The uptake of alphaproteobacterium-like cells by ancestors of eukaryotic cells and subsequent conversion of these alphaproteobacterium-like cells to mitochondria are thought to be key steps in the evolution of the first eukaryotic cells. The identification of acidocalcisomes in two alphaproteobacterial species some years ago and the presence of homologs of the vacuolar proton-translocating pyrophosphatase HppA, a marker protein of the acidocalcisome membrane in eukaryotes, in virtually all species within the alphaproteobacteria suggest that eukaryotic acidocalcisomes might also originate from related structures in ancestors of alphaproteobacterial species. Accordingly, alphaproteobacterial acidocalcisomes and eukaryotic acidocalcisomes should have similar features. Since hardly any information is available on bacterial acidocalcisomes, this study aimed at the characterization of organelle-like structures in alphaproteobacterial cells, with A. tumefaciens as an example.
KEYWORDS: polyphosphate, alphaproteobacteria, polyphosphate kinase, acidocalcisomes
ABSTRACT
Acidocalcisomes are membrane-enclosed, polyphosphate-containing acidic organelles in lower Eukaryota but have also been described for Agrobacterium tumefaciens (M. Seufferheld, M. Vieira, A. Ruiz, C. O. Rodrigues, S. Moreno, and R. Docampo, J Biol Chem 278:29971–29978, 2003, https://doi.org/10.1074/jbc.M304548200). This study aimed at the characterization of polyphosphate-containing acidocalcisomes in this alphaproteobacterium. Unexpectedly, fluorescence microscopic investigation of A. tumefaciens cells using fluorescent dyes and localization of constructed fusions of polyphosphate kinases (PPKs) and of vacuolar H+-translocating pyrophosphatase (HppA) with enhanced yellow fluorescent protein (eYFP) suggested that acidocalcisomes and polyphosphate are different subcellular structures. Acidocalcisomes and polyphosphate granules were frequently located close together, near the cell poles. However, they never shared the same position. Mutant strains of A. tumefaciens with deletions of both ppk genes (Δppk1 Δppk2) were unable to form polyphosphate but still showed cell pole-located eYFP-HppA foci and could be stained with MitoTracker. In conclusion, A. tumefaciens forms polyP granules that are free of a surrounding membrane and thus resemble polyP granules of Ralstonia eutropha and other bacteria. The composition, contents, and function of the subcellular structures that are stainable with MitoTracker and harbor eYFP-HppA remain unclear.
IMPORTANCE The uptake of alphaproteobacterium-like cells by ancestors of eukaryotic cells and subsequent conversion of these alphaproteobacterium-like cells to mitochondria are thought to be key steps in the evolution of the first eukaryotic cells. The identification of acidocalcisomes in two alphaproteobacterial species some years ago and the presence of homologs of the vacuolar proton-translocating pyrophosphatase HppA, a marker protein of the acidocalcisome membrane in eukaryotes, in virtually all species within the alphaproteobacteria suggest that eukaryotic acidocalcisomes might also originate from related structures in ancestors of alphaproteobacterial species. Accordingly, alphaproteobacterial acidocalcisomes and eukaryotic acidocalcisomes should have similar features. Since hardly any information is available on bacterial acidocalcisomes, this study aimed at the characterization of organelle-like structures in alphaproteobacterial cells, with A. tumefaciens as an example.
INTRODUCTION
Despite the detection of a surprising variety of prokaryotic subcellular structures (1), only a few intracellular compartments have been identified as membrane-enclosed, organelle-like structures in prokaryotic species (2). Well-known examples of intracellular compartments that are surrounded by a phospholipid bilayer membrane are the magnetosomes in magnetotactic bacteria (3). Anammoxosomes in Planctomycetes also represent intracellular compartments but have a ladderane-containing membrane instead of conventional phospholipid membranes (4). A third example of membrane-enclosed intracellular compartments in bacteria are the acidocalcisomes, which have been described in two alphaproteobacterial species, Agrobacterium tumefaciens (5) (also Agrobacterium fabrum and Rhizobium radiobacter) and Rhodospirillum rubrum (6). Acidocalcisomes are widespread in lower eukaryotes such as unicellular parasites (e.g., Trypanosoma species) but have been also found in higher eukaryotes, including humans (7, 8). Acidocalcisomes are related to lysosomes; they represent acidic compartments and are rich in calcium ions and phosphorus compounds such as phosphate, pyrophosphate, and polyphosphate (9). Obvious functions of acidocalcisomes include the storage of phosphate and divalent cations (Ca2+), but many other functions, such as osmoregulation, blood coagulation, and inflammation, have been ascribed to acidocalcisomes in eukaryotes (10).
Acidocalcisomes in bacterial species have not been further analyzed since their discovery in A. tumefaciens and R. rubrum (5, 6), and the function of acidocalcisomes in bacteria is unclear. We therefore initiated experiments to determine the properties of acidocalcisomes in bacteria and to reveal their function, using A. tumefaciens as an example.
RESULTS AND DISCUSSION
Isolation of acidocalcisomes.
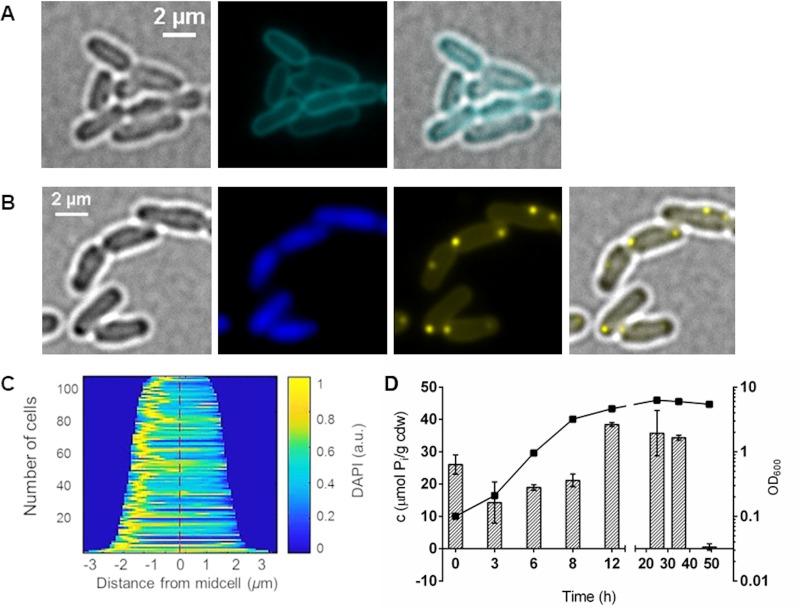
Characteristic features of (bacterial) acidocalcisomes are the interior low pH, the accumulation of polyphosphate (polyP) in the lumen, and the presence of the vacuolar proton-translocating pyrophosphatase HppA. Bacterial acidocalcisomes can be therefore detected by staining the cells with (i) LysoSensor dyes, which show good fluorescence in the protonated state under acidic conditions but only a little fluorescence in the nonprotonated state at neutral pH; (ii) 4′,6-diamidino-2-phenylindole (DAPI) and imaging at DAPI-polyP-specific emission wavelengths; or (iii) antibodies against HppA (5). We first stained A. tumefaciens with LysoSensor green DND-189 (Fig. 1A; see also Fig. S1 in the supplemental material). Surprisingly, staining of A. tumefaciens with this dye resulted in a clear staining of the cell membrane, but no intracellular structures were detected that could represent the expected acidocalcisomes. Since A. tumefaciens acidocalcisomes should contain polyP, we stained A. tumefaciens with DAPI and inspected the cells via fluorescence microscopy using a DAPI-polyP-specific emission wavelength of around 520 nm (Fig. 1B). The presence of DAPI-stainable polyP-containing globular structures was clearly confirmed in the exponential- and early-stationary-growth phases (see the next paragraph for details). polyP-containing cells were harvested, and the procedure for acidocalcisome isolation from A. tumefaciens (5) by enzymes/detergent/French press treatment, followed by a iodixanol density gradient centrifugation, was applied. Several bands were detected after centrifugation (Fig. S2A). Fluorescence microscopic inspection of the gradient fractions for the enrichment of DAPI-polyP-stainable material and determination of the polyP content of each fraction revealed the presence of polyP in all fractions without a clear enrichment of polyP in a specific fraction (Fig. S2B and S3). Also, in the (invisible) pellet fraction, a few globular structures were identified that showed DAPI-polyP-specific fluorescence in addition to cell debris and not completely lysed cells. When we additionally stained samples of each fraction with dyes suitable to detect intact phospholipid-membrane-surrounded organelle-like structures (MitoTracker or LysoTracker), we found much stained cell debris but no structures that showed fluorescence both in the red and in the DAPI-polyP channel and thus could represent acidocalcisomes. Repeated variations of the cell lysis method and iodixanol gradient centrifugation conditions did not lead to a substantial improvement. None of the six independent attempts gave evidence for the presence of intact acidocalcisomes after cell lysis and gradient centrifugation. Only polyP (without a MitoTracker signal) could be reproducibly detected in all gradient fractions. The isolation of intact acidocalcisomes from cells expressing the eypf-hppA fusion (see below for details) also failed. These results indicated that acidocalcisomes are extremely labile after cell lysis. We therefore decided to characterize acidocalcisomes by fluorescence microscopy of intact, living cells.
FIG 1.
