Many natural and synthetic microbial communities are composed of not only species whose biological properties are consistent with their corresponding communities but also ones whose chemophysical characteristics do not directly contribute to the performance of their communities. Even though the latter species are often essential to the microbial communities, their roles are unclear. Here, by investigation of an artificial two-member microbial consortium in n-alkane biodegradation, we showed that the microbial member without the n-alkane-degrading capability had a cross-feeding interaction with and metabolic regulation to the leading member for the synergistic n-alkane biodegradation. Our study improves the current understanding of microbial interactions. Because “assistant” microbes showed importance in communities in addition to the functional microbes, our findings also suggest a useful “assistant-microbe” principle in the design of microbial communities for either bioremediation or chemical production.
KEYWORDS: flux balance analysis, interspecies metabolite transfer, metabolomics, proteomics, synergistic biodegradation of n-alkanes
ABSTRACT
Biodegradation of alkanes by microbial communities is ubiquitous in nature. Interestingly, the microbial communities with high hydrocarbon-degrading performances are sometimes composed of not only hydrocarbon degraders but also nonconsumers, but the synergistic mechanisms remain unknown. Here, we found that two bacterial strains isolated from Chinese oil fields, Dietzia sp. strain DQ12-45-1b and Pseudomonas stutzeri SLG510A3-8, had a synergistic effect on hexadecane (C16 compound) biodegradation, even though P. stutzeri could not utilize C16 individually. To gain a better understanding of the roles of the alkane nonconsumer P. stutzeri in the C16-degrading consortium, we reconstructed a two-species stoichiometric metabolic model, iBH1908, and integrated in silico prediction with the following in vitro validation, a comparative proteomics analysis, and extracellular metabolomic detection. Metabolic interactions between P. stutzeri and Dietzia sp. were successfully revealed to have importance in efficient C16 degradation. In the process, P. stutzeri survived on C16 metabolic intermediates from Dietzia sp., including hexadecanoate, 3-hydroxybutanoate, and α-ketoglutarate. In return, P. stutzeri reorganized its metabolic flux distribution to fed back acetate and glutamate to Dietzia sp. to enhance its C16 degradation efficiency by improving Dietzia cell accumulation and by regulating the expression of Dietzia succinate dehydrogenase. By using the synergistic microbial consortium of Dietzia sp. and P. stutzeri with the addition of the in silico-predicted key exchanged metabolites, diesel oil was effectively disposed of in 15 days with a removal fraction of 85.54% ± 6.42%, leaving small amounts of C15 to C20 isomers. Our finding provides a novel microbial assembling mode for efficient bioremediation or chemical production in the future.
IMPORTANCE Many natural and synthetic microbial communities are composed of not only species whose biological properties are consistent with their corresponding communities but also ones whose chemophysical characteristics do not directly contribute to the performance of their communities. Even though the latter species are often essential to the microbial communities, their roles are unclear. Here, by investigation of an artificial two-member microbial consortium in n-alkane biodegradation, we showed that the microbial member without the n-alkane-degrading capability had a cross-feeding interaction with and metabolic regulation to the leading member for the synergistic n-alkane biodegradation. Our study improves the current understanding of microbial interactions. Because “assistant” microbes showed importance in communities in addition to the functional microbes, our findings also suggest a useful “assistant-microbe” principle in the design of microbial communities for either bioremediation or chemical production.
INTRODUCTION
Bioremediation of petroleum-contaminated environments and microbially enhanced oil recovery are two environment-friendly, cost-effective techniques to solve serious oil-related problems in current society (1, 2). Clearly, these two biotechniques have opposite aims: one for oil removal and the other for oil recovery. However, both of them preferably induce microbes or microbial consortia with the function of n-alkane degradation (3–5). Microbial communities are usually more advantageous for hydrocarbon treatment than the individual species by providing a wider substrate spectrum, higher robustness, and better adaptation to the complicated environments (6, 7). Interestingly, it has been observed that outstanding performance of petroleum alkane biodegradation could be achieved by some microbial communities composed of not only alkane consumers but also nonconsumers (8–11). What are the roles of the microbial members who do not appear to contribute to the main function of their communities? First, they might be dormant cells in the microbial communities under current conditions, acting like seeds waiting for suitable environmental conditions (12). Second, the alkane nonconsumers might have some close affiliations (13) or contact-independent interactions (14) with the alkane-degrading members. If the microbial members without the objective function of the microbial communities are active instead of dormant, metabolic exchange is considered to be a key reason for their survival in these habitats, and their activities should have neutral or positive effects on their functional partners possibly through division of labor (15). However, in some cases, either microbial dormancy or labor division patterns are not sufficient to explain some phenomena. For example, Ghazali et al. (16) screened out six bacterial isolates from oil-contaminated soil to construct bacterial consortium 1 (including three isolates) and consortium 2 (including all six isolates). They found that consortium 2 was more effective than consortium 1 for the removal of medium- and long-chain alkanes, especially n-tetradecane, in the diesel-contaminated soil, but unexpectedly the three isolates unique to consortium 2 were unable to utilize n-tetradecane or its downstream intermediates individually. Zanaroli et al. (17) isolated two microbial consortia from cow manure with remarkable diesel biodegradation activities, especially for the degradation of C10 to C24 n-alkanes. However, except for the fungus Trametes gibbosa, none of the isolated cultivable bacteria grew well on n-alkanes or degraded n-alkanes individually. Hence, several questions about those microbial communities remain to be answered, such as how the alkane degraders and nonconsumers cooperate with each other to achieve synergistic alkane biodegradation and what the roles of these alkane nonconsumers within the communities are. The newly developed multiple “omics” technologies (18) and mathematical modeling approaches, such as the genome-scale metabolic model (GEM) reconstruction (19), provide promising ways to better understand the microbial communities in nature and bioprocesses.
