC | 3 | 12 | 42 | 1.12 | 2.17 | 0.46 | X | Translation; regulation of translational fidelity | |
| D0R6B7 | Deoxynucleoside kinase | dgk1 | 10 | 25 | 60 | 1.05 | 2.07 | 0.48 | X | Nucleobase-containing compound metabolic process; deoxyribonucleoside monophosphate biosynthetic process; nucleotide biosynthetic process; phosphorylation | |
| D0R362 | Ribokinase | rbsK | 16 | 33 | 146 | 1.04 | 2.05 | 0.49 | X | Carbohydrate metabolic process; d-ribose metabolic process; phosphorylation; d-ribose catabolic process; carbohydrate phosphorylation | |
| D0R1J3 | 30S ribosomal protein S12 | rpsL | 9 | 15 | 134 | 0.93 | 1.90 | 0.53 | X | X | Translation |
| D0R2C4 | Recombination protein RecR | recR | 2 | 22 | 44 | 0.89 | 1.85 | 0.54 | X | DNA repair; DNA recombination; cellular response to DNA damage stimulus | |
| D0R4S3 | Isoleucine-tRNA ligase | ileS | 16 | 107 | 100 | 0.81 | 1.75 | 0.57 | X | Translation; tRNA aminoacylation for protein translation; isoleucyl tRNA aminoacylation; aminoacyl-tRNA metabolism involved in translational fidelity | |
| D0R1L4 | 30S ribosomal protein S5 | rpsE | 15 | 19 | 278 | 0.77 | 1.71 | 0.59 | X | Translation | |
| D0R5T0 | Uncharacterized protein | FI9785_1588 | 10 | 21 | 53 | 0.76 | 1.70 | 0.59 | X | − | |
| D0R434 | Asparagine-tRNA ligase | asnS | 18 | 50 | 72 | 0.74 | 1.67 | 0.60 | X | Translation; tRNA aminoacylation for protein translation; asparagyl tRNA aminoacylation | |
| D0R3G2 | 30S ribosomal protein S16 | rpsP | 5 | 11 | 227 | 0.73 | 1.66 | 0.60 | X | Translation | |
| D0R5D2 | 50S ribosomal protein L35 | rpmL | 7 | 8 | 80 | 0.69 | 1.61 | 0.62 | X | Translation | |
| D0R4W9 | ATP synthase subunit b | atpF | 7 | 18 | 42 | 0.65 | 1.57 | 0.64 | X | ATP biosynthetic process; ion transport; ATP synthesis coupled proton transport; ATP hydrolysis coupled cation transmembrane transport | |
| D0R5D1 | 50S ribosomal protein L20 | rplT | 8 | 13 | 101 | 0.63 | 1.55 | 0.65 | X | Ribosomal large subunit assembly; translation | |
| D0R608 | Chromosome partitioning protein ParB | parB | 6 | 33 | 51 | −3.26 | 0.10 | 9.60 | X | ||
| D0R4H4 | Pseudouridine synthase | FI9785_1123 | 5 | 27 | 115 | −3.02 | 0.12 | 8.09 | X | Psuedouridine synthesis; RNA modification | |
| D0R5U7 | Elongation factor P | efp | 8 | 21 | 105 | −2.99 | 0.13 | 7.96 | X | Translation; translational elongation; peptide biosynthetic process | |
| D0R1R2 | Putative glycosyl transferase | FI9785_242 | 8 | 35 | 46 | −2.50 | 0.18 | 5.64 | X | X | − |
| D0R254 | Extracellular solute-binding protein PhnD | phnD | 4 | 34 | 86 | −2.26 | 0.21 | 4.78 | X | Transmembrane transport | |
| D0R5M6 | Aggregation promoting factor | apf2 | 3 | 33 | 190 | −2.04 | 0.24 | 4.12 | X | − | |
| D0R1L2 | 50S ribosomal protein L6 | rplF | 12 | 19 | 205 | −2.01 | 0.25 | 4.04 | X | Translation | |
| D0R588 | Peptide chain release factor 3 | prfC | 10 | 59 | 63 | −1.99 | 0.25 | 3.97 | X | Translation; translational termination; regulation of translational termination | |
| D0R5Z4 | Tagatose-6-phosphate kinase | fruB | 4 | 33 | 153 | −1.96 | 0.26 | 3.88 | X | Carbohydrate metabolic process; lactose metabolic process; phosphorylation; carbohydrate phosphorylation | |
| D0R4W2 | MreB-like protein | mbl | 8 | 35 | 318 | −1.83 | 0.28 | 3.56 | X | Cell morphogenesis | |
| D0R4I9 | Ribonuclease Z | rnZ | 4 | 35 | 17 | −1.82 | 0.28 | 3.52 | X | tRNA processing; tRNA 3′-trailer cleavage, endonucleolytic; tRNA 3′-trailer cleavage; RNA phosphodiester bond hydrolysis, endonucleolytic | |
| D0R1L5 | 50S ribosomal protein L30 | rpmD | 2 | 6 | 37 | −1.81 | 0.29 | 3.50 | X | X | Translation |
| D0R1U3 | Tryptophan-tRNA ligase | trpS | 8 | 39 | 269 | −1.80 | 0.29 | 3.47 | X | Translation; tRNA aminoacylation for protein translation; tryptophanyl tRNA aminoacylation | |
| D0R5K3 | ATP-dependent DNA helicase | pcrA | 18 | 84 | 294 | −1.60 | 0.33 | 3.04 | X | DNA unwinding involved in DNA replication | |
| D0R2Q2 | 30S ribosomal protein S6 | rpsF | 9 | 12 | 255 | −1.60 | 0.33 | 3.03 | X | Translation | |
| D0R268 | Putative secreted protein | FI9785_210 | 21 | 102 | 323 | −1.55 | 0.34 | 2.93 | X | − | |
| D0R3I4 | Proline-tRNA ligase | proS | 16 | 63 | 297 | −1.38 | 0.39 | 2.60 | X | Translation; tRNA aminoacylation for protein translation; prolyl tRNA aminoacylation; aminoacyl-tRNA metabolism involved in translational fidelity | |
| D0R4U6 | Valine-tRNA ligase | valS | 20 | 101 | 323 | −1.37 | 0.39 | 2.59 | X | Translation; tRNA aminoacylation for protein translation; valyl tRNA aminoacylation; aminoacyl-tRNA metabolism involved in translational fidelity | |
| D0R662 | Phosphoenolpyruvate-dependent sugar phosphotransferase system EIIAB, probably mannose specific | manL | 13 | 36 | 323 | −1.16 | 0.45 | 2.24 | X | Phosphoenolpyruvate-dependent sugar phosphotransferase system; carbohydrate transmembrane transport | |
| D0R1P7 | Muramidase | FI9785_225 | 8 | 64 | 323 | −1.03 | 0.49 | 2.04 | X | Metabolic process; peptidoglycan catabolic process; cell wall macromolecule catabolic process | |
| D0R3J0 | Translation initiation factor IF-2 | infB | 13 | 99 | 122 | −1.02 | 0.49 | 2.03 | X | Translation; translational initiation | |
| D0R383 | Methionine aminopeptidase | pepM | 10 | 30 | 323 | −0.96 | 0.52 | 1.94 | X | Proteolysis; protein initiator methionine removal | |
| D0R501 | Probable transcriptional regulatory protein FI9785_1304 | FI9785_1304 | 5 | 27 | 99 | −0.88 | 0.54 | 1.84 | X | Regulation of transcription, DNA-templated | |
| D0R4I0 | Pyruvate kinase | pyk | 35 | 64 | 323 | −0.88 | 0.54 | 1.84 | X | Glycolytic process; phosphorylation | |
| D0R395 | Aminopeptidase | pepN | 13 | 96 | 140 | −0.87 | 0.55 | 1.83 | X | Proteolysis | |
| D0R2B3 | 50S ribosomal protein L10 | rplJ | 9 | 21 | 191 | −0.85 | 0.56 | 1.80 | X | Translation; ribosome biogenesis | |
| D0R3F4 | Oligopeptide-binding protein OppA | oppA | 7 | 65 | 50 | −0.81 | 0.57 | 1.76 | X | Transmembrane transport | |
| D0R1L8 | Adenylate kinase | adK | 13 | 24 | 323 | −0.74 | 0.60 | 1.67 | X | Nucleobase-containing compound metabolic process; nucleotide biosynthetic process; phosphorylation; AMP salvage; nucleoside monophosphate phosphorylation | |
| D0R5L2 | NH(3)-dependent NAD(+) synthetase | nadE | 6 | 31 | 149 | −0.71 | 0.61 | 1.63 | X | NAD biosynthetic process |
WT, L. johnsonii FI9785; D, Δeps_cluster; GO, gene ontology; –, no process identified.