Formation of polyP in A. tumefaciens. (A) Bacterial cells from exponential growth were stained with 10 μM LysoSensor green DND-189 and analyzed with a CFP filter. Merged images show an overlay of the CFP and bright-field channel. (B) LB-grown cells during exponential growth were stained with 0.5 μg/ml DAPI and imaged in bright-field fluorescence microscopy with a DAPI-DNA-specific or DAPI-polyP-specific filter. The microscopic image on the right is an overlay of bright-field microscopy and DAPI-polyP. (C) Demograph showing the localization of DAPI-polyP fluorescence by cell length. (D) The growth (OD600) and the amount of intracellular polyP on LB medium (measured as total inorganic phosphate in μmol per cell dry weight [cdw]) were determined at different growth stages. Error bars represent standard deviations of the results from three technical replicates. The deviation values for the OD values are too small to visualize.
Fluorescence microscopic detection of acidocalcisomes in A. tumefaciens.
To determine under which conditions acidocalcisomes were formed, we cultivated A. tumefaciens and investigated the cells via fluorescence microscopy (Fig. 1). Since LysoSensor green DND-189 stained only the cell membrane (using the dye at a concentration of 1 μM or 10 μM; see above and Fig. 1A and S1), we stained the cells routinely with DAPI. Fluorescent DAPI-polyP foci and polyP levels between 10 and 40 μmol inorganic phosphate residues per gram of cellular dry weight (μmol Pi/cdw) were detected during the exponential-growth phase and the early stationary phase (Fig. 1B and D). However, in the late-stationary-growth phase (50 h), polyP was hardly detectable (Fig. 1D). Most cells imaged in samples of the exponential-growth phase had one DAPI-polyP focus (Fig. 1B and C). Occasionally, cells with two DAPI-polyP foci and, in rare cases, cells with three or even four foci were detected. The foci were usually located at or near one of the poles but sometimes could also be detected in other parts of the cell. The frequent observation of foci at/near the cell poles was in agreement with the data from Seufferheld and coworkers, who observed DAPI-stainable and electron-dense, phosphorus- and calcium-containing globular structures (acidocalcisomes) also at/near the cell poles by fluorescence or electron microscopy, respectively (5). We conclude that polyP-containing globular structures are present during growth and are located predominantly near the cell pole(s).
The frequently observed close distance of the polyP-DAPI foci to the cell pole and/or to the inner surface of the cells suggested that the acidocalcisomes could originate from an invagination process of the cell membrane as has been described for the formation of magnetosomes (3). To test this possibility, we stained samples of A. tumefaciens that had been taken at various stages of growth and had been cultivated in the presence of the fluorescent dye FM1-43 or with FM4-64, with and without DAPI. As shown in a representative example in Fig. 2A, the dye FM1-43 stained the cell membrane, but polyP-DAPI foci were never costained with FM1-43 or with FM4-64 (not shown). When the cells were stained with LysoSensor green DND-189 and FM4-64, both dyes clearly stained the cytoplasm membrane, but structures inside the cells were not stained at all (Fig. 2B). The same results were obtained for samples of cultures in which the FM dyes had been added after sampling (not shown). Evidence for the origin of polyP-harboring membrane-enclosed acidocalcisomes by an invagination process of the cell membrane was not obtained, and a connection with acidocalcisomes is therefore unlikely.
FIG 2.
(A and B) Staining of A. tumefaciens with FM1-43 and DAPI after 24 h (A) and with FM4-64 and LysoSensor green DND-189 after 48 h (B). (A) Cells were grown in LB medium in the presence of 0.1 μg/ml FM1-43. Samples were taken at 0, 2, 4, 6, 24, 32, 48, 52, and 72 h after inoculation, stained with 0.5 μg/ml DAPI, and (from left to right) imaged with bright-field microscopy or with fluorescence microscopy with an FM1-43-specific or DAPI-polyP-specific filter, with the rightmost being an overlay of the green and yellow channels. The same result was obtained if FM4-64 was used instead of FM1-43 together with an FM4-64-specific filter set (not shown). (B) A. tumefaciens wild-type cells were stained after 48 h with 1 μM LysoSensor green DND-189 and 0.1 μg/ml FM4-64 and analyzed with an FM4-64-specific or a LysoSensor green-specific filter. The experiment was performed in three biological replicates. A typical result is shown.
Localization of the acidocalcisome membrane with phospholipid-sensing proteins.
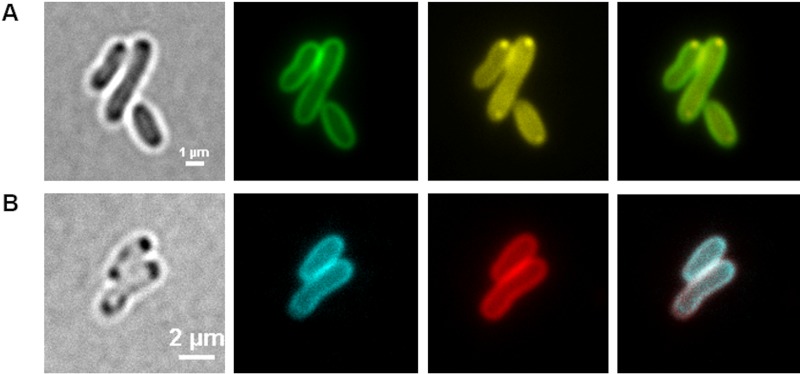
Since LysoSensor green DND-189 clearly stained the cell membrane of A. tumefaciens but failed to detect the acidocalcisomes, we tried to detect the acidocalcisome membrane by using phospholipid-specific proteins. The C2 domain of the bovine lactadherin protein (LactC2) is able to bind to phosphatidylserine and related membrane phospholipids (11–14). In a recent study, we showed that a fusion of the LactC2 domain with a fluorescent protein (dsRed2EC-LactC2) is able to bind to and visualize the cytoplasmic membranes of several proteobacteria (15). Remarkably, dsRed2EC-LactC2 fluorescence also colocalized with the magnetosome chains in Magnetospirillum gryphiswaldense, thus confirming the presence of a phospholipid layer around magnetosomes. We therefore thought that the same construct could be useful to stain the membrane of acidocalcisomes in A. tumefaciens. When we imaged cells harboring a plasmid coding for the dsRed2EC-LactC2 fusion protein (pBBR1MCS2-dsred2EC-lactC2), cap-like fluorescent foci near the cell poles or near the future cell poles in cells shortly before cell division were observed (Fig. 3), whereas the cell membrane of A. tumefaciens was only weakly stained with dsRed2EC-LactC2. However, globule-shaped fluorescent structures in the cells that could be indicative of the localization of acidocalcisomes were not detected (all stages between 0 and 48 h of growth on LB medium tested). Remarkably, when cells harboring the dsRed2EC-LactC2 fusion were stained with DAPI, granule-like DAPI-polyP foci were detected that did, however, not colocalize with the cap-like structures of dsRed2EC-LactC2 fluorescence. This indicated that the DAPI-stained polyP-containing structures do not contain substantial amounts of phospholipids and most likely are not covered by a phospholipid membrane. The nature of the cap-like structures at the cell poles remains obscure.
FIG 3.
A. tumefaciens cells harboring pBBR1MCS2-Pphac-dsred2EC-lactC2 after growth on LB medium for 24 h. From left to right are bright-field, dsRed channel, DAPI-polyP channel, and merged images. The dsRed2EC-LactC2 protein formed a cap-like fluorescence signal at one cell pole (white arrow head), whereas the DAPI-polyP-specific foci (arrows) appeared at both poles. The overlay of the dsRed2EC and DAPI-polyP signals showed no colocalization. The experiment was performed in three biological replicates. A typical result is shown.
Localization of acidocalcisomes with fluorescent dyes.
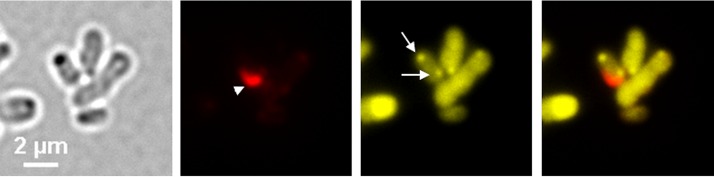
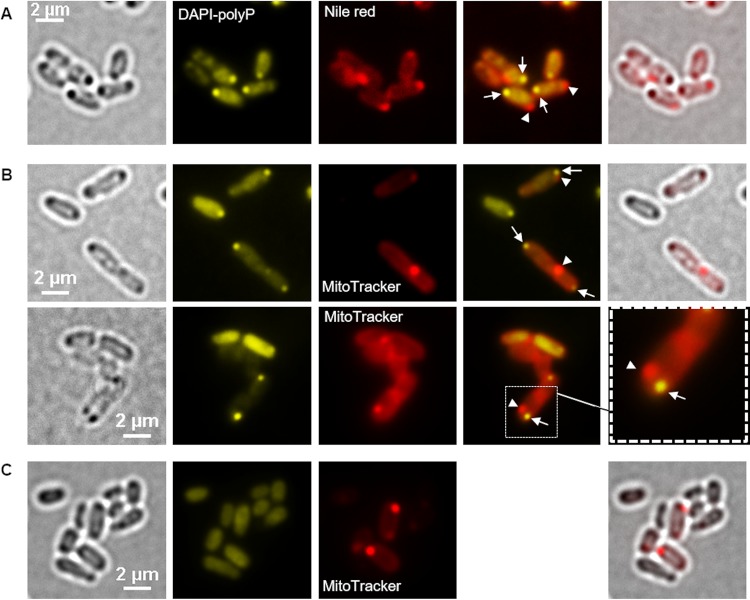
Hydrophobic compounds such as polyhydroxybutyrate (PHB) and membrane-enclosed compartments (e.g., magnetosomes in magnetotactic bacteria or mitochondria in eukaryotes) can be stained with fluorescent dyes that are enriched in a hydrophobic environment. Nile red and MitoTracker are examples of such compounds that are frequently used to stain hydrophobic structures (PHB and lipids) and compartments with electrochemical potential in prokaryotic and eukaryotic cells, respectively (for examples, see references 15–18). We therefore tested Nile red and MitoTracker for their use in detecting the acidocalcisomes in A. tumefaciens. Figure 4A shows A. tumefaciens wild-type cells that had been stained both with Nile red and with DAPI. Present in the DAPI-polyP channel were fluorescent foci near one of the cell poles that colocalized with globular dark structures in bright field. We conclude that the cells produced polyP in the form of granule-like structures and that at least some of the dark structures visible in bright field represented these polyP granules. The same cells also revealed Nile red foci (white arrowheads in Fig. 4A). However, these Nile red foci were located clearly apart from the DAPI-polyP fluorescence (white arrows in Fig. 4A, mostly near the other cell pole), and a dark structure in bright field was present only in some cases at the same position. The presence of PHB, which can be well stained by Nile red (16), was excluded by gas chromatography (GC) analysis of samples regardless of the time point of harvest (Fig. S4). Furthermore, Nile red staining of A. tumefaciens cells in which the PHB synthase gene (Atu_1604) had been deleted resulted in images (not shown) comparable to those for the wild type. The nature of the Nile red signals remains unclear, but the presence of PHB granules could be excluded in cells that were grown on LB medium (detection limit, <1% PHB of cellular weight).