In this study, we constructed a simple microbial consortium consisting of an alkane-degrading bacterial strain, Dietzia sp. strain DQ12-45-1b, and a potential non-alkane-degrading bacterial strain, Pseudomonas stutzeri SLG510A3-8, both of which were isolated from Chinese oil fields, to investigate the communication mechanism in the synthetic microbial consortium via GEM and omics analyses. The genera Dietzia and Pseudomonas were widely detected in oil fields (20–22). Dietzia species are Gram-positive bacteria that are widely distributed in diverse environments, and some Dietzia members have been shown to have the potential for hydrocarbon biodegradation (23). Among them, Dietzia sp. DQ12-45-1b was one of the few strains that were reportedly able to utilize a broad spectrum of n-alkanes (C6 to C40), and this property was attributed to the collaboration of a CYP153 alkane hydroxylase and an integral membrane alkane monooxygenase-rubredoxin fusion protein, AlkW1, in the strain (24–26). P. stutzeri, a Gram-negative bacterium, was seldom reported to have the capability to degrade hydrocarbons. For instance, P. stutzeri SLG510A3-8 lacked essential alkane degradation genes such as alkane hydroxylase genes (27), indicating its incompetence for growth on n-alkanes. However, in line with the fact that P. stutzeri is one of the most ubiquitous and abundant species in hydrocarbon-related environments (28–30), its strains are considered to interact with the hydrocarbon degraders when they are exposed to hydrocarbons and to play important roles in their microbial communities. Here, to investigate the microbial interaction between the hydrocarbon-degrading microbes and nonconsumers when they are exposed to hydrocarbons as the sole carbon source, we reconstructed a two-species GEM of Dietzia sp. DQ12-45-1b and P. stutzeri SLG510A3-8 and analyzed the metabolic flux distribution between the two “compartments” in the stoichiometric model using the flux balance analysis (FBA) algorithm. Afterward, we did in vitro cultivation, comparative proteomics analysis, and extracellular metabolomics analysis based on the in silico prediction of the metabolic exchange between the two strains on hexadecane (C16) to understand the mechanism of communication between the two strains. Our finding on the mutualistic interaction between the leading and supporting members reveal a novel assemblage strategy to construct synthetic microbial consortia with high-performing functions.
RESULTS
C16 degradation by the synthetic microbial consortium.
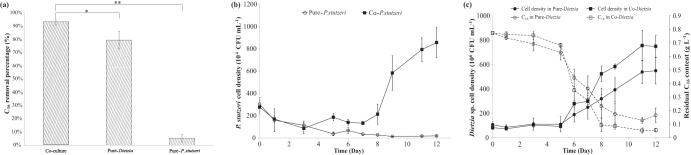
Bacterial strains Dietzia sp. strain DQ12-45-1b and P. stutzeri SLG510A3-8 were aerobically cocultivated on C16. When P. stutzeri was exposed to C16 individually, the percentage of C16 removed by P. stutzeri was low in a 12-day culture period (5.21% ± 2.88%) (Fig. 1a), and the P. stutzeri cell density decreased to a very low level (Fig. 1b). This indicated that even if P. stutzeri could adhere to a small amount of C16, it was unable to utilize C16, which was consistent with the fact that no alkane-oxidizing gene was identified in its genome. For Dietzia sp., C16 was easily available, since Dietzia sp. cells grew well on C16 (Fig. 1c), with 79.24% ± 6.71% of the total C16 being removed in 12 days in its monoculture (Fig. 1a). Nevertheless, the percentage of C16 removal by the microbial consortium (92.94% ± 1.67%) was significantly higher than those by each individual strain (P < 0.05). On the other hand, both P. stutzeri and Dietzia sp. in the coculture system grew much better than those in monocultures, even though the cell density of P. stutzeri was an order of magnitude less than that of Dietzia sp. (Fig. 1b and c). By analyzing the time courses of Dietzia cell density and residual C16 data, it was seen that the specific C16 removal rate of Dietzia sp. at its highest specific growth rate was significantly higher in the coculture system (1.38 ± 0.10 pg day−1 CFU−1) than in its monoculture system (0.71 ± 0.16 pg day−1 CFU−1) (P < 0.05). Based on the efficient performance in C16 degradation by the coculture of P. stutzeri and Dietzia sp., a synergistic effect on C16 biodegradation through metabolic communication was proposed for the coculture system, in which P. stutzeri might survive on the by-products of Dietzia sp. under the C16 exposure condition and, in return, secretory metabolites of P. stutzeri are able to induce a higher C16 recovery efficiency of the Dietzia strain.
FIG 1.
Growth of Dietzia sp. DQ12-45-1b and P. stutzeri SLG510A3-8 on hexadecane (C16) individually (pure-Dietzia sp. and pure-P. stutzeri) or together (co-Dietzia sp. and co-P. stutzeri). (a) C16 degradation efficiencies, a 12-day growth period, of the microbial community (coculture), and Dietzia sp. DQ12-45-1b (pure-Dietzia sp.), and P. stutzeri SLG510A3-8 (pure-P. stutzeri). (b) Time courses of P. stutzeri SLG510A3-8 cell densities (CFU ml−1) in its monoculture (pure-P. stutzeri) or the coculture system with Dietzia sp. DQ12-45-1b (co-P. stutzeri). (c) Time courses of Dietzia sp. DQ12-45-1b cell density (CFU ml−1) and the residual C16 content (g liter−1) in its monoculture (pure-Dietzia) or the coculture system (co-Dietzia). *, P < 0.05; **, P < 0.01.
Reconstruction and characterization of the two-species model.
To elucidate the interspecific metabolic communication between P. stutzeri and Dietzia sp., two-species metabolic network reconstruction and consortium-level flux balance analysis (FBA) were carried out. First, we reconstructed the single-species GEM of Dietzia sp. DQ12-45-1b, namely, iBH925 (see File S2 in the supplemental material), and the GEM of P. stutzeri SLG510A3-8, namely, iBH983 (see File S3 in the supplemental material). The accuracy of these two models was validated by the consistency between in vitro experimental data and in silico prediction of growth rates and available carbon source spectra (see File S1 in the supplemental material). Afterward, iBH925 and iBH983 were reconciled to unify the metabolites and reaction formats, followed by network integration. The resulted model for the microbial consortium, namely, iBH1908, contained 1,908 genes, 1,273 metabolites, and 2,670 reactions (see File S4 in the supplemental material). In iBH1908, the metabolic networks of Dietzia sp. and P. stutzeri were connected to each other through 68 shared metabolites (see Table S1 in File S1). Details of the modeling and analyzing processes of iBH925, iBH983, and iBH1908 are described in File S1.