X, protein identified as having significantly different abundances between D and WT with this treatment.
FIG 1.
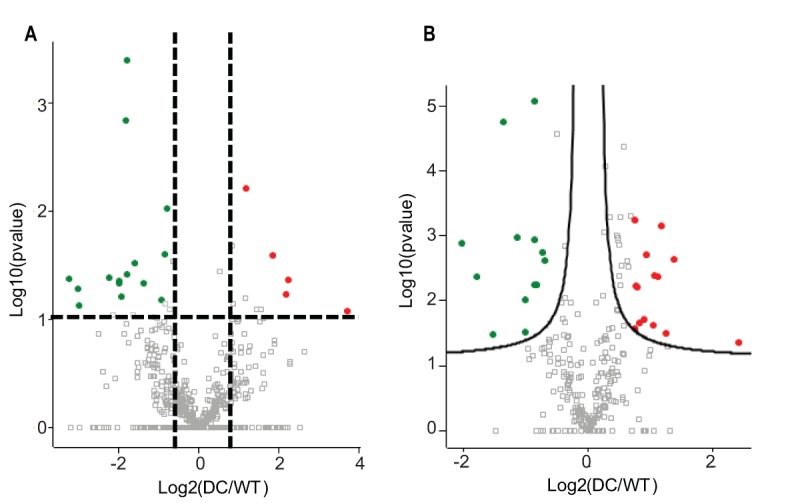
Volcano plots of differentially expressed proteins. Results compare L. johnsonii Δeps_cluster (DC) versus FI9785 (WT) obtained using a two-sided t test in panels A (iTRAQ) and B (label-free experiments). Red indicates abundance higher in DC than WT; green indicates abundance lower in DC than WT (using a P value of less than 0.05).
FIG 2.
Gene Ontology analyses of differentially expressed proteins. On the x axis, the Gene Ontology enriched terms are shown, and on the y axis, the percentage of enrichment is shown. Up, processes enriched in the Δeps_cluster mutant strain compared to the WT; Down, processes enriched in the WT compared to the mutant.
One protein found at a lower level in the Δeps_cluster strain—D0R1R2, encoded by FI9785_242 (242; renamed FI9785_RS00855)—was identified using Rapid Annotations using Subsystems Technology (RAST) analysis as a bactoprenol glycosyltransferase, which is involved in cell wall biosynthesis. This was one of the three proteins identified by both iTRAQ and the label-free protocol. Blastp analysis indicated homology to the glycosyltransferase 2 superfamily, particularly to domains cd04187 (DPM1-like bac; expected value [E-value], 7.24e−81), the PRK10714 superfamily (undecaprenyl-phosphate 4-deoxy-4-formamido-l-arabinose transferase; E-value, 1.28e−33), pfam00535 (glycosyltransferase family 2; E-value, 6.63e−28), and COG0463 (glycosyltransferase involved in cell wall biosynthesis; E-value, 2.2e−26). This protein was selected for gene deletion to investigate a possible role in EPS1 biosynthesis.
Deletion of 242 prevents biosynthesis of homopolysaccharide EPS1.
The coding sequence for FI9785_242 (242) was deleted from the L. johnsonii FI9785 genome to create strain Δ242. Comparison of proton nuclear magnetic resonance (1H-NMR) profiles of EPS extracted from the WT and strain Δ242 showed that EPS1 production was undetectable in samples extracted both from cell pellets and from supernatants (Fig. 3; Fig. S1A), indicating that 242 is essential for EPS1 production. NMR analysis of EPS extracted from a derivative of strain Δ242 containing a plasmid expressing the 242 gene under the regulation of a strong constitutive promoter (Δ242-p242) showed that complementation restored EPS1 expression, with an increased ratio of EPS1 to EPS2 compared to the WT (Fig. 3; Fig. S1A).
FIG 3.
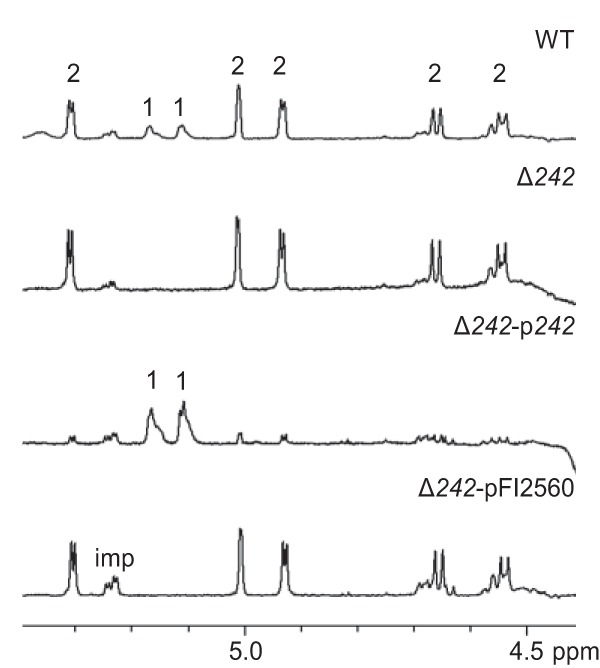
NMR analysis of pellet-associated EPS. 600 MHz 1H NMR spectra of EPS from WT and modified L. johnsonii (pellet samples, D2O, 338°K). Anomeric signals of EPS1 and EPS2 are labeled 1 and 2, respectively; imp, impurities. The presence of EPS1 is indicated by two H1 signals of equal intensity at 5.17 ppm [(1,2,6)-α-Glc] and 5.11 ppm (t-α-Glc). There are multiple H1 signals associated with EPS2 as indicated at the chemical shifts listed previously (20, 23).
Previous NMR analysis of EPS extracted from the Δeps_cluster and ΔepsA strains and then purified by trichloroacetic acid (TCA) precipitation failed to detect EPS1 or EPS2 (20, 23). However, our analysis here of crude EPS preparations prior to TCA purification, using an increased temperature and higher number of scans, revealed the presence of EPS1 in both strains (Fig. S1B). This indicates that the genes in the eps cluster which produce EPS2 are not required for EPS1 production.
241-242 show homology to GtrA-GtrB and have homologues in Gram-positive bacteria.