FIG 4.
Fluorescence microscopy of A. tumefaciens. (A and B) LB-grown A. tumefaciens wild type was stained with Nile red and DAPI (A) or with MitoTracker and DAPI (B). (C) Images showing a polyP-free mutant (A. tumefaciens Δppk1 Δppk2 mutant) stained with MitoTracker and DAPI. From left to right are bright-field, DAPI-polyP channel, red channel, merged DAPI-polyP channel and red channel, and merged bright-field and red channel or enlargement images. (A) The white arrows point to DAPI-polyP foci and the arrowheads to Nile red foci. (A and B) The MitoTracker or Nile red foci (arrowheads) and the DAPI-polyP signals (arrows) occurred in close proximity in the cells, but no colocalization was observed. (C) The deletion of both ppk genes caused the absence of DAPI-polyP foci but had no effect on the formation of MitoTracker foci. The experiments were performed in six biological replicates. A representative result is shown.
Next, we stained A. tumefaciens cells with MitoTracker and with DAPI (Fig. 4B and S5). Again, the DAPI-polyP-specific signals colocalized with dark globular structures visible in bright field. However, despite being closely located to each other, the MitoTracker foci were clearly separated from the DAPI-polyP foci. Since the presence of PHB could be excluded (see above), this result suggested the presence of two different structures near the cell pole; first, there is the polyP-containing structure that is also visible in bright field, and second, there is a MitoTracker-stainable structure that is different from the polyP-containing structure. When cells in which the two polyP kinase genes present in A. tumefaciens (ppk1 and ppk2; for details, see below) had been deleted were stained with MitoTracker, the same foci as in wild-type cells were detected (Fig. 4C). However, DAPI-polyP foci were absent. The nature of the MitoTracker-stainable material is unknown.
We also investigated cells that had been stained with LysoTracker, a dye similar to LysoSensor that shows fluorescence only under acidic conditions and that is used to stain lysosomes and acidocalcisomes. However, no reproducible results were obtained with this dye. Inhomogeneous staining was detected despite several independent attempts (Fig. S6). In summary, we were not able to provide evidence for the presence of membrane-enclosed polyP-containing acidocalcisomes. Rather, our results indicated the presence of two separate subcellular structures located closely together near the cell pole. One structure obviously represents polyP granules, and the other seems to contain or consist of a hydrophobic material. We speculate that the Nile red- and MitoTracker-stainable structures at the cell pole correspond to a hydrophobic or membrane-enclosed compartment that is free of DAPI-stainable polyP and could represent organelles that have been previously described as acidocalcisomes. The LactC2-stainable structure (Fig. 3) most likely corresponds to a region of the cytoplasmic membrane at one cell pole that apparently has a phospholipid composition that is better attracted by LactC2 than are other parts of the membrane. A similar complex staining behavior of the cytoplasmic membrane has been previously described for FM dyes, which enrich preferentially in the cytoplasmic membrane around the old cell pole in A. tumefaciens cells (19) if the concentration of the FM dye is limiting. On the other hand, the DAPI-stainable material might correspond to “ordinary” polyP granules that are not surrounded by a hydrophobic membranous structure and that are located in the close vicinity of the putative acidocalcisomes near the cell pole. To find additional evidence for this assumption, we decided to localize the positions of key enzymes of acidocalcisomes (the vacuolar proton-translocating pyrophosphatase HppA) and of polyP granules (the polyP kinases [PPKs]).
Localization of the acidocalcisome-specific vacuolar proton-translocating pyrophosphatase HppA.
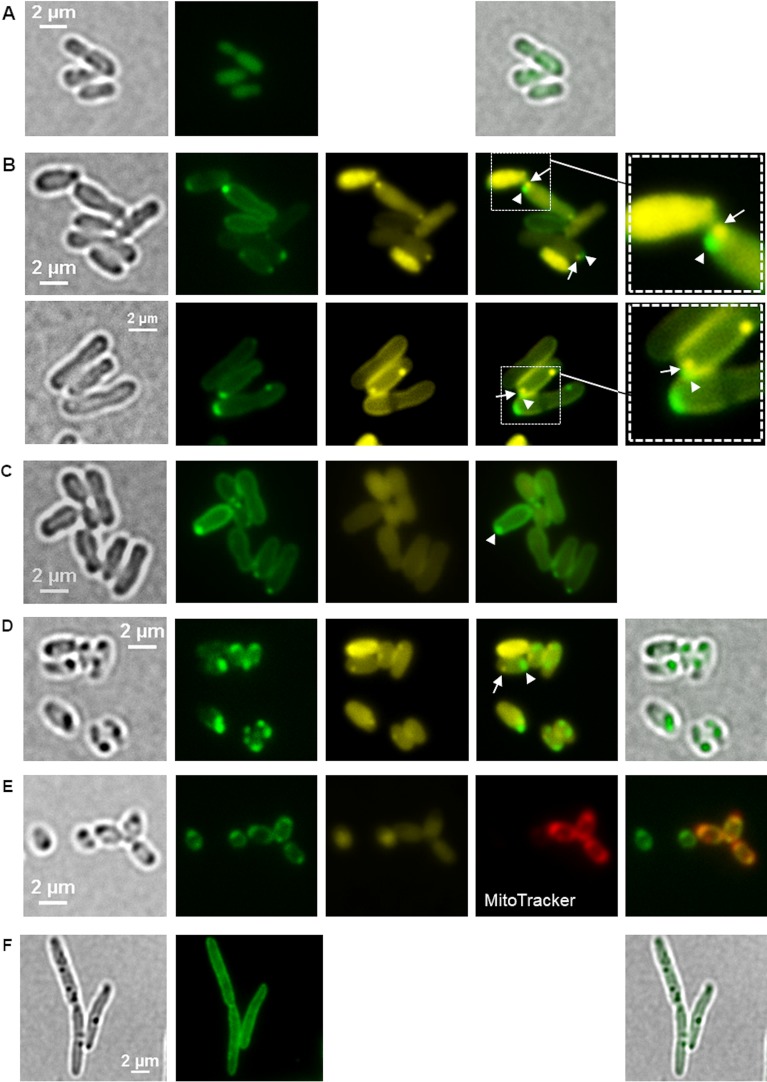
The vacuolar proton-translocating pyrophosphatase HppA is the characteristic enzyme of acidocalcisomes and is preferentially localized in the acidocalcisome membrane (6, 20–22). The vacuolar proton-translocating function of HppA is thought to cause an acidification of acidocalcisomes. Inspection of the A. tumefaciens genome revealed the presence of one hppA gene (Atu_1174). The genomic context of hppA suggested that it is a singly transcribed gene. Furthermore, the amino acid sequence of the A. tumefaciens HppA protein shows sequence similarities to the HppA proteins of other species. BLAST analysis revealed sequence identities of 37% with the HppA of Trypanosoma cruzi and 42% with the HppA of Arabidopsis thaliana. Analysis of the predicted HppA amino acid sequence (712 amino acids) with the TMHMM server v. 2.0 (DTU Bioinformatics) suggested that the 72.5-kDa protein forms 16 transmembrane helices, similar to other HppAs, and is most likely a membrane-integrated protein in A. tumefaciens. We thought that labeling of HppA by fusion with a fluorescent protein and fluorescence microscopic detection of the fusion protein would be a suitable tool to indirectly determine the subcellular localization of acidocalcisomes in A. tumefaciens. To this end, we constructed a fusion of hppA with the gene for the enhanced yellow fluorescent protein (eyfp) on a broad-host-range plasmid (pBBR1MCS2) (for proper positioning of eyfp within hppA, see Materials and Methods) and transferred the resulting plasmid to A. tumefaciens. The expression of eyfp alone as a control yielded soluble eYFP fluorescence in the cells (Fig. 5A). Figure 5B and Movies S7A and S7B show fluorescence microscopy images of A. tumefaciens cells expressing the eyfp-hppA fusion from the pBBR1MCS2 plasmid. A colocalization of eYFP-HppA with the cytoplasmic membrane was observed in all cells. In addition, most cells had one or two fluorescent foci at or near the cell pole(s). This result is similar to data from R. rubrum showing that HppA is present at the position of the suspected acidocalcisomes and in the cytoplasm membrane (6). The same localization pattern of eYFP-HppA at the cell membrane and as a focus near one cell pole was observed when the eyfp-hppA fusion was expressed in a background in which the two polyP kinase genes (ppk1 and ppk2) had been deleted (Fig. 5C). This mutant is unable to form polyP granules or to synthesize detectable amounts of polyP (see below). This result demonstrated that the localization of eYFP-HppA is independent of the presence of polyP.