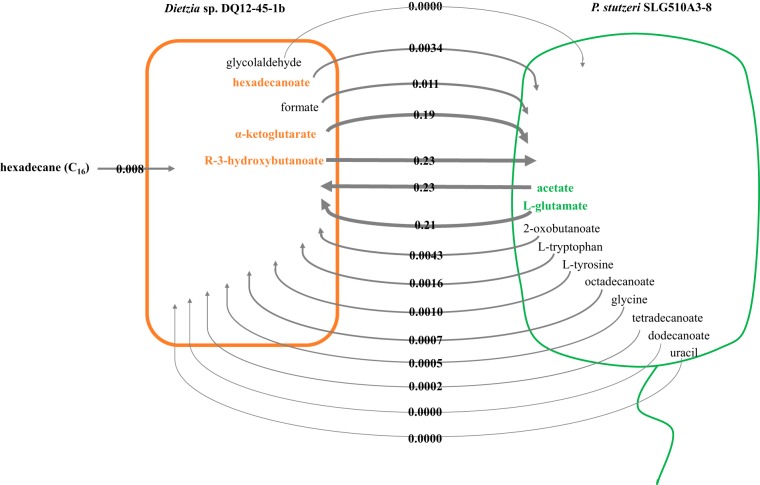
By using FBA, the flux distribution in Dietzia sp. and P. stutzeri was simulated for individual growth or coculture in C16-exposed minimal medium. A maximum growth rate μ0.0718 = 2.38 × 10−2 h−1 (see Table S2 in File S1) for Dietzia sp. in monoculture was predicted by FBA on Dietzia sp. GEM iBH925 at the C16 uptake rate of 7.18 × 10−2 mmol g−1 h−1, which was determined by batch culture (see Fig. S1 in File S1). Similarly, with the same nutrient constraints, a maximum growth rate of 0 h−1 for P. stutzeri was predicted by FBA on P. stutzeri GEM iBH983. In contrast, FBA on the two-species GEM iBH1908, in which the Dietzia sp. growth rate was constrained to be 2.38 × 10−2 h−1 while the C16 uptake rate was fixed at 8.00 × 10−2 mmol g−1 h−1 based on our previous measurement (File S1), predicted a maximum growth rate of 5.60 × 10−3 h−1 for P. stutzeri. Taken together, these predictions were in agreement with our previous findings that no P. stutzeri cell grew on C16 individually but its cell density gradually increased in the presence of Dietzia sp. (Fig. 1b). The global-level flux distribution in iBH1908 reflected that Dietzia sp. provided five metabolites to P. stutzeri, while P. stutzeri secreted another 10 compounds for Dietzia sp. utilization (Fig. 2). P. stutzeri GEM iBH983 and Dietzia sp. GEM iBH925 were utilized to predict the importance of each individual interspecific metabolite. We simulated the growth rates of P. stutzeri on iBH983 via FBA when the absorption rates of the five compounds transferred from Dietzia sp. to P. stutzeri, i.e., R-3-hydroxybutanoate, α-ketoglutarate, formate, hexadecanoate, and glycolaldehyde, were set at the responding values in Fig. 2. Under these conditions, the maximum predicted growth rate of P. stutzeri increased from 0 to 3.00 × 10−2 h−1, confirming that the metabolites clearly enhanced P. stutzeri growth. Afterward, the five compounds were taken out one by one in turn, and the growth rates of P. stutzeri were recalculated. The maximum predicted P. stutzeri growth rate (2.99 × 10−2 h−1) was almost unaffected when formate and glycolaldehyde uptake rates were both constrained to 0 mmol g−1 h−1. However, the drops were significant when each of the other three compounds was constrained, suggesting that R-3-hydroxybutanoate, α-ketoglutarate, and hexadecanoate were key compounds secreted by Dietzia sp. to support P. stutzeri growth in the C16-exposed minimal medium. Similarly, the 10 P. stutzeri secreted metabolites in Fig. 2 were tested in iBH925. Addition of the 10 compounds induced a cell growth rate rise from 2.38 × 10−2 to 5.53 × 10−2 h−1. Acetate and l-glutamate were found to be the key metabolites responsible for the significant rise, as the maximum predicted growth rate of Dietzia sp. decreased to 2.56 × 10−2 h−1 if the two metabolites were excluded.
FIG 2.
In silico-predicted interspecific metabolites that induced the growth of P. stutzeri SLG510A3-8 and Dietzia sp. DQ12-45-1b on C16 using iBH1908.
In vitro validation of in silico-predicted key exchanged compounds.
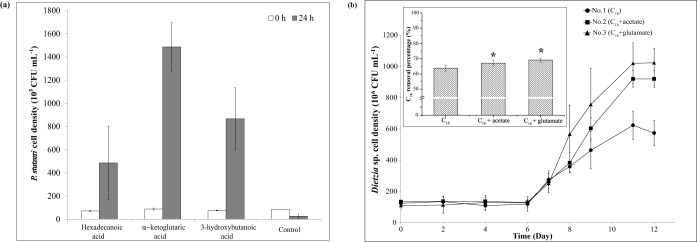
Cells of P. stutzeri SLG510A3-8 were inoculated in the minimal medium supplemented with R-3-hydroxybutanoic acid, α-ketoglutaric acid, or hexadecanoic acid. As shown in Fig. 3a, P. stutzeri biomass accumulated on each of the three compounds after 24 h of incubation, of which α-ketoglutaric acid was preferred by P. stutzeri more than the other two compounds. The in vitro result confirmed that in silico-predicted key exchange compounds were able to be utilized by P. stutzeri and suggested that the three compounds might be the reason why P. stutzeri could survive in the coculture system when C16 was the sole exogenous carbon source.
FIG 3.
In vitro cultivation of P. stutzeri SLG510A3-8 and Dietzia sp. DQ12-45-1b on in silico-predicted key exchanged compounds. (a) Cell densities of P. stutzeri grown on hexadecanoic acid, α-ketoglutaric acid, R-3-hydroxybutyric acid, or no carbon source (control). (b) Time courses of Dietzia sp. DQ12-45-1b cell density (graph) and C16 removal rates (histogram) in treatments 1, 2, and 3, in which the cultures were supplemented with C16 alone, a mixture of C16 and acetate, and a mixture of C16 and glutamate, respectively. *, P < 0.05.
In order to test if acetate and glutamate promoted C16 biodegradation by Dietzia sp. strain DQ12-45-1b, Dietzia sp. cells were cultivated in the C16-exposed minimal medium with the addition of low concentrations of sodium acetate or sodium glutamate, respectively. Higher Dietzia sp. biomass and lower residual C16 contents in the cultures supplemented with acetate and glutamate (Fig. 3b) indicated the importance of these two compounds in supporting Dietzia sp. growth and C16 biodegradation. Furthermore, the specific C16 removal rate of Dietzia sp. at its highest growth rate in the treatment containing either acetate or glutamate was significantly higher than that in the control (P < 0.05) (Table 1), which was in agreement with above in vitro batch culture result that the specific C16 removal rate of Dietzia sp. in the coculture system was significantly higher than that in the Dietzia sp. monoculture system. At the same time, we tested the effects of the other three compounds, which were in silico predicted to be very actively flowing at the interspecific region in iBH1908 (R-3-hydroxybutanoic acid, α-ketoglutaric acid, and hexadecanoic acid), on Dietzia sp. cell growth and C16 biodegradation. The specific C16 removal rates in Dietzia sp. monoculture systems with the addition of these three compounds were not significantly different from those in the monoculture system without any additives (Table 1), which was inconsistent with our in vitro data mentioned above. This result indicated that C16 biodegradation by Dietzia sp. could be enhanced through the in silico-predicted P. stutzeri secretions but not by some other compounds.
TABLE 1.