Blastp analysis showed that amino acid homologues of 242 are widely distributed among Lactobacillus spp., with a high conservation of amino acid sequence (71 to 100% in the first 70 matches). Alignment of 242 with GtrB proteins from Shigella phage SfII and Escherichia coli, a putative bactoprenol glycosyltransferase CsbB from Bacillus subtilis, and a polyisoprenyl-phosphate glycosyltransferase from a Synechocystis sp. whose crystal structure has been solved (25) shows areas of homology across the whole sequence, including the motifs DXD and DXSXD, which have previously been identified as being conserved in glycosyltransferases (25–27) (Fig. 4A). Mutation of selected amino acids in the Synechocystis sp. GtrB was previously shown to affect enzymatic activity (25); all but one of these amino acids are conserved in 242 (Fig. 4A).
FIG 4.
Amino acid alignments with GtrA and GtrB proteins. (A) Translation of 242 coding sequence (NCBI reference sequence WP_012845545) aligned with GtrB proteins from Shigella phage SfII (NCBI Protein accession number YP_008318506 [52]), E. coli K-12 (NCBI Protein accession number P77293 [53]), B. subtilis CsbB (NCBI Protein accession number Q45539 [54]) and Synechocystis sp. strains (NCBI Protein accession number Q55487 and Protein Data Bank number 5EKP [25]). Conserved motifs DXD and DXSXD are underlined, and residues affecting activity in 5EKP are marked with a #. (B) Translation of the 241 coding sequence (NCBI reference sequence WP_004896037) aligned with GtrA family proteins from Shigella phage SfII (NCBI Protein accession number YP_008318507 [52]) and E. coli K-12 (NCBI Protein accession number P77682 [53]). Black, dark gray, and light gray indicate 100%, 80%, and 60% homology, respectively.
Blastp analysis of the translated product of the gene upstream of 242, FI9785_241 (241; renamed FI9785_RS00850), shows homology to domains pfam04138 (GtrA-like protein; E-value, 3.04e−18) and COG2246 (putative flippase GtrA; E-value, 2.78e−07). When aligned to the GtrA sequence pairing the SfII and E. coli GtrBs, 241 shows some conservation of sequence but less than that seen with the GtrB counterparts (Fig. 4B). GtrAB pairs have been identified in a range of Gram-negative bacteria and their bacteriophages and are commonly found with a glycosyltransferase GtrX, with the three-protein complex engineering the glycosylation of O-antigens with a single sugar moiety (28). However, we could not identify any further glycosyltransferases in the L. johnsonii FI9785 genome in the immediate vicinity of 241 and 242.
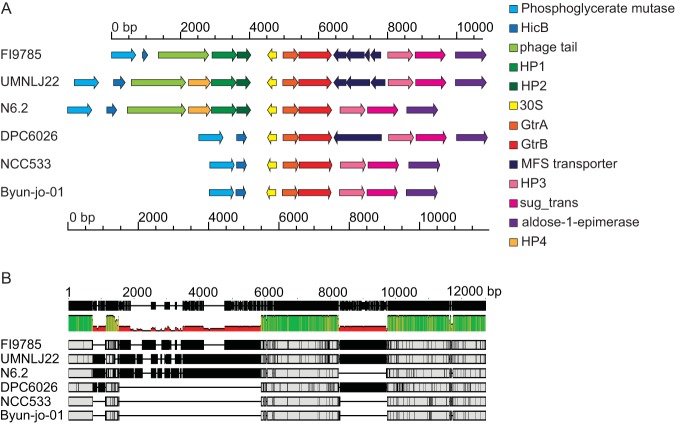
The 241-242 pair and surrounding genes show strong nucleotide conservation in other strains of L. johnsonii isolated from different sources. A surrounding 11.1-kb section encompassing 15 open reading frames (ORFs) from L. johnsonii FI9785 was compared with equivalent regions from annotated genomes of strains isolated from the human gut (NCC533), pig intestine (DPC6026), rat feces (N6.2), turkey (UMNLJ22), and mouse feces (Byun-jo-01), selecting the area between homologues of 2,3-diphosphoglycerate-dependent phosphoglycerate mutase and an aldose 1-epimerase family protein (Fig. 5). The conservation of ORFs surrounding the gtrAB pair varies among strains, with some ORFs being present but interrupted by stop codons. The section encoding the 30S ribosomal protein, 241 and 242, is present in all genomes. Translated sequences of ORFs which are present in more than one genome show high amino acid similarity between strains; the 242 sequence (NCBI reference sequence WP_012845545) shows 99 to 100% identity with the equivalent sequences in the other genomes (NCBI reference sequences WP_012845545, WP_011161379, and WP_014567007). Alignment of the surrounding nucleotide region showed high conservation of the region covering the 241-242 pair, and analysis of these two genes in the 6 genomes showed between 97.1 and 99.8% nucleic acid identity with the FI9785 sequence (Fig. 5B). The central region of strong nucleotide conservation stretches from upstream of the 30S ribosomal gene to the noncoding sequence after 242.
FIG 5.
Conservation of genes with L. johnsonii strains from different environments. (A) ORFs are shown from genomes of L. johnsonii strains FI9785 (GenBank accession number FN298497 [49], nucleotides 184194 to 194938, loci FI9785_RS00820 to FI9785_RS00875), UMNLJ22 (GenBank accession number NZ_CP021704 [T. J. Johnson and B. Youmans, unpublished], nucleotides 699996 to 711750, loci A3P32_RS03290 to A3P32_RS03350); N6.2 (GenBank accession number NC_022909 [55], nucleotides 210473 to 221016, loci T285_RS00860 to T825_RS00915); DPC6026 (GenBank accession number NC_017477 [56], nucleotides 202698 to 210932, loci LJP_RS00920 to LJP_RS00960); NCC533 (GenBank accession number NC_005362 [57], nucleotides 196136 to 202659, loci LJ_RS00845 to LJ_RS00880), and Byun-jo-01 (GenBank accession number NZ_CP029614 [D. Kim, unpublished], nucleotides shown in complement 1111505 to 1117990, loci C0060_RS05265 to C0060_RS05300) with the GtrA-GtrB pairs aligned. (B) Nucleotide alignment of the sequences in panel A performed with Mauve to indicate areas of high sequence conservation. HicB, Hic B family antitoxin; phage tail, putative phage tail-related protein; HP, hypothetical protein; 30S, 30S ribosomal protein S14; MFS transporter, major facilitator family transporter; sug-trans, sugar transporter.
241 is required for EPS1 biosynthesis.
To confirm the involvement of the putative flippase 241 in EPS1 production, a deletion mutant (Δ241) and its derivatives containing a 241 expression plasmid (Δ241-p241) or an empty plasmid control (Δ241-pQI0001) were constructed and their EPS analyzed using NMR. As with Δ242, gene deletion prevented EPS1 production while complementation restored biosynthesis (Fig. 6; Fig. S1C). A mutant where both 241 and 242 were deleted also showed production of EPS2 only (data not shown).
FIG 6.