FIG 5.
Localization of eYFP-HppA in A. tumefaciens. (A) Homogeneous fluorescence in cells expressing eyfp without fusion confirmed that eYFP is a soluble, cytoplasmic protein in A. tumefaciens. Samples were taken from three biological replicates. (B) A. tumefaciens cells harboring pBBR1MCS2-PphaC-eyfp-hppA were stained with DAPI and imaged (from left to right) by bright field and fluorescence microscopy using an eYFP-specific filter and a DAPI-polyP-specific filter. The image second from the right shows an overlay of the eYFP channel and the DAPI-polyP channel with a magnification shown in the rightmost image. Arrowheads and arrows point to eYFP-HppA foci and DAPI-polyP foci, respectively, located side by side at the cell pole. Microscopy images were taken from four biological replicates. (C) polyP-free mutant of A. tumefaciens (Δppk1 Δppk2 mutant) harboring pBBR1MCS2-PphaC-eyfp-hppA. The experiment was performed in two biological replicates. (D) LB-grown cells of A. tumefaciens (48 h) with a genomic eyfp-hppA integration. (E) eYFP-HppA foci were strongly visible, and additional staining with MitoTracker showed a colocalization of the MitoTracker and eYFP-HppA signals. (F) Introduction of the pBBR1MCS2-PphaC-eyfp-hppA plasmid into R. eutropha resulted in only membrane staining and no formation of fluorescent foci. Samples were analyzed in three (D and E) and two (F) biological replicates.
When cells in which the hppA gene was replaced by a genome-integrated eyfp-hppA fusion under the native hppA promoter to avoid a gene copy effect of the plasmid-derived expression of eyfp-hppA were investigated, the intensity of the eYFP-HppA fluorescence was weaker than that of the plasmid-expressed construct. Hardly any distinct fluorescence foci were detectable in cells that were imaged during the growth phase (not shown). However, eYFP-HppA fluorescence became readily detectable during the stationary-growth phase (24 to 48 h after inoculation) in the form of fluorescent foci at the cell poles (Fig. 5D), while the eYFP-HppA fluorescence at the cell membrane was less intense and hardly detectable. Interestingly, bright-field microscopy revealed dark-stained structures at the same positions as the cell pole-located eYFP-HppA fluorescence foci. Moreover, these structures could be stained with MitoTracker but not with DAPI when imaged at a DAPI-polyP-specific wavelength (Fig. 5E). DAPI-stainable polyP granules were regularly identified during growth but were only rarely detectable in the late-stationary-growth phase. If DAPI-stained polyP-specific foci were detected in the stationary phase, they were clearly separated from the position of eYFP-HppA fluorescence (see below for details).
For comparison, eyfp-hppA was also expressed in two non-acidocalcisome-forming species, Ralstonia eutropha (Fig. 5F) and Escherichia coli. In these species, eYFP-HppA generally colocalized with the cytoplasm membrane.
Acidocalcisomes of A. tumefaciens do not colocalize with DAPI-stainable polyP.
When we stained A. tumefaciens cells that expressed the eyfp-hppA fusion (either from pBBR1MCS2-PphaC-eyfp-hppA or from the genomic eyfp-hppA integration) with DAPI and imaged the cells at DAPI-polyP-specific and eYFP-specific wavelengths, in most cases, we detected cells with either one or two DAPI-polyP-specific foci (in the lag phase and the exponential-growth phase). However, cells harboring the pBBR1MCS2-PphaC-eyfp-hppA plasmid showed one or two eYFP-HppA-specific fluorescence foci in all growth phases (Fig. S8), whereas the cells with genomic eyfp-hppA integration formed distinct fluorescence foci only in the stationary-growth phase (Fig. 5B and D). Only rarely did we detect cells with both DAPI-polyP-specific and eYFP-HppA-specific fluorescence foci in cells with the genomic eyfp-hppA integration. In such cells, the DAPI-polyP-specific foci and the eYFP-HppA-specific foci did not colocalize (see magnifications in Fig. 5B) despite being located closely together. An optical artifact caused by the differential diffraction of short and longer wavelengths could be excluded (Fig. S9). We conclude that the DAPI-polyP foci and the eYFP-HppA foci represent different subcellular structures in A. tumefaciens. Since no DAPI-polyP signal could be detected at the position of the eYFP-HppA foci, the structures representing the eYFP-HppA foci do not contain detectable amounts of DAPI-stainable polyP. Our data suggest that acidocalcisomes and polyphosphate granules are located close together but are different structures in A. tumefaciens. Deletion of the hppA gene had no effect on the growth of A. tumefaciens (Fig. S10A) or on the formation of polyP granules and MitoTracker foci; likewise, the PPK1-mCherry signals were still present in A. tumefaciens ΔhppA mutant cells (Fig. S10B and C).
polyP kinases are responsible for polyP granule formation and utilization but do not affect the formation of HppA-containing acidocalcisome-like structures.
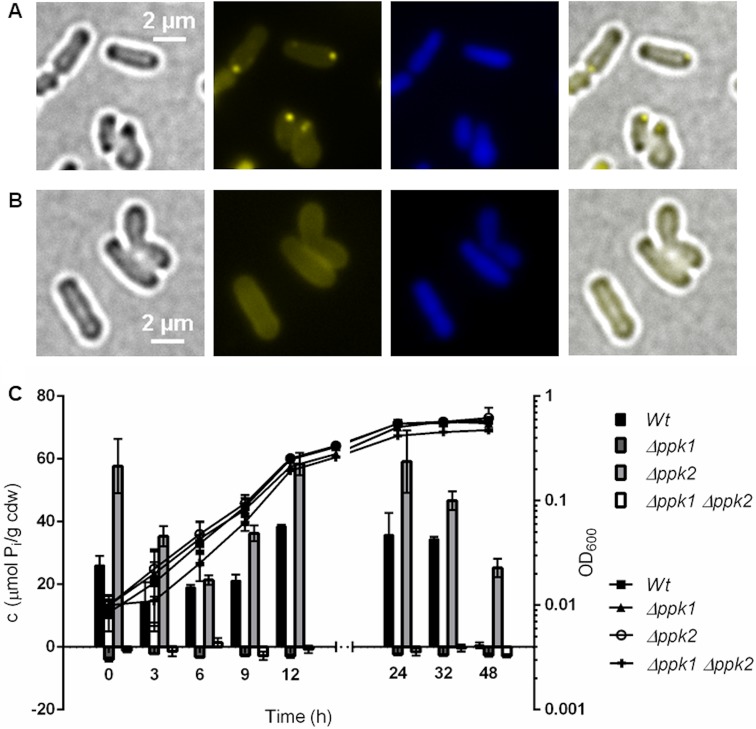
As shown above, A. tumefaciens has polyP granules that seemed to be distinct from the structures that harbor HppA and that possibly represent acidocalcisomes. polyP is synthesized by the action of polyP kinases (PPKs) (23). Two genes coding for PPKs are present in the A. tumefaciens genome. The first (Atu1144, ppk1) is a member of class 1 PPKs (≈80-kDa proteins with N, H, C1, and C2 domains [24]) and is located next to the gene for an exopolyphosphatase (PPX) as in many other prokaryotic species. The second PPK (Atu0418, ppk2) is a member of class 2 PPKs (one PPK2 motif, 34 kDa) and is located in the neighborhood of an operon encoding the high-affinity phosphate transporter (Pst) components. We constructed A. tumefaciens cells with precise deletions of ppk1, ppk2, or both ppk genes. Growth was not detectably affected in either the Δppk1 or Δppk2 single mutant or the Δppk1 Δppk2 double mutant on LB medium. The Δppk1 mutant and the Δppk1 Δppk2 double mutant were unable to produce DAPI-detectable polyP granules, and no polyP could be extracted from cells lacking ppk1 (Fig. 6A to C). The Δppk2 mutant was not impaired in polyP formation; instead, the Δppk2 mutant formed even more polyP than did the wild type (Fig. 6C). These findings suggest that PPK1 is responsible for polyP formation and that PPK2 in vivo might work toward nucleotide synthesis at the expense of polyP rather than work in the direction of polyP formation.
FIG 6.
PPK1 is responsible for polyP formation in A. tumefaciens. (A and B) LB-grown A. tumefaciens Δppk2 mutant cells (A) still formed DAPI-polyP foci, but DAPI-stained A. tumefaciens Δppk1 mutant cells (B) showed no characteristic DAPI-polyP fluorescence signal. From left to right are bright-field, DAPI-polyP channel, DAPI-DNA channel, and merged images. Microscopy images were taken from three biological replicates. (C) Growth on LB medium (OD600) and polyP content in μmol Pi per cell dry weight (cdw) of A. tumefaciens wild type and Δppk1, Δppk2, and Δppk1 Δppk2 mutants. The growth experiment was performed in a plate reader with five technical replicates and the polyP quantification with three technical replicates.