C16 biodegradation kinetics of Dietzia sp. DQ12-45-1b in monocultures with C16 as the sole carbon source or with the addition of sodium acetate, sodium glutamate, R-3-hydroxybutanoic acid, α-ketoglutaric acid, or hexadecanoic acid
| Treatment | Carbon source | Highest specific growth rate (day−1)c | P valuea | Responding time (days)c | Responding specific C16 removal rate (pg CFU−1 day−1)c | P valueb |
|---|---|---|---|---|---|---|
| 1 | C16 | 0.47 ± 0.02 | 6.99 ± 0.19 | 0.34 ± 0.04 | ||
| 2 | C16 + sodium acetate | 0.55 ± 0.03 | 8.50 × 10−2 | 7.60 ± 0.25 | 0.54 ± 0.06 | 1.11 × 10−2 |
| 3 | C16 + sodium glutamate | 0.76 ± 0.26 | 0.17 | 7.19 ± 0.28 | 0.46 ± 0.05 | 1.27 × 10−2 |
| 4 | C16 + R-3-hydroxybutanoic acid | 0.47 ± 0.02 | 0.80 | 7.35 ± 0.28 | 0.37 ± 0.03 | 0.28 |
| 5 | C16 + α-ketoglutaric acid | 0.49 ± 0.06 | 0.52 | 7.43 ± 0.25 | 0.35 ± 0.04 | 9.46 × 10−2 |
| 6 | C16 + hexadecanoic acid | 0.82 ± 0.12 | 4.93 × 10−2 | 7.56 ± 0.19 | 0.37 ± 0.10 | 0.55 |
P value between the highest specific growth rates of Dietzia sp. DQ12-45-1b in treatment 1 and the responding treatment.
P value between the specific C16 removal rates at the times when Dietzia sp. DQ12-45-1b had the highest specific growth rates in treatment 1 and the responding treatment.
Values are ± standard deviation.
Label-free proteomics and extracellular metabolomics analyses.
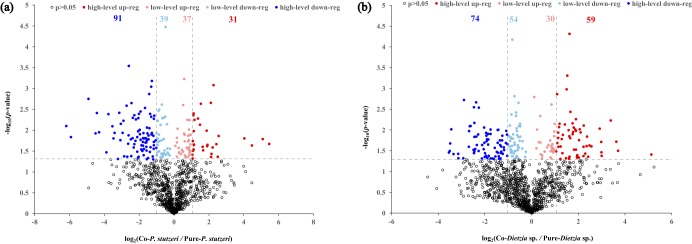
To learn about the metabolic adjustment details of Dietzia sp. and P. stutzeri from their monoculture to the coculture states, Dietzia sp. and P. stutzeri cells were separately inoculated in each side of a two-chamber bioreactor (see Fig. S2 in File S1), cultivated aerobically on C16-exposed minimal medium, and harvested at the mid-exponential phase for proteomics analysis, with the monocultures of each strain as the controls. The results showed that a total of 1,541 Dietzia sp. proteins and 1,292 P. stutzeri proteins were detected from trypsin-digested protein mixtures of the coculture and monoculture samples, with a false-discovery rate (FDR) of <0.05. As shown in Fig. 4a, in the case of cocultivation, a total of 68 P. stutzeri proteins were significantly upregulated compared with those in P. stutzeri monoculture system (P < 0.05), among which 31 proteins showed at least a 2-fold increase in expression values and were termed high-level upregulated proteins, while the remaining 37 proteins were designated low-level upregulated proteins. On the other hand, a total of 91 and 39 P. stutzeri proteins were significantly high-level and low-level downregulated, respectively, in the coculture system (P < 0.05). Similarly, we found that 59 and 30 Dietzia proteins were high- and low-level upregulated in the coculture system, respectively, while 74 and 54 Dietzia proteins were high- and low-level downregulated, respectively (Fig. 4b). The functions of the differently expressed proteins for the two strains were identified in NCBI and are listed in Tables S3 and S4 in File S1.
FIG 4.
Volcano plot analyses of 1,292 P. stutzeri proteins (a) and 1,541 Dietzia sp. proteins (b) identified in coculture triplicates and pure culture triplicates of the two strains by label-free proteomics analysis. In each diagram, the hollowed diamonds below the horizontal dashed line indicate undifferentially expressed proteins in the two treatments (P > 0.05), and the solid diamonds above the dashed line represent differentially expressed ones (P < 0.05), of which erythrine represents high-level upregulated proteins (fold change > 2), pale red represents low-level upregulated proteins (1 < fold change < 2), pale blue represents low-level downregulated proteins (0.5 < fold change < 1), and royal blue represents high-level downregulated proteins (fold change < 0.5).
Among the differentially expressed proteins, we focused on proteins relating to the metabolism of C16 and the five important interspecific metabolites mentioned above (hexadecanoic acid, α-ketoglutaric acid, R-3-hydroxybutanoic acid, acetate, and l-glutamate). Among the differentially expressed proteins, nine proteins in Dietzia sp. and 13 proteins in P. stutzeri were involved in the metabolism of the six compounds; the proteins and their fold changes are listed in Tables S3 and S4. As shown in Fig. 5, the up- and downexpression of the 22 enzymes in the microbial consortium could be interpreted as if the five interspecific metabolites indeed transferred between Dietzia sp. and P. stutzeri in the in silico-predicted directions. Generally, the central metabolism of P. stutzeri became relatively less active in the coculture system than in its monoculture system. In contrast, the tricarboxylic acid (TCA) cycle of Dietzia sp. turned out to be more active when the cells were incubated together with P. stutzeri. This indicated that the P. stutzeri cells grown on Dietzia sp. secretions might reorganize the flux distribution to produce more acetate and l-glutamate for their partner via stripping their intracellular active metabolism unrelated to acetate and l-glutamate biosynthesis down to essentials while expressing more enzymes relating to acetate and l-glutamate biosynthesis.
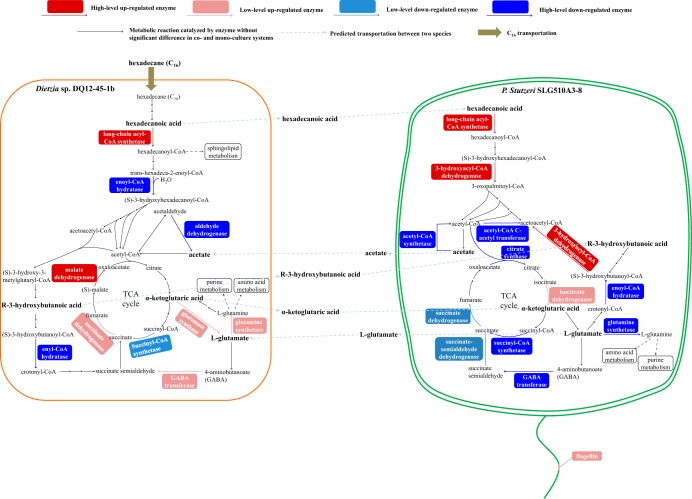
FIG 5.