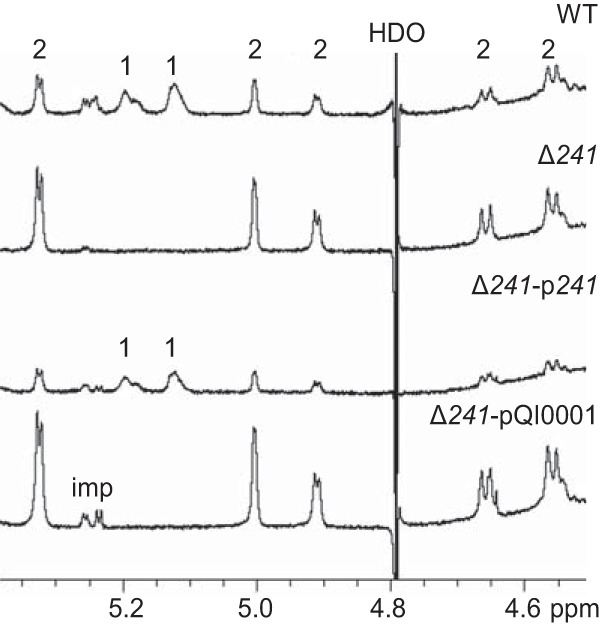
NMR analysis of pellet-associated EPS showing the effect of 241 deletion and complementation. The 600 MHz 1H NMR spectra of EPS from WT and engineered L. johnsonii (pellet samples, D2O, 300°K) are shown. Anomeric signals of EPS1 and EPS2 are labeled 1 and 2, respectively; imp, impurities.
EPS1 production affects growth.
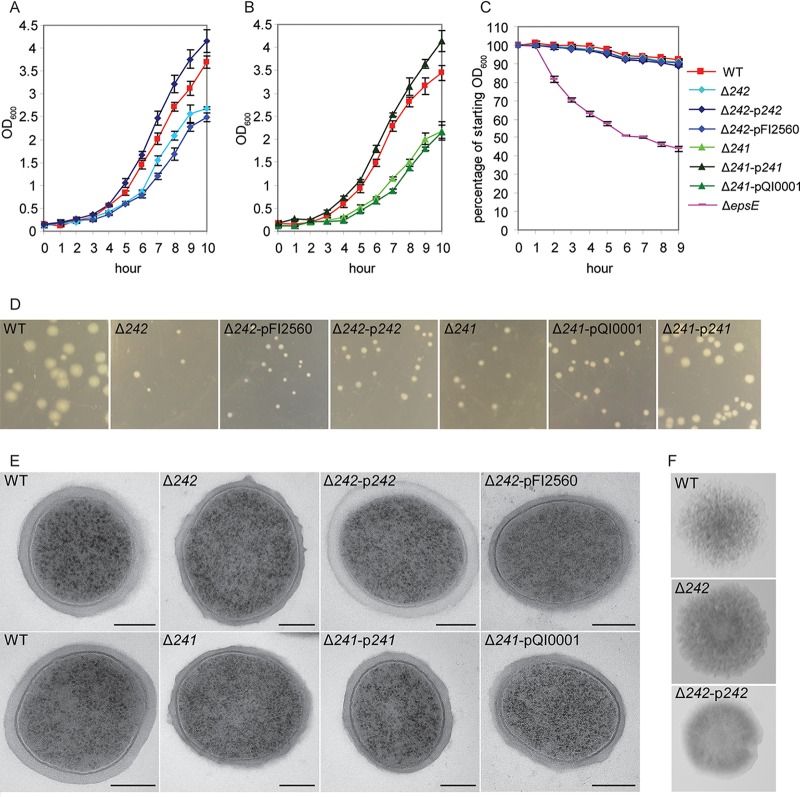
The Δ242 and Δ241 strains both showed a slower growth phenotype than the wild type, both in liquid and on solid media (Fig. 7). This phenotype was similar when the strain contained an empty vector control, but normal growth was restored in liquid by overexpression of the 242 or 241 gene, although plate growth remained slightly retarded in the 242 complemented mutant. Mutant colonies did reach the size of typical 1-day WT colonies after further incubation within 2 days. The slow-growth phenotype was maintained during growth in anaerobic conditions and at a lower temperature (30°C). The presence or absence of EPS1 did not seem to affect aggregation, while as noted previously, nonproduction of EPS2 in ΔepsE caused a strong aggregation phenotype (21), suggesting that EPS2 is a primary contributor to low aggregation of the WT (Fig. 7C). Deletion of 242 also did not have a strong effect on colony phenotype, with colonies retaining a rough and crinkled appearance, although overexpression of 242 resulted in a smoother colony upon longer incubation. Transmission electron microscopy (TEM) showed that the Δ242 and Δ241 mutants retained a visible EPS layer; this was more frequently irregular than in WT samples (Fig. 7E). Cells overexpressing 242 or 241 also exhibited a thick EPS layer, and in the case of Δ242p242, this layer was consistently paler, suggesting a different response to the osmium staining.
FIG 7.
Phenotypic characterization of 241 and 242 deletion. (A and B) Growth of L. johnsonii strains in liquid at 37°C showing an increase in optical density. (C) Aggregation of overnight cultures. (D) Differences in colony size in strains given the same incubation time at 37°C. (E) TEM analysis of cells from overnight cultures (bar = 200 nm); WT, wild type. (F) Colony phenotypes.
DISCUSSION
Effect of EPS2 loss on the L. johnsonii FI9785 proteomic profile.
Apart from variations in proteins associated with ribosome structure, translation, and protein synthesis, very few biological processes seemed strongly affected in the soluble protein content by the loss of EPS2 synthesis in the Δeps_cluster mutant strain. Comparative analysis of proteins from Lactobacillus plantarum grown at two temperature conditions, which gave a 10-fold difference in EPS production, also showed few changing proteins (29). It is interesting that loss of EPS2 production correlated with lower abundance of 242 in the Δeps_cluster mutant strain than in the WT. We have now determined that this mutant is able to produce EPS1, but its biosynthesis is affected, either by the absence of the eps cluster genes or EPS2 itself or in response to changed cell conditions responding to reduction of a protective layer. The regulation of EPS synthesis has been linked to external signal and quorum sensing in a range of bacteria, including L. plantarum (30). BLAST analysis of a putative transcriptional regulator, D0R501, which was also less abundant in the Δeps_cluster strain, showed a relationship to the YebC/PmpR family; regulators of this family are involved in a range of processes, including quorum sensing (31). Further investigation of the regulation of EPS1 and EPS2 genes, proteins, and polymers and how they relate to each other will be an interesting area for future study.
Involvement of putative flippase and bactoprenol glycosyltransferase in homopolysaccharide biosynthesis in L. johnsonii.
The evidence from EPS NMR profiles from deletion and complementation strains indicates that putative bactoprenol glycosyltransferase 242 and neighboring putative flippase 241 are key components in the production of the branched glucan EPS1. In lactic acid bacteria, α-glucans such as dextran are commonly synthesized by glucansucrases, which cleave sucrose and then add glucose to a growing chain (17). Three other mechanisms of EPS and O-antigen polysaccharide (O-PS) biosynthesis have been described in bacteria—the Wzx/Wzy-dependent pathway, the ATP-binding cassette (ABC) transporter-dependent pathway, and the synthase-dependent pathway (32). The first two mechanisms begin with the addition of a phosphorylated monosaccharide from a UDP-sugar to a lipid carrier, commonly thought to be undecaprenyl phosphate (5, 33, 34), while the synthase pathway utilizes cytosolic nucleotide-activated sugars (35, 36). Guan and coworkers described a three-gene operon, gtrABX, involved in O-antigen glycosylation in a bacteriophage infecting Shigella flexnerii and demonstrated that bactoprenol glucose transferase GtrB transferred [14C]glucose to decaprenyl phosphate in vitro (28). They proposed a model where GtrB catalyzes the transfer of glucose from UDP-glucose to bactoprenol, GtrA flips the complex across the cytoplasmic membrane, and specific glycosyltransferase GtrX transfers the glucose to a specific residue on the O-antigen repeating unit (28). More recently, GtrB homologues have been shown to be involved in glycosylation of lipoteichoic and wall teichoic acids, and a similar 3-component mechanism has been proposed (37–39).