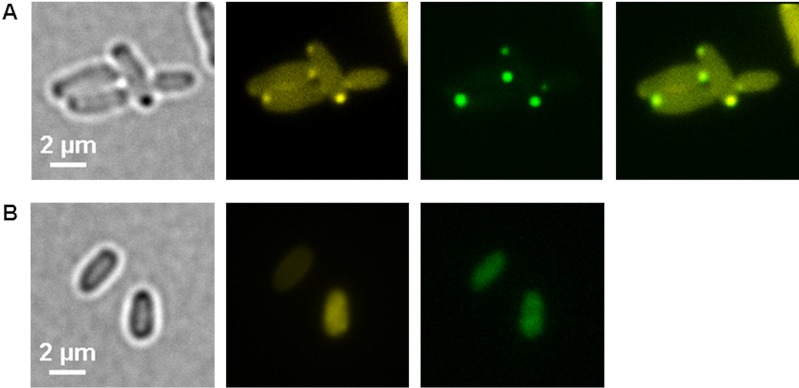
Additionally, we used the R. eutropha phosin PptA to confirm the absence of polyP in a Δppk1 Δppk2 mutant of A. tumefaciens. Phosins are polyP-binding proteins characterized by the presence of a so-called conserved histidine α-helical domain (CHAD). Phosins were always attached to polyP in Gram-negative and Gram-positive bacteria (25–27) and presumably also bind to polyP in Archaea and plants (28). The expression of an eyfp-pptA fusion in the A. tumefaciens wild type revealed eYFP-PptA foci that colocalized with DAPI-polyP foci, whereas in a Δppk1 Δppk2 background, homogeneous fluorescence of eYFP-PptA was observed (Fig. 7A and B), confirming the absence of polyP to which PptA could bind.
FIG 7.
Expression of eyfp-pptA from R. eutropha in A. tumefaciens. (A and B) A. tumefaciens wild-type strain (A) and A. tumefaciens Δppk1 Δppk2 mutant strain (B) each harboring pBBR1MCS2-PphaC-eyfp-pptA. From left to right are bright-field, DAPI-polyP channel, eYFP-PptA channel, and merged images. Note the colocalization of eYFP-PptA with DAPI-polyP in the wild type and soluble eYFP-PptA fluorescence in the polyP-free A. tumefaciens Δppk1 Δppk2 mutant cells. The experiment was performed in two biological replicates.
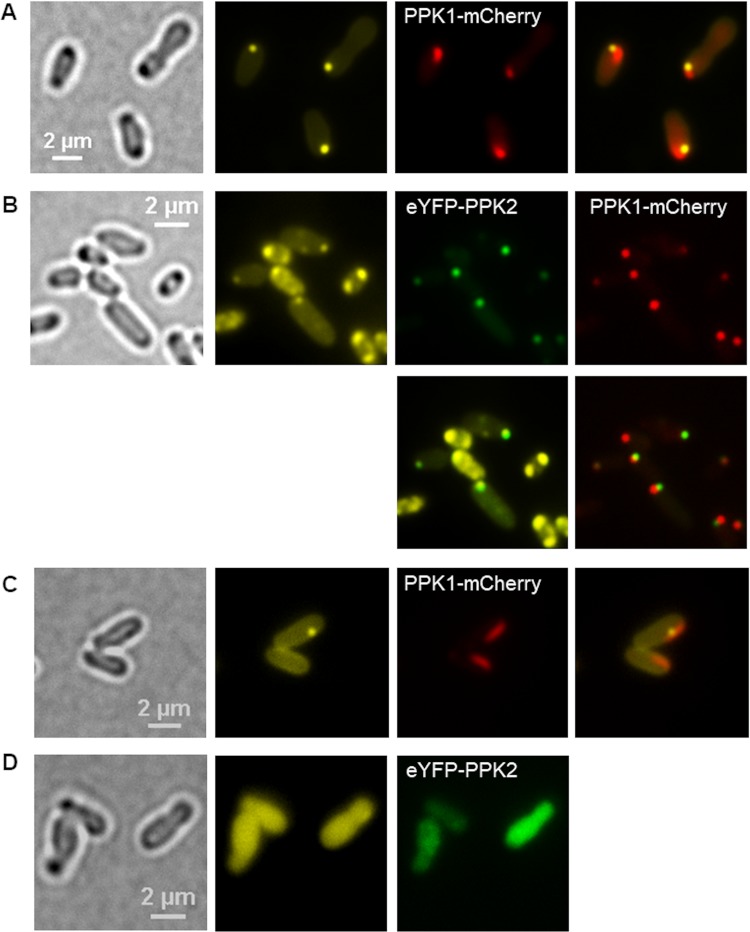
To localize the subcellular positions of PPK1 and PPK2, fusions of the ppk genes with mcherry and with eyfp were constructed (ppk1-mcherry and eyfp-ppk2, respectively). The expression of ppk1-mcherry from pBBR1MCS2 in the wild type (Fig. 8A) yielded elliptically shaped fluorescence foci that were located side by side with DAPI-polyP foci but did not (completely) colocalize with polyP granules. The same ellipse- to rod-shaped fluorescence foci were detected when ppk1-mcherry was expressed from the pBBR1MCS2 plasmid in an A. tumefaciens background in which both ppk genes had been deleted (Fig. 8C) or in an A. tumefaciens ΔhppA mutant (Fig. S10C). The formation of DAPI-polyP foci side by side with the PPK1-mCherry foci confirmed that PPK1 is sufficient to complement the polyP-negative phenotype. When a genome-integrated ppk1-mCherry fusion was expressed in the A. tumefaciens wild type, globular instead of elliptical PPK1-mCherry foci were observed side by side with DAPI-polyP foci (Fig. 8B), suggesting that the elliptical form of the plasmid-expressed construct was caused by overexpression of the gene. When additional eypf-ppk2 was expressed, eYFP-PPK2 foci were detected that clearly colocalized with DAPI-polyP (Fig. 8B).
FIG 8.
Localization of PPK1 and PPK2 in A. tumefaciens. (A) LB-grown cells of A. tumefaciens harboring pBBR1MCS2-PphaC-ppk1-mcherry were stained with DAPI and imaged (from left to right) in bright field, in the DAPI-polyP channel, and in the mCherry channel. The experiment was performed in five biological replicates. (B) A. tumefaciens cells with a genome-integrated ppk1-mcherry gene and harboring pBBR1MCS2-PphaC-eyfp-ppk2 are shown. (C and D) Cells of the A. tumefaciens Δppk1 Δppk2 mutant into which pBBR1MCS2-PphaC-ppk1-mcherry plasmid (C) or pBBR1MCS2-PphaC-eyfp-ppk2 (D) had been transferred. Note that ppk1-mcherry, but not eyfp-ppk2, was able to complement the formation of DAPI-polyP granules. (B to D) Samples were taken from three biological replicates.
Complementation of the Δppk1 Δppk2 mutant with a plasmid harboring eyfp-ppk2 did not lead to the formation of DAPI-polyP foci and confirmed that PPK2 in A. tumefaciens is not able to form polyP. Instead, only soluble fluorescence of eYFP-PPK2 was detected (Fig. 8D). In conclusion, PPK1 seems to be located in the form of a protein cluster closely attached to polyP granules and is responsible for polyP formation, whereas PPK2 is presumably important for consuming polyP and is always attached to polyP granules (if polyP is present).
Conclusions.
This study showed that A. tumefaciens is able to form two separate subcellular structures. polyP granules and acidocalcisome-like structures are located closely together and were predominantly detected at or near the cell poles. The formation of one of these structures is dependent on the presence of ppk1, and this structure contains DAPI-stainable polyP. PPK2 is attached to the polyP granule but does not form it. No evidence could be obtained that the polyP-containing structure is enclosed by a phospholipid membrane or harbors the vacuolar proton-translocating pyrophosphatase HppA, a marker enzyme of acidocalcisomes in eukaryotes. We therefore conclude that the polyP-containing structures are “ordinary” polyP granules as seen in other bacterial species, such as the alphaproteobacteria Caulobacter crescentus (29) and Magnetospirillum gryphiswaldense (30), the betaproteobacterium R. eutropha (31), the gammaproteobacteria Pseudomonas aeruginosa (32) and Pseudomonas putida (26), and others. The second subcellular structure of A. tumefaciens harbors the vacuolar proton-translocating pyrophosphatase HppA (in the stationary-growth phase) and may be surrounded by a membrane or consist of a hydrophobic material but is free of DAPI-stainable polyP. This structure probably corresponds to the structures that had been previously described for A. tumefaciens and R. rubrum and were referred to as acidocalcisomes in these studies (5, 6). The contents and putative functions of these acidocalcisome-like structures await clarification.