Framework showing the regulation of expression of proteins relating to hexadecanoic acid, 3-hydroxybutanoic acid, α-ketoglutaric acid, acetate, and l-glutamate metabolic pathways in Dietzia sp. DQ12-45-1b and P. stutzeri SLG510A3-8 in the artificial consortium when they were exposed to C16. C16 was consumed by Dietzia sp. DQ12-45-1b and was converted to hexadecanoic acid and then to β-oxidation products. Meanwhile, two P. stutzeri enzymes involved in hexadecanoic acid β-oxidation, the long-chain acyl coenzyme A (acyl-CoA) synthetase and the 3-hydroxyacyl-CoA dehydrogenase, had high-level upregulation in the coculture system, indicating that hexadecanoic acid produced by Dietzia sp. was assimilated by P. stutzeri in the coculture system. The higher expression of P. stutzeri enzymes involved in hexadecanoic acid β-oxidation and the high-level downexpression of three P. stutzeri enzymes involved in acetate consumption, acetyl-CoA synthetase, acetyl-CoA C-acetyltransferase, and citrate synthase, would lead to the increased production of acetate. Coincidentally, the Dietzia sp. enzyme aldehyde dehydrogenase responding for acetate production was high-level downregulated when the cells coexisted with P. stutzeri. The lower expression of this protein could be interpreted as if Dietzia sp. indeed utilized extracellular acetate produced by its partner. If this was true, R-3-hydroxybutanoate, a downstream product of acetate, might accumulate in Dietzia sp. and then be secreted to the surrounding environment for P. stutzeri utilization. This inference was confirmed by the fact that an enoyl-CoA hydratase in P. stutzeri, whose function was the hydrolysis of crotonyl-CoA to the R-3-hydroxybutanoate derivative (S)-3-hydroxybutanoyl-CoA, was high-level downregulated in the coculture system, while a P. stutzeri 3-hydroxyacyl-CoA dehydrogenase, whose function was the dehydrogenation of (S)-3-hydroxybutanoyl-CoA to acetoacetyl-CoA, was high-level upregulated. In addition, if acetate was supplied by P. stutzeri to Dietzia sp., α-ketoglutaric acid, a second downstream product of acetate, might accumulate in Dietzia sp. and then be converted to l-glutamate in Dietzia sp. or be secreted to the surrounding environment for P. stutzeri utilization. This inference was confirmed by the facts that the expression of succinyl-CoA synthetase in Dietzia, which used the downstream product of α-ketoglutaric acid as the substrate, was low-level downregulated, while the expression of Dietzia glutamate synthase, which catalyzed l-glutamate production from α-ketoglutaric acid, was low-level upregulated. In P. stutzeri, the high-level downregulation of enzymes relating to l-glutamate consumption, such as glutamine synthetase and gamma-aminobutyric acid (GABA) transferase, suggested that l-glutamate might be accumulated and then be secreted to the surrounding environment for Dietzia sp. utilization. This hypothesis could explain why the glutamine synthetase and GABA transferase in Dietzia sp. were low-level upregulated in the coculture system.
In addition, culture samples taken from the two-chamber bioreactors were processed to obtain the cell-free supernatant for determination of the extracellular metabolomic composition. Overall, 43 compounds were detected in the coculture and monoculture systems (see Table S5 in File S1). Among the 43 metabolites, the in silico-predicted five key exchanged compounds were all detected in their original or derivatized forms in the supernatant from the coculture system, which was in agreement with above-described proteomic analysis (Fig. 5). Some other in silico-predicted interspecific metabolites were also identified in the metabolome profiles, such as dodecanoate, octadecanoate, and tetradecanoate, as well as their derivatized products, which was in agreement with our in silico prediction (Table 1). However, the remaining identified extracellular metabolites, such as d-lactic acid, glycolic acid, oxalate, inositol, and the derivatized product of ethanol, were out of the computational prediction. This suggested that the two-species metabolic model could be refined further for a better predictive capability.
In silico prediction of the key Dietzia enzyme responding for enhanced C16 biodegradation.
According to our proteome data, the relative amounts of the known Dietzia alkane hydroxylases AlkW1, AlkW2, and CYP153 showed no significant difference between the monoculture and coculture systems (P ≥ 0.05) (see Table S6 in File S1), indicating that the synergistic C16 biodegradation did not depend on the expression level of Dietzia alkane hydroxylases. The key metabolic pathways affecting the C16 consumption rate of Dietzia sp. in the coculture system should be among the 10 metabolic reactions which were catalyzed by the nine differentially expressed Dietzia sp. enzymes (Fig. 5). FBA on iBH925 was applied again to determine the key reactions. The model was modified at the nine metabolic reactions and was analyzed using FBA. The modification principle was as follows: first, we obtained the global-level flux distribution of iBH925 at its C16 uptake rate of 7.18 × 10−2 mmol g−1 h−1 via FBA (V0.0718), in which the biomass growth rate μ0.0718 (2.38 × 10−2 h−1) (Table S2) was present; after that, the flux rates of the 10 metabolic reactions mentioned above were constrained to 2-fold (for high-level upregulation), 1.5-fold (for low-level upregulation), 1/1.5 (for low-level downregulation), or 1/2 (for high-level downregulation) of the responding flux values in V0.0718; next, the C16 uptake rate in iBH925 was modified to be equal to that in iBH1908 (8.00 × 10−2 mmol g−1 h−1) as mentioned above; and finally, the maximum predicted biomass growth rate μ0.08 was computed via FBA again, and μ0.08 was compared with μ0.0718. If μ0.08 with some special reaction flux constraints was similar to μ0.0718, the enzymes which catalyzed the special reactions would be considered the key enzymes for the synergistic C16 biodegradation in the synthetic microbial consortium. It was found that the value of μ0.08 (2.43 × 10−2 h−1) was similar to that of μ0.0718 when the flux rate of Rxn13 catalyzed by succinate dehydrogenase was constrained, but the value was obviously different from that of μ0.0718 when the flux rate modification was operated on any one of the remaining nine metabolic reactions. This suggested that the synergistic C16 biodegradation in the synthetic microbial consortium might be due to the higher-level expression of succinate dehydrogenase induced by the substrates produced from P. stutzeri. The potential molecular mechanism under which the succinate dehydrogenase positively affects C16 biodegradation by Dietzia sp. was then examined.
Enhanced diesel biodegradation by using the synergistic microbial consortium with the addition of in silico-predicted key exchanged compounds.