Our hypothesis is that 242 acts as a GtrB homologue, adding a glucose molecule to a lipid carrier, while the product of neighboring gene 241 functions as a flippase. However, the full process of chain and branch formation, and the possible involvement of glycosyltransferases elsewhere in the genome, remains to be determined. 241-242 may be involved in the decoration of a linear chain synthesized by other enzymes or may be an integral part of a biosynthetic cluster. The ability of bacterial glycosyltransferases to act on different substrates and even in different pathways has been noted (40). The genes encoding the three-component system involved in Staphylococcus aureus lipoteichoic acid glycosylation are not all located together on the chromosome (38), so it would not be unprecedented for a distant gene(s) to be involved in a three- or four-component EPS biosynthetic pathway. The genome of L. johnsonii FI9785 contains several other glycosyltransferase genes which may be involved in synthesis of a linear chain, acting in concert with 241-242 to produce the final external EPS1. It is hoped that further examination of these genes will lead to a clearer model for the synthesis of this unusual EPS.
Effect of 242 or 241 deletion on L. johnsonii.
Mutations affecting L. johnsonii FI9785 EPS synthesis have been shown to affect aggregation, biofilm formation, adhesion to human HT29 cells and chicken gut explants, and resistance to stress, suggesting that EPS has a protective capacity (20, 21, 23, 41). We found that gene deletion of 242 or 241 slowed bacterial growth. The slow-growth phenotype is still seen at lowered temperatures or in the absence of oxygen, suggesting that it is not caused by increased sensitivity of cells to these conditions due to a reduction of the EPS layer. Further, removal of EPS2 did not seem to have the same effect, as the Δeps_cluster mutant strain showed a similar growth rate to the wild type when grown for proteomic analysis. It has been noted that mutations which might prevent the release of undecaprenyl phosphate by blocking the full EPS biosynthetic process affect cell viability, either by reducing the amount of undecaprenyl available for other processes or by membrane destabilization in the presence of lipid intermediates (5, 42). However, it is not obvious why deletion of a protein proposed to glycosylate the lipid carrier might have a similar effect unless there are other components of EPS1 biosynthesis that might also interact with the carrier.
In conclusion, we found that a putative glycosyltransferase, 242, was less abundant in the Δeps_cluster strain and that deletion of its gene prevented the accumulation of EPS1, while plasmid complementation restored production. In silico analysis indicated that 242 and its preceding gene, 241, show similarity to two members of a three-component system, gtrABX, shown to mediate O-antigen glycosylation in Gram-negative bacteria and, more recently, to be involved in teichoic acid glycosylation in Gram-positive species. Further deletion and complementation studies showed that 241 was also essential for EPS1 production. High conservation of nucleotide sequence with other L. johnsonii strains and the presence of analogous genes in other lactobacilli suggest that this might be part of a novel mechanism for homopolysaccharide EPS biosynthesis in Gram-positive bacteria. EPS/O-PS biosynthetic pathways have been studied in detail, but many questions remain unanswered, and new enzymes are still being discovered (43). Given the potential technological applications of EPS, there is significant interest in engineering novel forms (32), and their important roles in protection and biofilm formation make EPS biosynthesis a valid target for novel strategies to control pathogens. Further discovery of alternative mechanisms may give future opportunities to both understand and exploit bacterial EPS synthesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. johnsonii strains were grown as described previously (41) in homemade De Man Rogosa Sharpe medium (MRS) using 2% glucose as a carbon source at 37°C. Lactococcus lactis MG1614 (44) was grown in GM17 (Oxoid) at 30°C. Plasmids were selected and maintained using chloramphenicol (pFI2560 and pQI0001) at 7.5 μg ml−1 or 5 μg ml−1 and erythromycin (pG+host9) at 10 μg ml−1 and 5 μg ml−1 for L. johnsonii and L. lactis, respectively. The L. johnsonii strains and plasmids produced and/or used in this study are listed in Table 2.
TABLE 2.
L. johnsonii strains created and used in this study
| Strain | Genotype | Description | Plasmid | Reference |
|---|---|---|---|---|
| FI9785 | Wild type | Poultry isolate | 18 | |
| FI10754 | Δeps_cluster | eps gene cluster deleted | 20 | |
| FI11504 | Δ242 | FI9785 with 242 gene deleted | This study | |
| FI11646 | Δ242-p242 | FI11504 complemented with the 242 gene in expression plasmid pFI2560 | pFI2843 | This study |
| FI11647 | Δ242-pFI2560 | FI11504 with pFI2560 empty vector control | pFI2560 | This study |
| FI11669 | Δ241 | FI9785 with 241 gene deleted | This study | |
| FI11670 | Δ241-p241 | FI11669 complemented with the 241 gene in expression plasmid pQI0001 | pQI0002 | This study |
| FI11671 | Δ241-pQI0001 | FI11669 with pQI0001a empty vector control | pQI0001a | This study |
| FI10785 | ΔepsA | epsA transcriptional regulator from eps gene cluster deleted | 23 | |
| FI10844 | ΔepsE | epsE priming glycosyltransferase from eps gene cluster deleted | 21 |
Plasmid pFI2560 with cloning site NcoI altered to NdeI-BamHI.
Isolation of proteins.
Soluble protein extracts were prepared from L. johnsonii FI9785 and Δeps_cluster strains inoculated from overnight cultures at 2% into prewarmed medium and grown to an optical density (OD600) of 2.0 (6 to 7 h). Cells from 15-ml aliquots were harvested by centrifugation at 3,000 × g for 15 min at 4°C, washed with 5 ml phosphate-buffered saline (PBS) containing 1× cOmplete protease inhibitor (Roche), recentrifuged, washed with 1 ml PBS/protease inhibitor, and recentrifuged at 13,000 × g for 2 min at 4°C before removal of the supernatant and freezing on dry ice. Three biological replicates and one technical replicate were prepared for each strain. Pellets were resuspended in 500 μl extraction buffer (50 mM HEPES [pH 7.7], 0.3% SDS, 1× protease inhibitor, 5 U ml−1 RNase-free DNase [Promega], 10 mM MgSO4, and 1 mM CaCl2) and then sonicated using a Soniprep 150 (Sanyo) for 7 cycles of 15 s with 30 s incubation on ice between cycles. After centrifugation at 13,000 × g for 25 min at 4°C to pellet debris, the supernatant was precipitated overnight with 5 volumes of cold acetone at –20°C. Proteins were collected by centrifugation at 14,000 × g for 10 min at 4°C and stored at –20°C. Total soluble protein was resuspended in 250 μl 0.5 M trimethylammonium bicarbonate buffer (Sigma), 0.05% SDS, and 1× protease inhibitor and stored in LoBind tubes (Eppendorf). Concentrations were measured using Bradford reagent (Bioline).
Quantitative mass spectrometry.