The “disappearance” of DAPI-stainable polyP granules in the late-stationary-growth phase and the appearance of eYFP-HppA foci in the stationary phase might suggest that membrane-free polyP granules could be the precursors of membrane-surrounded acidocalcisomes. However, in this case, one would expect the presence of detectable amounts of polyP in these structures, as Seufferheld and coworkers have clearly identified elevated concentrations of phosphorus at the positions of electron-dense structures in comparison to other positions in the cytoplasm (5) by energy-dispersive X-ray (EDX) analysis. However, cells in late-stationary phase did not contain DAPI-stainable polyP, and polyP could not be extracted from such cells. We therefore think that A. tumefaciens is able to form only “ordinary” polyP granules that are not surrounded by a membrane. Unfortunately, we have not yet performed transmission electron microscopy (TEM) to find independent support for our assumption. However, if we carefully examine the TEM image of A. tumefaciens in Fig. 1 of reference 5, the evidence for the presence of a membrane around the globular, electron-dense structures near the cell pole is not without doubt. Furthermore, the assumption that polyP and acidocalcisomes are part of the same organelle-like structures was deduced by individual staining of cells on separate microscopic slides with either LysoSensor, cycloprodigiosin (an uncoupler of HppA activity with affinity to acidocalcisomes), or DAPI (see Fig. 3 of reference 5). Therefore, a colocalization of DAPI-polyP and acidocalcisomes cannot be well differentiated from close localization of the two structures. Further studies are necessary to examine the nature of the acidocalcisome-like structures in A. tumefaciens and related alphaproteobacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains, plasmids, and oligonucleotides used in this study are shown in Table 1. Escherichia coli and A. tumefaciens C58 strains were grown in lysogeny broth (LB; 88 rpm agitation) medium supplemented with the appropriate antibiotics at 37°C or 30°C, respectively. For additional growth experiments, A. tumefaciens was grown in AT minimal medium with glucose and (NH4)2SO4 (ATGN), as described elsewhere (33), with the exception that 0.19 mM Fe(NH4) citrate was used instead of 0.125 M FeSO4−.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotide used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristics or sequencea | Reference or source |
|---|---|---|
| Strains | ||
| Agrobacterium tumefaciens C58 | Wild type, Kms | |
| A. tumefaciens ΔphaC mutant | Deletion of PHB synthase gene phaC (Atu_1604) | This study |
| A. tumefaciens ΔhppA mutant | Deletion of H+-pyrophosphatase hppA (Atu_1174) | This study |
| A. tumefaciens Δppk1 mutant | Deletion of polyP kinase PPK1 gene ppk1 (Atu_1144) | This study |
| A. tumefaciens Δppk2 mutant | Deletion of polyP kinase PPK2 gene ppk2 (Atu_0418) | This study |
| A. tumefaciens Δppk1 Δppk2 mutant | Deletion of ppk1 and ppk2 | This study |
| A. tumefaciens eyfp-hppA | Chromosomal replacement of hppA by eyfp-hppA | This study |
| Ralstonia eutropha H16 | Wild type, forms PHB and polyP granules but no acidocalcisomes (no hppA gene) | DSMZ428 |
| Plasmids | ||
| pBBR1MCS2-PphaC-eyfp-c1 | Broad-host-range vector for construction of gene fusions with eyfp, confers Kmr, constitutive expression from PphaC | 34 |
| pBBR1MCS2-PphaC-dsred2EC-c1 | Broad-host-range vector for construction of gene fusions with Dsred2EC constitutive expression from PphaC | 15 |
| pBBR1MCS2-PphaC-eyfp-hppA | Constitutive expression of eyfp-hppA | This study |
| pBBR1MCS2-PphaC-eyfp-ppk2 | Constitutive expression of eyfp-ppk2 | This study |
| pBBR1MCS2-PphaC-ppk1-mcherry | Constitutive expression of ppk1-mcherry | This study |
| pBBR1MCS2-PphaC-dsred2EC-lactC2 | Constitutive expression of dsred2EC-lactC2 | 15 |
| pBBR1MCS2-PphaC-eyfp-pptA | Constitutive expression of eyfp-pptA | 23 |
| pHis_EcPPX1 | Source of PPX | A. Saiardi, UC London |
| Oligonucleotides | ||
| Sacl_upF_atu1144 | CGATCAGAGCTCGACCGGCGGGGTTTTGACGATACG | |
| upR_atu1144 | GCCGTTAATTAAGCCGGCCCTGTTCCGTCCCCTGAC | |
| downF_atu1144 | CGGCTTAATTAACGGCATGACTCGATCAGAAGCACAG | |
| SpeI_downR_atu1144 | GCACTAGTCGAAGGATTTGACGTGTTTTCTGGC | |
| Sacl_upF_atu0418 | CGATCAGAGCTCCCTTACGCCGGAAATGACGTT | |
| upR_atu0418 | GCCGTTAATTAAGCCGGGTTTCCTCCGGATGTTGT | |
| downF_atu0418 | CGGCTTAATTAACGGCGACCGGGAGACG | |
| SpeI_downR_atu0418 | GCACTAGTTGACTGGGTCGAGGTCTTTAATGC | |
| Sacl_upF_atu1604 | CGATCAGAGCTCCTTATATTCGGCTTCCGAA | |
| upR_atu1604 | GCCGTTAATTAAGCCGGCACGATCCTCCA | |
| downF_atu16004 | CGGCTTAATTAACGGCTCTCATCCGCCACTCTC | |
| SpeI_downR_atu1604 | GCACTAGTCGCCACGTTTGGTGGCTGTA | |
| 500bp_up_1174_fwd | tcgagctcggtacccgggggcgcgccTCTGGGCAGCGCTCATCC | |
| 500bp_up_1174_rev | actatgggtattacggtcatGCGCACTTACCTCCCAAAAG | |
| eYFP_atu1174_fwd | cttttgggaggtaagtgcgcATGACCGTAATACCCATAGTAATTTTATGCG | |
| eYFP_atu1174_rev | cagtctagctatcgccatgactagtTCAGTGGGCAAGAACTGC | |
| PPK1_mcherry_fwd | tcgagctcggtacccgggggcgcgccATGGACGCTATCGCGCAG | |
| PPK1_mcherry_rev | cccctgtgcttctgatcgagTTACTTGTACAGCTCGTCCATG | |
| 500bp_down_ppk1_fwd | tggacgagctgtacaagtaaCTCGATCAGAAGCACAGGGG | |
| 500bp_down_ppk1_rev | cagtctagctatcgccatgactagtCGGCAGGGTGATGCCCTC |
Uppercase letters indicate bases complementary to the respective gene sequence. Kms, kanamycin sensitive; Kmr, kanamycin resistant.
Isolation of acidocalcisomes.
The method for acidocalcisome isolation was performed as closely as possible to the previously described method for acidocalcisome isolation from A. tumefaciens and R. rubrum (5, 6); bacteria were cultivated in LB medium for 18 h after inoculation under agitation. A second preculture was inoculated 1:10, cultivated for 24 h, and used to inoculate the main culture (400 ml) at an optical density at 600 nm (OD600) of 0.1. The main culture was harvested after growth for 18, 24, or 48 h by centrifugation at 3,900 × g (Beckman rotor JLA 8.1) and 4°C. Each pellet was resuspended in 20 ml lysis buffer {125 mM sucrose, 50 mM KCl, 4 mM MgCl2, 10 mM EDTA, 20 mM HEPES-KOH [4-(2-hydroxyethyl)-1-piperazine-ethane-sulfonic acid], 5 mM dithiothreitol (DTT), 0.1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 10 μM pepstatin, 10 μM leupeptin, 10 μM trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane, and 10 μM N-tosyl-l-lysine-chloromethyl-ketone (pH 7.2)} containing 2 mg/ml lysozyme. After incubation on ice for 30 min, 1 μl/ml DNase and RNase (each 10 mg/ml) were added to the lysate and subsequently passed through a French press (SLM-Aminco; Spectometric Instruments) twice at 1,000 lb/in2. Lysis of the cells was verified by microscopy. The lysate was incubated on ice under agitation for 1 h with an equal volume of silica/silicon carbide (400-mesh particle size), followed by centrifugation at 1,000 × g for 5 min to remove DNA and RNA. After two washings of the pellet with lysis buffer, the supernatant fractions were combined and centrifuged at 14,500 × g for 10 min. The pellet was resuspended in 2 ml of lysis buffer, and the resulting suspension was diluted with one volume of OptiPrep (60% [wt/vol] iodixanol) and applied on top of a discontinuous iodixanol gradient consisting of 4-ml volumes of 24, 28, 30, 35, and 40% iodixanol in a 35-ml polycarbonate centrifugation tube. Subsequently, the gradient was centrifuged at 50,000 × g in a Beckman SW 32 Ti rotor for 60 min. Gradient fractions were isolated and stored at –20°C or used directly.
Alternatively, we followed the OptiPrep application sheet S48 protocol provided by Alere Technologies (http://www.axis-shield-density-gradient-media.com/S48.pdf). Here, the lysis buffer (buffer B) consisted of 0.125 M sucrose, 50 mM KCl, 4 mM MgCl2, 0.5 mM EDTA, 5 mM dithiothreitol (DTT), and 20 mM HEPES-KOH (pH 7.2). Solution C (0.125 M sucrose, 0.3 M KCl, 24 mM MgCl2, 3.0 mM EDTA, 30 mM DTT, 120 mM HEPES-KOH [pH 7.2]) was used for the dilution of 50% (wt/vol) iodixanol. Working solution D was prepared by mixing 5 volumes of OptiPrep with 1 volume of solution C. Furthermore, the cell suspension was diluted in iodixanol and applied as the 24% (vol/vol) concentration step of a discontinuous gradient as described in the OptiPrep application sheet mentioned above (http://www.axis-shield-density-gradient-media.com/S48.pdf).
Construction of gene fusions with fluorescent protein genes.