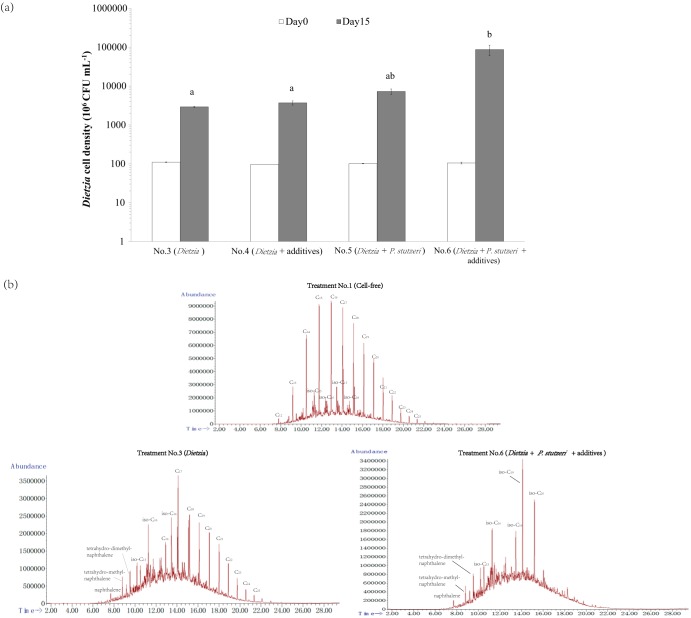
Accordingly, the synergistic biodegradation capability of the microbial consortium was not limited to C16 but also included the synthetic n-alkane mixture of C14, C16, and C28 (see Fig. S3 in File S1), and the synergistic effect of the consortium on various n-alkanes was proved in silico to be enhanced by the sustained supply of acetate and glutamate from P. stutzeri (see Table S7 in File S1). Here, we inferred that the microbial consortium of Dietzia sp. and P. stutzeri might have synergistic biodegradation capability on diesel oil, a specific fractional distillate of petroleum composed mainly of alkanes with chain lengths between C8 and C25 (31), and that acetate and glutamate might be able to improve the microbial consortium-based diesel biodegradation. In order to verify the inferences, Dietzia sp. without or with the addition of sodium acetate and sodium glutamate was aerobically cultivated on the minimal medium supplemented with 10 g liter−1 diesel oil (treatments 3 and 4, respectively), taking the Dietzia sp.-P. stutzeri coculture without or with the additives as two positive controls (treatments 5 and 6, respectively) and cell-free culture and the P. stutzeri monoculture as two negative controls (treatments 1 and 2, respectively). All the cultures were harvested on day 15 for Dietzia sp. cell density measurement and determination of the diesel removal percentage. As shown in Fig. 6a, the cell density of Dietzia sp. had a significantly higher increase in the coculture systems than in the Dietzia sp. monoculture system, especially in treatment 6. As shown in Table 2, P. stutzeri was likely incapable of diesel biodegradation, while Dietzia sp. was effective for diesel removal. Notably, the coculture system with the addition of slight amounts of acetate and glutamate had the highest removal percentage (85.54% ± 6.42%), which was over 10% higher than that of the Dietzia sp. monoculture (73.53% ± 3.27%). Afterwards, we analyzed the hydrocarbon compositions of the disposed diesel oil and found that the composition profile in the coculture system with additives was much simpler than those in the cell-free culture and Dietzia sp. monoculture (Fig. 6b), as well as those in P. stutzeri monoculture and Dietzia sp. monoculture with additives (see Fig. S4 in File S1). According to the composition profiles, n-alkanes and the isomers with chain lengths between C12 and C14 were completely removed from the diesel samples in treatments 3 and 6, suggesting a very effective removal of the medium-chain-length n-alkanes and their isomers by Dietzia sp.; for the n-alkanes with chain lengths between C15 and C25, they were partially removed from the diesel sample in treatment 3 but were completely disposed of in treatment 6, leaving small amounts of C15 to C20 isomer residues. Our results suggested that the strategy using the synergistic microbial consortium plus in silico-predicted interspecific regulators was very efficient for diesel disposal.
FIG 6.
Diesel biodegradation using Dietzia sp. DQ12-45-1b, P. stutzeri SLG510A3-8, or their consortium with or without the addition of sodium acetate and sodium glutamate. (a) Dietzia cell densities on day 0 (white bars) and day 15 (black bars) in the minimal medium supplemented with 10 g liter−1 diesel (treatment 3), with the mixture of sodium acetate and sodium glutamate (treatment 4), with P. stutzeri (treatment 5), and with both the additive mixture and P. stutzeri (treatment 6). Letters above the bars represent significant differences. (b) TIC-GC/MS chromatograms of diesel oil samples after a 15-day disposal in the cell-free culture (treatment 1), treatment 3, and treatment 6.
TABLE 2.
Diesel removal in treatments 2 to 6 after a 15-day disposal
| Treatment | Bacterium and additives | Diesel removal (%)a |
|---|---|---|
| 2 | P. stutzeri | 1.82 ± 0.09 |
| 3 | Dietzia sp. | 73.53 ± 3.27 |
| 4 | Dietzia sp. + additives | 76.37 ± 4.13 |
| 5 | Dietzia sp. + P. stutzeri | 83.56 ± 5.66 |
| 6 | Dietzia sp. + P. stutzeri + additives | 85.54 ± 6.42 |
Values are ± standard deviation.
DISCUSSION
It has been reported that the fraction of hydrocarbon-degrading microorganisms is as high as 1% to 10% of the total population in hydrocarbon-polluted environments, which was usually less than 0.1% in unpolluted environments (32, 33). However, the functions of the remaining 90% to 99% of the microbial populations in these hydrocarbon-related ecosystems were unusually ignored. To reveal a potential contribution of these remaining microbial populations, a microbial consortium consisting of an alkane degrader, Dietzia sp. DQ12-45-1b, and a potential alkane nonconsumer, P. stutzeri SLG510A3-8, was investigated. Surprisingly, the microbial consortium was found to have a greater C16 removal rate than Dietzia sp. alone. The synergistic effect was not limited only to C16 degradation but also included degradation of synthetic and natural n-alkane mixtures. This finding was in agreement with cases of bioaugmentation with fungal-bacterial consortia, in which the bacterial members acted as the pollutant degraders while the fungal members did not (34, 35). However, the synergistic interaction between our two bacterial strains should be different from that in these cases, since fungi were referred to as the physical vectors for bacterial transport in these consortia because of use of their hyphae. Accordingly, Dietzia sp. was realized to convert C16 into not only its biomass components but also some metabolic by-products available to P. stutzeri. In return, P. stutzeri fed back some other organic compounds to support the further growth of Dietzia sp. and to regulate the expression of some Dietzia enzymes, such as succinate dehydrogenase, for improving its C16 uptake.