Bacterial protein samples, three biological replicates of mutants and controls and one technical replicate, were digested by trypsin, and the tryptic peptides were labeled using the iTRAQ 8-plex kit (AB Sciex Pte. Ltd., USA) following the manufacturer’s instructions. The samples of each experiment were pooled and fractionated using a high-pH reversed-phase peptide fractionation kit (Pierce, Thermo Fisher Scientific). Each single fraction was analyzed using an nLC MS/MS Orbitrap Fusion trihybrid mass spectrometer coupled with a nano flow ultrahigh-performance liquid chromatography (UHPLC) system (Thermo Fisher Scientific). The peptides were separated after being trapped on a C18 precolumn, using a gradient of 3 to 40% acetonitrile in 0.1% formic acid over 50 min at a flow rate of 300 nl min−1 at 40°C. The peptides were fragmented in the linear ion trap by a data-dependent acquisition method, selecting the 40 most intense ions. For label-free experiments, each tryptic peptide sample was analyzed in triplicate as described above. All analyses were performed in triplicate. The raw data were analyzed with MaxQuant version 1.6.2.3 (Resource Identification Portal [RRID]:SCR_014485) using Andromeda software and consulting the Uniprot_ Lactobacillus johnsonii (strain FI9785) (1,726 sequences) protein database; the tolerance on parents was 10 ppm and on fragments was 0.02 ppm. The variable modifications allowed were oxidation on methionine and carboxyamidomethylation on cysteine as fixed modifications. The false discovery rate was below 1% using a decoy and reverse database, and the identified proteins contained at least 2 peptides with at least 6 amino acids sequenced. iTRAQ and label-free quantitative analyses were also performed using MaxQuant software and evaluated using Perseus statistical software (RRID:SCR_015753) with a two-sided t test, setting a P value of less than 0.05 and false-discovery rate (FDR) less than 0.01. Gene Ontology analyses were performed using the QuickGO algorithm European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI; RRID:SCR_004608).
Plasmid construction and gene deletion.
Genes were deleted from the L. johnsonii FI9785 chromosome as described previously (21) using the thermosensitive vector pG+host9 (45) containing a knockout cassette of the partial upstream and downstream genes, amplified and joined by splice overlap extension PCR using primers designed to generate restriction sites for cloning and to create spliced products (see supplemental text and Table 3). Initial cloning was performed using electrocompetent Lactococcus lactis MG1614 (46) with growth at 28°C. After sequence confirmation, plasmids were transformed into electrocompetent L. johnsonii FI9785 (47), and gene replacement was performed as described previously (45) using 30°C as the permissive temperature and 42°C as the nonpermissive temperature. For recovery of Δ242 and Δ241, it was necessary to recover deletions at 30°C, and excised plasmids were cured by successive subculturing. For complementation, the 242 gene was cloned into the L. johnsonii expression plasmid pFI2560 (21), and the 241 gene was cloned into pFI2560-derivative pQI0001 (see supplemental text); the ligation products and control vector were transformed into electrocompetent L. johnsonii FI9785 as before.
TABLE 3.
Oligonucleotide primers used for creation of deletion constructs and plasmids and assessment of sequence integrity, integration, and excision
| Primer | Sequence 5′–3′a |
|---|---|
| 241Eco_F | GATGAATTCACGCTGCTTAG |
| 241splice243_R | CGGCTTTTTGTCATATACTTTAACAGTCTTTCTTAT |
| 243Spe_R | CTACTAGTCATGATTGATTTTGGT |
| 243splice241_F | AGAAAGACTGTTAAAGTATATGACAAAAAGCCGA |
| 241_IF | GCTTCTACGTCACCAGCTTCT |
| 243_IR | TCCACAGTTTCGAACTGGTG |
| 240_F | ATGTCTAAAGTGTGACTATATGTT |
| 240splice242_R | TACTTTAACAGTCTTTCTTAGGCTTATTTTCCCTTCT |
| 242splice240_F | AGAAGGGAAAATAAGCCTAAGAAAGACTGTTAAAGTA |
| 242Spe_R | CATTTGACTAGTCATCATTCGGTAGTC |
| 240_IF | GAATGTCTAAAGTGTGACTATATGTT |
| 242_IR | ACGGTTGTATTCAGGCATATTC |
| pGhost1 | AGTCACGACGTTGTAAAACGACG |
| pGhostR | TACTACTGACAGCTTCCAAGG |
| pForVec | ACAGCAATGTTACAAGTTGAAAT |
| p181 | GCGAAGATAACAGTGACTCTA |
| 242_COD2F | AAAAAATTATCAATTATAGTTCCTTG |
| 242_C_R | GAAGCTCCACGTGAACTTC |
| 241_NdeF | TAACATATGGGTATTTTTAAAAGAATAC |
| 241_BamR | TTTGGATCCTTTAACAGTCTTTCTTATTAC |
Mismatching base pairs to insert restriction sites or for splice overlap extension are in bold.
Bioinformatic analysis.
Translated gene sequence homologies and domain searches were performed using Blastp (RRID:SCR_001010) (48). The L. johnsonii genome FN298497 (49) was reanalyzed using RAST (RRID:SCR_014606) (50). Amino acid alignments were performed using the clustalW algorithm (RRID:SCR_002909) in Vector NTI (Invitrogen; RRID:SCR_014265) and visualized using GeneDoc. Nucleotide alignments were performed using Geneious (Biomatters Ltd., New Zealand; RRID:SCR_010519); short sequences were aligned using Geneious alignment, and larger genome segments were aligned using Mauve (RRID:SCR_012852) (51).
Isolation and NMR spectroscopy of EPS.
Crude EPS was isolated from 2-day 500-ml cultures grown in MRS at 37°C as described previously (21), except that the initial extraction of capsular EPS from the washed bacterial pellet was performed by sonication in 50 ml 1 M NaCl for 7 cycles of 45 s with 30 s incubation on ice between cycles, followed by centrifugation at 6,000 × g and 4°C for 30 min to remove bacterial debris before the rounds of ethanol precipitation, the initial ethanol precipitation was for 3 days instead of overnight, and crude EPS was not further purified by TCA precipitation. EPS samples were analyzed using NMR as before (20) but with heating to 338°K (Δ242 series) and an increased number of scans (1,024). Samples in the Δ241 series were measured at 300°K.
Growth, aggregation, and phenotype studies.
Overnight (15-h) cultures of WT, ΔepsE, Δ242-p242, and Δ241-p241 strains and 20-h cultures of Δ242, Δ242-pFI2560, Δ241, and Δ241-pQI0001 strains were used as inocula for growth and aggregation studies. For liquid growth, 20-ml broths were inoculated at 2%, and the OD600 of 10-fold diluted samples was measured every hour during aerobic growth at 37°C. Colony size on plates was monitored aerobically at 30°C and 37°C and anaerobically at 37°C. All liquid growth of plasmid-containing strains was supplemented with chloramphenicol, while plate growth was nonselective. For aggregation, triplicate 1-ml samples from vortexed overnight cultures were transferred to cuvettes, and the OD600 was measured hourly during incubation at room temperature. Growth and aggregation assays were each performed three times, and representative curves are shown. TEM images were taken from overnight cultures as described previously (20).
Data availability.
All strains reported in this work are deposited in the Quadram Institute Bioscience culture collection and are available from the corresponding author upon request. For accession numbers, see Table 2.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from Biotechnology and Biological Sciences Research Council Institute Strategic Programme grants Gut Health and Food Safety BB/J004529/1 and Gut Microbes and Health BB/R012490/1, project BBS/E/F/000PR10356.
TEM images were prepared by Kathryn Cross and Catherine Booth (Quadram Institute Bioscience). We are grateful to Emmanuelle Maguin for kind provision of the pG+host9 vector and thank Nathalie Juge for critical reading of the manuscript.