For the construction of in-frame gene fusions, PCR-generated fragments of the respective gene were digested with appropriate restriction enzymes and ligated into pBBR1MCS2-pphaC-eyfp (harboring the gene for enhanced yellow fluorescent protein [eYFP]) under the control of the constitutive PHB synthase gene (phaC) promoter of Ralstonia eutropha (34). The DNA sequences of the PCR-amplified regions were determined by using commercial DNA sequencing services for each construct. Only constructs with verified DNA sequences were used. Alternatively, Gibson cloning was done as follows. First, 15 μl of the Gibson Assembly buffer (5× isothermal [ISO] reaction buffer [final concentration in Gibson mix of 25% polyethylene glycol 800 {PEG-800}, 500 mM Tris-HCl {pH 7.5}, 50 mM MgCl2, 50 mM DTT, 1 mM each dinucleoside triphosphates {dNTPs}], 5 mM NAD, 0.08 U T5 exonuclease, 0.5 U Phusion DNA polymerase, and 80 U Taq ligase, with water added to 15 μl) together with 5 μl of PCR-generated DNA fragments (insert-to-backbone ratio, 2:1 to 8:1) were incubated at 50°C for 1 h, at 25°C for 3 min, and at 4°C for 3 min. E. coli TOP10 cells were transformed with 5 μl of a 1:4 dilution of the Gibson batch by heat shock. Recombinant clones were selected by plating on agar medium containing the appropriate antibiotic. The isolated plasmids were subsequently transferred to A. tumefaciens C58 via electroporation at 2.5 kV, 500 Ω, and 25 μF. Selection of transformants was performed after regeneration of the cells on LB medium at 30°C for 75 min by plating and subsequent growth on LB agar containing 50 μg/ml kanamycin.
The positioning of the fusion of eyfp into the hppA-DNA sequence was done in a way such that the formed eYFP protein was fused into the cytoplasm-exposed loop after the first transmembrane helix of HppA. A similar procedure has been previously successfully used to detect the in vivo position of an HppA-green fluorescent protein (HppA-GFP) fusion in Arabidopsis thaliana (35).
Construction of gene deletions and insertions.
The sacB-containing pLO3 suicide vector was used for the construction of markerless gene deletion mutants. For construction of ppk1 (Atu_1144), ppk2 (Atu_0418), hppA (Atu_1174), or phaC (Atu_1604) deletion mutants, the respective 500-bp up- and downstream regions were amplified using the oligonucleotide pairs listed in Table 1. After cloning of the PCR-generated inserts into pLO3 via the SacI and Spel restriction sites, the respective plasmids (pLO3_Δppk1, pLO3_Δppk2, pLO3_ΔhppA, or pLO3_ΔphaC) were transferred into A. tumefaciens via electroporation (see above). Clones with the genome-integrated plasmid were verified by PCR and cultivated in LB medium in the absence of kanamycin overnight at 30°C. Then, 1 ml of the culture was used to inoculate 9 ml of fresh LB medium containing 1% (wt/vol) sucrose. After an incubation period of 3 h at 30°C, 100 μl of a 10−1 dilution (in LB medium) was plated on LB agar containing 10% (wt/vol) sucrose and grown at 30°C. Arising colonies with the deleted gene of interest were identified and verified by PCR amplification of the genomic region around the gene deletion site and DNA sequence determination of the generated PCR product. The construction of genomic gene fusions and exchange of the parental gene by the gene fusion were performed in an analogous manner. To verify the DNA sequence of each construct, commercial DNA sequencing of the entire length of the PCR-amplified region was determined.
Microscopic methods.
Fluorescence microscopic visualization of the polyanion polyP in A. tumefaciens was conducted by staining the cells with 0.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) for at least 15 min. polyP was detected with the aid of a DAPI-polyP-specific filter set (excitation, 415/20 nm; emission, 520/60 nm). Fluorescence microscopy and detection of fluorescent proteins (mCherry, dsRed2EC, and eYFP) were performed on a Nikon Ti-E microscope (model number MEA53100) by using F36-503 tetramethylrhodamine (TRITC) high contrast (HC) (excitation, 543/22 nm; emission, 593/40 nm) and F36-528 YFP HC (excitation, 500/24 nm; emission, 542/27 nm) filter sets for analysis. Cells that had been stained with LysoTracker red DND-99 (4 μg/ml in H2O), MitoTracker redFM (5 μg/ml in dimethyl sulfoxide [DMSO]), Nile red (0.05 μg/ml in 60% ethanol), and FM4-64 (0.1 μg/ml in DMSO) were visualized using standard filter sets with 562/40-nm excitation and 594-nm emission (long-pass filter), whereas FM1-43 (0.1 μg/ml in DMSO) was detected at 500/24-nm excitation and at 542/27-nm emission. Samples stained with LysoSensor green DND-189 (0.05, 1, or 10 μM in DMSO) were analyzed with fluorescence microscopy using an F31-044 enhanced cyan fluorescent protein (eCFP)/CFP filter set (excitation, 436/20 nm; emission, 480/40 nm). Cells were imaged with a digital camera (Hamamatsu Orca-Flash 4.0 scientific complementary metal-oxide semiconductor [sCMOS] camera) and processed with Nikon imaging software and GIMP 2.10.14. polyP granules could be also visualized by bright-field microscopy. To image the cells, 3-μl portions of the sample were immobilized on agarose pads (1% [wt/vol] in phosphate-buffered saline) and covered with a coverslip. For time-lapse experiments, 3-μl portions of a cell suspension were added to a Nunc glass-base dish (12 mm) and covered with LB agar pads (1% [wt/vol] in LB medium). During microscopy, cells were incubated at 30°C in an incubation chamber and imaged every 5 min for 8 to 12 h.
polyP extraction and purification.
The purification of polyP from lyophilized cell samples was done by neutral phenol-chloroform extraction and ethanol precipitation following a protocol described elsewhere (36). Two milligrams of dried cells was resuspended in AE buffer (50 mM sodium acetate [pH 5.3], 10 mM EDTA) at 4°C. The sample was transferred to a 1.5-ml Eppendorf tube containing 300 μl phenol and 40 μl SDS (10% [wt/vol]). After incubation at 65°C for 5 min, followed by 1 min on ice, 300 μl of trichloromethane was added. Subsequently, the tube was mixed and centrifuged at room temperature for 2 min at 13,000 × g. The top aqueous phase (around 450 μl) containing the polyP was transferred to a prepared 1.5-ml tube with 350 μl trichloromethane, mixed, and centrifuged again at room temperature for 2 min at 13,000 × g. Two microliters of RNase A (10 mg/ml) and 2 μl of DNase I (10 mg/ml) were incubated together with the top aqueous phase for 1 h at 37°C in a new 1.5-ml tube. Subsequently, the sample was transferred to a precooled (–20°C) 1.5-ml tube containing 1 ml of ethanol and 40 μl of 3 M sodium acetate (pH 5.3) and incubated for 3 h or overnight at –20°C to precipitate polyP. Finally, the sample was centrifuged at 13,000 × g and 4°C for 20 min, and the supernatant was discarded by decantation. The polyP precipitate was washed with 500 μl of 70% ethanol, followed by centrifugation for 5 min at 13,000 × g and 4°C. The supernatant was discarded by decantation, and the dried pellet was dissolved in 1 ml distilled water and stored at –20°C.
Phosphate quantification by malachite green assay.
Cell-extracted polyP was hydrolyzed to monophosphate using exopolyphosphatase from Saccharomyces cerevisiae (ScPPX) at 37°C for 15 min. To this end, 150 μl polyP extract was mixed with 40 μl of 5× ScPPX1 reaction buffer (100 mM Tris-HCl [pH 7.5], 25 mM MgCl2, 250 mM ammonium acetate), 9 μl H2O, und 1.6 μl ScPPX1 (1 μg) (purified from E. coli/pHis_EcPPX1). Each polyP sample was processed in triplicate, and controls without PPX1 were used to calculate the background phosphate levels. Fifty microliters per sample was pipetted into the wells of a 96-well plate together with 35 μl solution A (4 volumes of 2 N HCl and 3 volumes of 0.1 M Na2MoO4) and 15 μl of solution B (0.042% malachite green [MG] in H2O). After incubation for 2 min at room temperature, 100 μl of 1.6 N H2SO4 was added to each well, and the formation of the malachite green-colored phosphomolybdate complex was allowed for 30 min at room temperature. The concentration of the complex was determined at A625 using a plate reader (Eon; BioTek, Bad Friedrichshall, Germany). The concentration of inorganic phosphate (Pi) was calculated by comparison of the A625 values with a calibration graph obtained with solutions of 5, 15, 30, 60, 90, and 120 μM KH2PO4. The data are expressed in μmol Pi/cdw.
Other techniques.