Succinate dehydrogenase catalyzes the electron transfer from succinate to quinone as shown in the following chemical reaction: succinate + quinone → fumarate + quinol. Succinate dehydrogenase is a central metabolic enzyme coupling the bacterial growth process controlled by the TCA cycle with the energy production resulting from the aerobic respiratory chain. Mammalian cells with succinate dehydrogenase deficiency are diagnosed to have syndromic disease (36). For microbials, there are many reports showing that this enzyme is effective for microbial adaptation under different conditions. Hartman et al. reported that succinate dehydrogenase was a governor of Mycobacterium tuberculosis cellular respiration in the adaptation to low-oxygen environments (37). Kiefler et al. significantly increased the carbon conversion efficiency of the industrial obligate aerobic bacterium Gluconobacter oxydans from its cytoplasmic oxidized compounds to biomass by knocking in a heterologous gene cluster expressing succinate dehydrogenase and the responding flavinylation factor (38). Considering that petroleum hydrocarbons cause oxidative stress to bacteria during their biodegradation (5), it is inferred that the upregulation of Dietzia sp. succinate reductase induced by the P. stutzeri secretion was used for the oxidative stress adaptation of Dietzia sp. during its C16 biodegradation, and then the degradation rates in the adaptive Dietzia sp. were enhanced. Further studies using molecular techniques on Dietzia sp. DQ12-45-1b will show whether succinate dehydrogenase regulates C16 biodegradation through oxidative stress adaptation.
Deeply understanding the metabolic interactions among members in microbial consortia with special functions helps to develop efficient microbial consortia or communities in industrial applications. Until now, almost all success stories about the rational design of microbial communities for efficient microbiological processes are based on the following two strategies. The first one is the aggregation method, in which every member could accomplish the tasks of its synthetic microbial consortium but with underperforming efficiencies (39, 40). The second strategy is the “division-of-labor” approach, in which the individual constituents perform complementary roles (41–43). Our study provides a new strategy: the boosting method, in which some members without the desired biological functions promote positive performances of other members who have the desired biological functions or stepwise functions. The microbiological functions of the communities consisting of the supporting and the leading members could be even further enhanced by strengthening their interactions. Taking the consortium of Dietzia sp. and P. stutzeri as an example, although its synergistic rate of biodegradation of diesel oil was as high as 83.56% ± 5.66%, we found that the cleanup percentage could be further improved by adding slight amounts of two exogenous compounds closely related to the synergistic n-alkane biodegradation (Table 2; see Fig. S3 in File S1 in the supplemental material).
In summary, in our study Dietzia sp. DQ12-45-1b and P. stutzeri SLG510A3-8 were found to have a synergistic effect on C16 biodegradation, even though P. stutzeri could not assimilate C16. By integrating in silico prediction and in vitro validation, it was found that Dietzia sp. and P. stutzeri had a cross-feeding interaction when they were coexposed to C16, in which Dietzia sp. was the nutrient supplier of P. stutzeri while P. stutzeri was the n-alkane degradation regulator donator to Dietzia sp. Our finding of the synergy mechanism between the two species provides a novel microbial community assemblage strategy for enhancing functions of microbiological processes and helps to deeply understand microbial behaviors in natural ecosystems in the future.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Dietzia sp. strain DQ12-45-1b and Pseudomonas stutzeri SLG510A3-8 were isolated in our laboratory from oil fields in China and deposited in the China General Microbiological Culture Collection Center (CGMCC) (Beijing, China) under accession numbers 1.10709 and 1.15316, respectively. After the two strains were grown in GPY (10 g liter−1 glucose, 5 g liter−1 yeast extract, 10 g liter−1 tryptone) and LB (10 g liter−1 NaCl, 5 g liter−1 yeast extract, 10 g liter−1 tryptone) media, respectively, at 150 rpm and 30°C until the mid-exponential phase, they were centrifuged and washed with the minimal medium (44) three times to make inoculating seed suspensions. Afterward, the two clean seeds were separately inoculated into two flasks or mixed in one flask, each containing 100 ml minimal medium supplemented with 0.7734 g liter−1 C16. The initial cell density of each strain was an optical density at 600 nm (OD600) of 0.1. The culture without inoculum was taken as the control. As described in File S1 in the supplemental material, cocultivation of the two strains was also done in minimal medium supplemented with a synthetic n-alkane mixture consisting of C14, C16, and C28 or with diesel oil. Since the colonies of Dietzia sp. were orange and smooth, being significantly different from those of P. stutzeri, which were brown and wrinkled, samples were taken every 1 to 2 days for cell density counting by CFU until the early stationary phase. The collected data were processed using nonlinear curve fitting followed by differentiation in Origin8.5 (OriginLab Co., MA, USA) to obtain the time courses of specific growth rates.
Analysis of C16 removal rates.
At the end of the cultivation period, the residual C16 in each flask was extracted using 20 ml n-hexane in a separating funnel and then was analyzed using a 5975C gas chromatograph (GC) coupled with a 7890A mass spectrometer (MS) detector (Agilent Technologies, Santa Clara, CA, USA). Helium was applied as the carrier gas at a flow rate of 25 ml min−1. The oven temperature was set to 100°C for 1 min and then increased to 260°C at a rate of 20°C min−1 and held at that temperature for 5 min. The external standard method was utilized to obtain the C16 concentrations in each sample. Afterward, the residual C16 concentration data were processed in Origin8.5 to obtain the time courses of specific C16 removal rates. The C16 removal rate was calculated as the difference in the percentage of the residual C16 concentration between the experimental and the control treatments in 0.7734 g liter−1 C16.
Reconstruction and constraint-based analysis of the two-species metabolic model.
The single-species GEMs of Dietzia sp. DQ12-45-1b and P. stutzeri SLG510A3-8 were reconstructed as described in File S1 and then were modified by unifying the format of reactions and metabolites before being integrated to form the two-species model following the approach of Klitgord and Segrè (45). In the two-species model, the metabolites were localized into a total of five compartments as follows: (i) [d] = cytoplasm of Dietzia sp., (ii) [t] = extracellular space of Dietzia sp., (iii) [p] = cytoplasm of P. stutzeri, (iv) [s] = extracellular space of P. stutzeri, and (v) [e] = environment shared by the two strains. FBA was used to calculate the metabolic flux distribution in the reconstructed GEMs, in which the biomass reactions accounting for all known biomass constituents and their fractional contributions to the overall cellular biomass were used as the objective to calculate the flux distribution solution in the feasible flux space for the maximal growth rate. System constraints were defined with the upper bound for each metabolic reaction as 1,000 and the lower bounds as −1,000 and 0 for the reversible and irreversible reactions, respectively, except for the exchange reactions. The lower bounds of exchange reactions relating to oxygen and the medium compounds were set to −1,000, while the remaining ones were set to 0. All calculations were performed under the linear programming algorithm using the COBRA toolbox (46) on the MATLAB platform (The MathWorks Inc., Natick, MA).