M.J.M., A.D., and A.N. contributed to the conception and design of the study; M.J.M., A.D., I.J.C., and G.L.G. performed the acquisition, analysis, and interpretation of the data; all authors contributed to the writing of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Caggianiello G, Kleerebezem M, Spano G. 2016. Exopolysaccharides produced by lactic acid bacteria: from health-promoting benefits to stress tolerance mechanisms. Appl Microbiol Biotechnol 100:3877–3886. doi: 10.1007/s00253-016-7471-2. [DOI] [PubMed] [Google Scholar]
- 2.Zeidan AA, Poulsen VK, Janzen T, Buldo P, Derkx PMF, Oregaard G, Neves AR. 2017. Polysaccharide production by lactic acid bacteria: from genes to industrial applications. FEMS Microbiol Rev 41:S168–S200. doi: 10.1093/femsre/fux017. [DOI] [PubMed] [Google Scholar]
- 3.Lebeer S, Claes IJJ, Verhoeven TLA, Vanderleyden J, De Keersmaecker SCJ. 2011. Exopolysaccharides of Lactobacillus rhamnosus GG form a protective shield against innate immune factors in the intestine. Microb Biotechnol 4:368–374. doi: 10.1111/j.1751-7915.2010.00199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donot F, Fontana A, Baccou JC, Schorr-Galindo S. 2012. Microbial exopolysaccharides: main examples of synthesis, excretion, genetics and extraction. Carbohydr Polym 87:951–962. doi: 10.1016/j.carbpol.2011.08.083. [DOI] [Google Scholar]
- 5.Yother J. 2011. Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol 65:563–581. doi: 10.1146/annurev.micro.62.081307.162944. [DOI] [PubMed] [Google Scholar]
- 6.Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, Motherway MO, Shanahan F, Nally K, Dougan G, van Sinderen D. 2012. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci U S A 109:2108–2113. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Górska S, Sandstrőm C, Wojas-Turek J, Rossowska J, Pajtasz-Piasecka E, Brzozowska E, Gamian A. 2016. Structural and immunomodulatory differences among lactobacilli exopolysaccharides isolated from intestines of mice with experimentally induced inflammatory bowel disease. Sci Rep 6:37613. doi: 10.1038/srep37613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones SE, Paynich ML, Kearns DB, Knight KL. 2014. Protection from intestinal inflammation by bacterial exopolysaccharides. J Immunol 192:4813–4820. doi: 10.4049/jimmunol.1303369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zivkovic M, Hidalgo-Cantabrana C, Kojic M, Gueimonde M, Golic N, Ruas-Madiedo P. 2015. Capability of exopolysaccharide-producing Lactobacillus paraplantarum BGCG11 and its non-producing isogenic strain NB1, to counteract the effect of enteropathogens upon the epithelial cell line HT29-MTX. Food Res Int 74:199–207. doi: 10.1016/j.foodres.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Lindstrom C, Xu J, Oste R, Holst O, Molin G. 2013. Oral administration of live exopolysaccharide-producing Pediococcus parvulus, but not purified exopolysaccharide, suppressed Enterobacteriaceae without affecting bacterial diversity in ceca of mice. Appl Environ Microbiol 79:5030–5037. doi: 10.1128/AEM.01456-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salazar N, Ruas-Madiedo P, Kolida S, Collins M, Rastall R, Gibson G, de Los Reyes-Gavilán CG. 2009. Exopolysaccharides produced by Bifidobacterium longum IPLA E44 and Bifidobacterium animalis subsp. lactis IPLA R1 modify the composition and metabolic activity of human faecal microbiota in pH-controlled batch cultures. Int J Food Microbiol 135:260–267. doi: 10.1016/j.ijfoodmicro.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 12.Guttenplan SB, Blair KM, Kearns DB. 2010. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet 6:e1001243. doi: 10.1371/journal.pgen.1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HS, Park SJ, Lee KH. 2009. Role of NtrC-regulated exopolysaccharides in the biofilm formation and pathogenic interaction of Vibrio vulnificus. Mol Microbiol 74:436–453. doi: 10.1111/j.1365-2958.2009.06875.x. [DOI] [PubMed] [Google Scholar]
- 14.Koo H, Xiao J, Klein MI, Jeon JG. 2010. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol 192:3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter J, Schwab C, Loach DM, Ganzle MG, Tannock GW. 2008. Glucosyltransferase A (GtfA) and inulosucrase (Inu) of Lactobacillus reuteri TMW1.106 contribute to cell aggregation, in vitro biofilm formation, and colonization of the mouse gastrointestinal tract. Microbiology 154:72–80. doi: 10.1099/mic.0.2007/010637-0. [DOI] [PubMed] [Google Scholar]
- 16.Freitas F, Alves VD, Reis MA. 2011. Advances in bacterial exopolysaccharides: from production to biotechnological applications. Trends Biotechnol 29:388–398. doi: 10.1016/j.tibtech.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Leemhuis H, Pijning T, Dobruchowska JM, van Leeuwen SS, Kralj S, Dijkstra BW, Dijkhuizen L. 2013. Glucansucrases: three-dimensional structures, reactions, mechanism, alpha-glucan analysis and their implications in biotechnology and food applications. J Biotechnol 163:250–272. doi: 10.1016/j.jbiotec.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 18.La Ragione RM, Narbad A, Gasson MJ, Woodward MJ. 2004. In vivo characterization of Lactobacillus johnsonii FI9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry. Lett Appl Microbiol 38:197–205. doi: 10.1111/j.1472-765x.2004.01474.x. [DOI] [PubMed] [Google Scholar]
- 19.Manes-Lazaro R, Van Diemen PM, Pin C, Mayer MJ, Stevens MP, Narbad A. 2017. Administration of Lactobacillus johnsonii FI9785 to chickens affects colonisation by Campylobacter jejuni and the intestinal microbiota. Br Poult Sci 58:373–381. doi: 10.1080/00071668.2017.1307322. [DOI] [PubMed] [Google Scholar]
- 20.Dertli E, Colquhoun IJ, Gunning AP, Bongaerts RJ, Le Gall G, Bonev BB, Mayer MJ, Narbad A. 2013. Structure and biosynthesis of two exopolysaccharides produced by Lactobacillus johnsonii FI9785. J Biol Chem 288:31938–31951. doi: 10.1074/jbc.M113.507418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horn N, Wegmann U, Dertli E, Mulholland F, Collins SRA, Waldron KW, Bongaerts RJ, Mayer MJ, Narbad A. 2013. Spontaneous mutation reveals influence of exopolysaccharide on Lactobacillus johnsonii surface characteristics. PLoS One 8:e59957. doi: 10.1371/journal.pone.0059957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maina NH, Tenkanen M, Maaheimo H, Juvonen R, Virkki L. 2008. NMR spectroscopic analysis of exopolysaccharides produced by Leuconostoc citreum and Weissella confusa. Carbohydr Res 343:1446–1455. doi: 10.1016/j.carres.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Dertli E, Mayer MJ, Colquhoun IJ, Narbad A. 2016. EpsA is an essential gene in exopolysaccharide production in Lactobacillus johnsonii FI9785. Microb Biotechnol 9:496–501. doi: 10.1111/1751-7915.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, Pfeiffer JK. 2018. Bacteria facilitate enteric virus co-infection of mammalian cells and promote genetic recombination. Cell Host Microbe 23:77–88. doi: 10.1016/j.chom.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ardiccioni C, Clarke OB, Tomasek D, Issa HA, von Alpen DC, Pond HL, Banerjee S, Rajashankar KR, Liu Q, Guan Z, Li C, Kloss B, Bruni R, Kloppmann E, Rost B, Manzini MC, Shapiro L, Mancia F. 2016. Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis. Nat Commun 7:10175. doi: 10.1038/ncomms10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue H, Suzuki D, Asai K. 2013. A putative bactoprenol glycosyltransferase, CsbB, in Bacillus subtilis activates SigM in the absence of co-transcribed YfhO. Biochem Biophys Res Commun 436:6–11. doi: 10.1016/j.bbrc.2013.04.064. [DOI] [PubMed] [Google Scholar]