PHB was determined by gas chromatography after conversion of PHB in lyophilized samples to 3-hydroxybutyrate methyl esters, as described previously (37). The optical density of cells was measured in a 1-ml microcuvette at 600 nm with 3 biological replicates. Additionally, cell growth was measured in 96-well plates using a plate reader (BioTek) at 600 nm in 200 μl of LB medium. To this end, 6 to 8 biological replicates were inoculated at an OD of ≈0.01.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant of the Deutsche Forschungsgemeinschaft to D.J. and by the GRK1708 (University of Tübingen).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Dobro MJ, Oikonomou CM, Piper A, Cohen J, Guo K, Jensen T, Tadayon J, Donermeyer J, Park Y, Solis BA, Kjær A, Jewett AI, McDowall AW, Chen S, Chang Y-W, Shi J, Subramanian P, Iancu CV, Li Z, Briegel A, Tocheva EI, Pilhofer M, Jensen GJ. 2017. Uncharacterized bacterial structures revealed by electron cryotomography. J Bacteriol 199:e00100-17. doi: 10.1128/JB.00100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant CR, Wan J, Komeili A. 2018. Organelle formation in bacteria and archaea. Annu Rev Cell Dev Biol 34:217–238. doi: 10.1146/annurev-cellbio-100616-060908. [DOI] [PubMed] [Google Scholar]
- 3.Uebe R, Schüler D. 2016. Magnetosome biogenesis in magnetotactic bacteria. Nat Rev Microbiol 14:621–637. doi: 10.1038/nrmicro.2016.99. [DOI] [PubMed] [Google Scholar]
- 4.van Teeseling MCF, Neumann S, van Niftrik L. 2013. The anammoxosome organelle is crucial for the energy metabolism of anaerobic ammonium oxidizing bacteria. J Mol Microbiol Biotechnol 23:104–117. doi: 10.1159/000346547. [DOI] [PubMed] [Google Scholar]
- 5.Seufferheld M, Vieira M, Ruiz FA, Rodrigues CO, Moreno S, Docampo R. 2003. Identification of organelles in bacteria similar to acidocalcisomes of unicellular eukaryotes. J Biol Chem 278:29971–29978. doi: 10.1074/jbc.M304548200. [DOI] [PubMed] [Google Scholar]
- 6.Seufferheld M, Lea CR, Vieira M, Oldfield E, Docampo R. 2004. The H+-pyrophosphatase of Rhodospirillum rubrum is predominantly located in polyphosphate-rich acidocalcisomes. J Biol Chem 279:51193–51202. doi: 10.1074/jbc.M406099200. [DOI] [PubMed] [Google Scholar]
- 7.Docampo R, Moreno S. 2011. Acidocalcisomes. Cell Calcium 50:113–119. doi: 10.1016/j.ceca.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Docampo R. 2016. The origin and evolution of the acidocalcisome and its interactions with other organelles. Mol Biochem Parasitol 209:3–9. doi: 10.1016/j.molbiopara.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lander N, Cordeiro C, Huang G, Docampo R. 2016. Polyphosphate and acidocalcisomes. Biochem Soc Trans 44:1–6. doi: 10.1042/BST20150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Docampo R, Huang G. 2016. Acidocalcisomes of eukaryotes. Curr Opin Cell Biol 41:66–72. doi: 10.1016/j.ceb.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen MH, Berglund L, Rasmussen JT, Petersen TE. 1997. Bovine PAS-6/7 binds alpha v beta 5 integrins and anionic phospholipids through two domains. Biochemistry 36:5441–5446. doi: 10.1021/bi963119m. [DOI] [PubMed] [Google Scholar]
- 12.Andersen MH, Graversen H, Fedosov SN, Petersen TE, Rasmussen JT. 2000. Functional analyses of two cellular binding domains of bovine lactadherin. Biochemistry 39:6200–6206. doi: 10.1021/bi992221r. [DOI] [PubMed] [Google Scholar]
- 13.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. 2008. Membrane phosphatidylserine regulates surface charge and protein localization. Science 319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 14.Del Vecchio K, Stahelin RV. 2018. Investigation of the phosphatidylserine binding properties of the lipid biosensor, lactadherin C2 (LactC2), in different membrane environments. J Bioenerg Biomembr 50:1–10. doi: 10.1007/s10863-018-9745-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bresan S, Sznajder A, Hauf W, Forchhammer K, Pfeiffer D, Jendrossek D. 2016. Polyhydroxyalkanoate (PHA) granules have no phospholipids. Sci Rep 6:26612. doi: 10.1038/srep26612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spiekermann P, Rehm BH, Kalscheuer R, Baumeister D, Steinbüchel A. 1999. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch Microbiol 171:73–80. doi: 10.1007/s002030050681. [DOI] [PubMed] [Google Scholar]
- 17.Alsford S, Kawahara T, Isamah C, Horn D. 2007. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol Microbiol 63:724–736. doi: 10.1111/j.1365-2958.2006.05553.x. [DOI] [PubMed] [Google Scholar]
- 18.Maslov I, Bogorodskiy A, Mishin A, Okhrimenko I, Gushchin I, Kalenov S, Dencher NA, Fahlke C, Büldt G, Gordeliy V, Gensch T, Borshchevskiy V. 2018. Efficient non-cytotoxic fluorescent staining of halophiles. Sci Rep 8:2549. doi: 10.1038/s41598-018-20839-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zupan JR, Cameron TA, Anderson-Furgeson J, Zambryski PC. 2013. Dynamic FtsA and FtsZ localization and outer membrane alterations during polar growth and cell division in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A 110:9060–9065. doi: 10.1073/pnas.1307241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz FA, Marchesini N, Seufferheld M, Govindjee, Docampo R. 2001. The polyphosphate bodies of Chlamydomonas reinhardtii possess a proton-pumping pyrophosphatase and are similar to acidocalcisomes. J Biol Chem 276:46196–46203. doi: 10.1074/jbc.M105268200. [DOI] [PubMed] [Google Scholar]
- 21.Seufferheld MJ, Kim KM, Whitfield J, Valerio A, Caetano-Anollés G. 2011. Evolution of vacuolar proton pyrophosphatase domains and volutin granules: clues into the early evolutionary origin of the acidocalcisome. Biol Direct 6:50. doi: 10.1186/1745-6150-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang G, Docampo R. 2015. Proteomic analysis of acidocalcisomes of Trypanosoma brucei uncovers their role in phosphate metabolism, cation homeostasis, and calcium signaling. Commun Integr Biol 8:e1017174. doi: 10.1080/19420889.2015.1017174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao NN, Gómez-García MR, Kornberg A. 2009. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem 78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y, Huang W, Lee SSK, Xu W. 2005. Crystal structure of a polyphosphate kinase and its implications for polyphosphate synthesis. EMBO Rep 6:681–687. doi: 10.1038/sj.embor.7400448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer LM, Aravind L. 2002. The catalytic domains of thiamine triphosphatase and CyaB-like adenylyl cyclase define a novel superfamily of domains that bind organic phosphates. BMC Genomics 3:33. doi: 10.1186/1471-2164-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tumlirsch T, Jendrossek D. 2017. Proteins with CHADs (conserved histidine α-helical domains) are attached to polyphosphate granules in vivo and constitute a novel family of polyphosphate-associated proteins (phosins). Appl Environ Microbiol 83:e03399-16. doi: 10.1128/AEM.03399-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werten S, Rustmeier NH, Gemmer M, Virolle M-J, Hinrichs W. 2019. Structural and biochemical analysis of a phosin from Streptomyces chartreusis reveals a combined polyphosphate- and metal-binding fold. FEBS Lett 593:2019–3468. doi: 10.1002/1873-3468.13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenzo-Orts L, Hohmann U, Zhu J, Hothorn M. 2019. Molecular characterization of CHAD domains as inorganic polyphosphate-binding modules. Life Sci Alliance 2:e201900385. doi: 10.26508/lsa.201900385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry JT, Crosson S. 2013. Chromosome replication and segregation govern the biogenesis and inheritance of inorganic polyphosphate granules. Mol Biol Cell 24:3177–3186. doi: 10.1091/mbc.E13-04-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raschdorf O, Plitzko JM, Schüler D, Müller FD. 2014. A tailored galK counterselection system for efficient markerless gene deletion and chromosomal tagging in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 80:4323–4330. doi: 10.1128/AEM.00588-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tumlirsch T, Sznajder A, Jendrossek D. 2015. Formation of polyphosphate by polyphosphate kinases and its relationship to poly(3-hydroxybutyrate) accumulation in Ralstonia eutropha strain H16. Appl Environ Microbiol 81:8277–8293. doi: 10.1128/AEM.02279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Racki LR, Tocheva EI, Dieterle MG, Sullivan MC, Jensen GJ, Newman DK. 2017. Polyphosphate granule biogenesis is temporally and functionally tied to cell cycle exit during starvation in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 114:E2440–E2449. doi: 10.1073/pnas.1615575114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tempé J, Petit A, Holsters M, Montagu M, Schell J. 1977. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc Natl Acad Sci U S A 74:2848–2849. doi: 10.1073/pnas.74.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeiffer D, Wahl A, Jendrossek D. 2011. Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol Microbiol 82:936–951. doi: 10.1111/j.1365-2958.2011.07869.x. [DOI] [PubMed] [Google Scholar]
- 35.Segami S, Makino S, Miyake A, Asaoka M, Maeshima M. 2014. Dynamics of vacuoles and H+-pyrophosphatase visualized by monomeric green fluorescent protein in Arabidopsis: artifactual bulbs and native intravacuolar spherical structures. Plant Cell 26:3416–3434. doi: 10.1105/tpc.114.127571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bru S, Martínez-Laínez JM, Hernández-Ortega S, Quandt E, Torres-Torronteras J, Martí R, Canadell D, Ariño J, Sharma S, Jiménez J, Clotet J. 2016. Polyphosphate is involved in cell cycle progression and genomic stability in Saccharomyces cerevisiae. Mol Microbiol 101:367–380. doi: 10.1111/mmi.13396. [DOI] [PubMed] [Google Scholar]
- 37.Juengert J, Bresan S, Jendrossek D. 2018. Determination of polyhydroxybutyrate (PHB) content in Ralstonia eutropha using gas chromatography and Nile red staining. Bio Protoc 8:e2748. doi: 10.21769/BioProtoc.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.