- 27.Mavris M, Manning PA, Morona R. 1997. Mechanism of bacteriophage SfII-mediated serotype conversion in Shigella flexneri. Mol Microbiol 26:939–950. doi: 10.1046/j.1365-2958.1997.6301997.x. [DOI] [PubMed] [Google Scholar]
- 28.Guan S, Bastin DA, Verma NK. 1999. Functional analysis of the O antigen glucosylation gene cluster of Shigella flexneri bacteriophage SfX. Microbiology 145:1263–1273. doi: 10.1099/13500872-145-5-1263. [DOI] [PubMed] [Google Scholar]
- 29.Vera Pingitore E, Pessione A, Fontana C, Mazzoli R, Pessione E. 2016. Comparative proteomic analyses for elucidating metabolic changes during EPS production under different fermentation temperatures by Lactobacillus plantarum Q823. Int J Food Microbiol 238:96–102. doi: 10.1016/j.ijfoodmicro.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Sturme MH, Nakayama J, Molenaar D, Murakami Y, Kunugi R, Fujii T, Vaughan EE, Kleerebezem M, de Vos WM. 2005. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J Bacteriol 187:5224–5235. doi: 10.1128/JB.187.15.5224-5235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang H, Li L, Dong Z, Surette MG, Duan K. 2008. The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J Bacteriol 190:6217–6227. doi: 10.1128/JB.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid J. 2018. Recent insights in microbial exopolysaccharide biosynthesis and engineering strategies. Curr Opin Biotechnol 53:130–136. doi: 10.1016/j.copbio.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Greenfield LK, Whitfield C. 2012. Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr Res 356:12–24. doi: 10.1016/j.carres.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 34.Islam ST, Lam JS. 2014. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can J Microbiol 60:697–716. doi: 10.1139/cjm-2014-0595. [DOI] [PubMed] [Google Scholar]
- 35.Hubbard C, McNamara JT, Azumaya C, Patel MS, Zimmer J. 2012. The hyaluronan synthase catalyzes the synthesis and membrane translocation of hyaluronan. J Mol Biol 418:21–31. doi: 10.1016/j.jmb.2012.01.053. [DOI] [PubMed] [Google Scholar]
- 36.Whitney JC, Howell PL. 2013. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol 21:63–72. doi: 10.1016/j.tim.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eugster MR, Haug MC, Huwiler SG, Loessner MJ. 2011. The cell wall binding domain of Listeria bacteriophage endolysin PlyP35 recognizes terminal GlcNAc residues in cell wall teichoic acid. Mol Microbiol 81:1419–1432. doi: 10.1111/j.1365-2958.2011.07774.x. [DOI] [PubMed] [Google Scholar]
- 38.Kho K, Meredith TC. 2018. Salt-induced stress stimulates a lipoteichoic acid-specific three component glycosylation system in Staphylococcus aureus. J Bacteriol 200:e00017-18. doi: 10.1128/JB.00017-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rismondo J, Percy MG, Grundling A. 2018. Discovery of genes required for lipoteichoic acid glycosylation predicts two distinct mechanisms for wall teichoic acid glycosylation. J Biol Chem 293:3293–3306. doi: 10.1074/jbc.RA117.001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tytgat HL, Lebeer S. 2014. The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol Mol Biol Rev 78:372–417. doi: 10.1128/MMBR.00007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dertli E, Mayer MJ, Narbad A. 2015. Impact of the exopolysaccharide layer on biofilms, adhesion and resistance to stress in Lactobacillus johnsonii FI9785. BMC Microbiol 15:8. doi: 10.1186/s12866-015-0347-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ou L, Ang L, Chujun Z, Jingyu H, Yongli M, Shenjing Y, Junhua H, Xu G, Yulong Y, Rui Y, Jinpan H, Bin D, Xiufang H. 2018. Identification and characterization of six glycosyltransferases involved in the biosynthesis of a new bacterial exopolysaccharide in Paenibacillus elgii. Appl Microbiol Biotechnol 102:1357–1366. doi: 10.1007/s00253-017-8673-y. [DOI] [PubMed] [Google Scholar]
- 43.Williams DM, Ovchinnikova OG, Koizumi A, Mainprize IL, Kimber MS, Lowary TL, Whitfield C. 2017. Single polysaccharide assembly protein that integrates polymerization, termination, and chain-length quality control. Proc Natl Acad Sci U S A 114:E1215–E1223. doi: 10.1073/pnas.1613609114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasson MJ. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 154:1–9. doi: 10.1128/JB.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maguin E, Prevost H, Ehrlich SD, Gruss A. 1996. Efficient insertional mutagenesis in lactococci and other Gram-positive bacteria. J Bacteriol 178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holo H, Nes IF. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123. doi: 10.1128/AEM.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horn N, Wegmann U, Narbad A, Gasson MJ. 2005. Characterisation of a novel plasmid p9785S from Lactobacillus johnsonii FI9785. Plasmid 54:176–183. doi: 10.1016/j.plasmid.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wegmann U, Overweg K, Horn N, Goesmann A, Narbad A, Gasson MJ, Shearman C. 2009. Complete genome sequence of Lactobacillus johnsonii FI9785, a competitive exclusion agent against pathogens in poultry. J Bacteriol 191:7142–7143. doi: 10.1128/JB.01182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: Rapid Annotations using Subsystems Technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.George DT, Stephenson DP, Tran E, Morona R, Verma NK. 2013. Complete genome sequence of SfII, a serotype-converting bacteriophage of the highly prevalent Shigella flexneri serotype 2a. Genome Announc 1:e00626-13. doi: 10.1128/genomeA.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 54.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Cordani JJ, Connerton IF, Cummings NJ, Daniel RA, Denziot F, Devine KM, Düsterhöft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 55.Leonard MT, Valladares RB, Ardissone A, Gonzalez CF, Lorca GL, Triplett EW. 2014. Complete genome sequences of Lactobacillus johnsonii strain N6.2 and Lactobacillus reuteri strain TD1. Genome Announc 2:e00397-14. doi: 10.1128/genomeA.00397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guinane CM, Kent RM, Norberg S, Hill C, Fitzgerald GF, Stanton C, Ross RP. 2011. Host specific diversity in Lactobacillus johnsonii as evidenced by a major chromosomal inversion and phage resistance mechanisms. PLoS One 6:e18740. doi: 10.1371/journal.pone.0018740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pridmore RD, Berger B, Desiere F, Vilanova D, Barretto C, Pittet AC, Zwahlen MC, Rouvet M, Altermann E, Barrangou R, Mollet B, Mercenier A, Klaenhammer T, Arigoni F, Schell MA. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc Natl Acad Sci U S A 101:2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains reported in this work are deposited in the Quadram Institute Bioscience culture collection and are available from the corresponding author upon request. For accession numbers, see Table 2.