In vitro validation of in silico-predicted key interspecific compounds.
Seeds of Dietzia sp. DQ12-45-1b were inoculated into 100 ml minimal medium supplemented with 0.7734 g liter−1 C16 alone or with the addition of 0.0446 g liter−1 sodium acetate, 0.0368 g liter−1 sodium glutamate, 0.0283 g liter−1 R-3-hydroxybutanoic acid, 0.0318 g liter−1 α-ketoglutaric acid, or 0.0174 g liter−1 hexadecanoic acid at an initial OD600 of 0.1. The carbon molar contents of the five additives in the experimental treatments were equal to 2.0% of the carbon amount in 0.7734 g liter−1 C16. The pH in each treatment was adjusted to 8.0 before medium sterilization. All the cultures were incubated at 150 rpm and 30°C. Samples were taken for CFU counting and measurement of the residual C16 amount until the early stationary phase. To obtain the time courses of the specific growth rates and the specific C16 removal rates in each treatment, the collected data were processed in Origin8.5 as described above.
Seeds of P. stutzeri SLG510A3-8 were inoculated into 100 ml minimal medium supplemented with 0.872 g liter−1 hexadecanoic acid, 1.590 g liter−1 α-ketoglutaric acid, or 1.416 g liter−1 R-3-hydroxybutanoic acid at an initial OD600 of 0.1. The carbon molar content in each of the three compounds was equal to that in 0.7734 g liter−1 C16. The culture without any carbon source was taken as the control. All the cultures were incubated at 150 rpm and 30°C. Samples were taken for CFU counting after 24 h of cultivation.
Label-free proteome identification and quantification.
Cells for proteomics analysis were cultivated in a 300-ml two-chamber bioreactor. As shown in Fig. S2 in File S1, the two chambers in the bioreactor were separated by a cellulose acetate filter (pore size = 0.2 μm; Sartorius Stedim Biotech GmbH, Göttingen, Germany). Seeds of Dietzia sp. DQ12-45-1b and P. stutzeri SLG510A3-8 were inoculated into different chambers, each of which contained 100 ml minimal medium supplemented with 0.7734 g liter−1 C16. The initial cell density for each strain was an OD600 of 0.1. Meanwhile, we developed the P. stutzeri monoculture control by inoculating P. stutzeri seeds into another bioreactor with an initial OD600 of 0.1 in each side and did the same for the Dietzia sp. monoculture control. All cultures were kept at 30°C with aeration. Bacterial cells were harvested at day 8 for proteome extraction. Details of proteomic sample preparation are described in File S1. The resulting tryptic peptide samples were loaded into an LC-LTQ/Orbitrap-MS (Thermo Fisher Scientific, San Jose, CA, USA). The mobile phase was comprised of solvent A (97% H2O, 3% acetonitrile, 1% formic acid) and solvent B (99.9% acetonitrile and 0.1% formic acid). The detection of peptide spectra was as described by Liu et al. (47). The proteome qualification was performed using the Mascot software (Matrix Science, Boston, MA, USA) against genome databases for the two strains, while the proteome quantification was based on comparison of the spectrometric spectral counts for selective proteolytic peptides (48).
Extracellular metabolomic profiling.
Cell-free cultures of Dietzia sp. DQ12-45-1b, P. stutzeri SLG510A3-8, and their mixture were obtained by centrifugation of 2-ml samples at 4°C and 10,000 rpm for 10 min to remove C16-exposed cells. For desalinization, 10 ml methanol was added to each cell-free culture, vortexed for 5 min, and kept at 4°C for 5 h. The mixtures were then centrifuged at 20,000 rpm and 4°C for 10 min, and the supernatant was lyophilized (Freeze-Dryer; Boyikang, Beijing, China) for 4 h. The lyophilized samples were derivatized as described by Mujahid et al. (49). GC-MS analysis of the derivatized samples was performed on a GCMS-QP2010 (Shimadzu, Japan). One microliter of derivatized sample was injected into an RTx-5MS column (30 m by 0.25 mm by 0.25 μm) with helium as the carrier gas at a constant flow of 1 ml min−1. The inlet temperature was set at 250°C. The oven temperature was held at 80°C for 2 min, ramped to 300°C at 20°C min−1, and then held at 300°C for 5 min. Electron impact ionization with an ionization energy of 70 eV and transfer line temperature of 220°C was used. Mass spectra were recorded at 50 to 700 m/z for 5 to 19 min. Metabolites were identified from the GC-MS data based on mass spectral comparison to the standard NIST library 2.0 (National Institute of Standards and Technology, 2008) and Wiley 9 (Wiley-VCH Verlag GmbH & Co. KgaA, Weinheim, Germany), with a match threshold of >80 (with a maximum match equal to 100). Relative metabolite abundances were calculated from peak areas (unique mass) of identified metabolites from GC-MS data and then were amended using the peak area of methyl tetradecanoate to minimize instrumental errors.
In vitro cultivation of the two-species consortium with the addition of predicted exchanged compounds for diesel removal efficiency test.
Seeds of Dietzia sp. DQ12-45-1b were inoculated into the minimal medium containing 10 g liter−1 diesel oil with or without the addition of P. stutzeri SLG510A3-8 and exogenous addition of 0.045 g liter−1 sodium acetate and 0.037 g liter−1 sodium glutamate. The culture without inoculum and the one inoculated with P. stutzeri SLG510A3-8 alone were taken as the two control treatments. Every treatment was performed in triplicates. All the cultures were incubated at 150 rpm and 30°C. Samples were taken weekly until the stationary phase or at the end of the culture period for Dietzia sp. cell density counting with CFU. At the end of the culture period, the oil removal rates were tested using the weight difference assay as described in EPA method 3500B, and diesel composition profiling was tested using GC-MS. Samples for diesel composition profiling were prepared as described previously. During the GC-MS processing, the oven temperature was set to 50°C for 1 min and then increased to 260°C at a rate of 10°C min−1 and held at that temperature for 5 min, after which it was sequentially increased to 280°C at 10°C min−1 and held at that temperature for 0.5 min.
Statistical analysis.
All in vitro experiments were performed in triplicates and subjected to statistical analysis. The standard deviations were calculated in Microsoft Excel to represent the errors for the triplicate data. The P values between two triplicates were also calculated in Microsoft Excel, and a P value of <0.05 was considered statistically significant. For the proteomics analysis, proteins with a |log2(fold change)| of >1 were defined as high-level regulated proteins, and those with a |log2(fold change)| of <1 were defined as low-level regulated proteins. The significantly changed proteins were annotated according to the KEGG database to identify metabolic pathways in each strain that were influenced by the other one.
Data availability.
The DNA sequencing data are deposited at GenBank under accession numbers CP046567.1, CP046568.1, and CP046569.1 for Dietzia sp. DQ12-45-1b and CP011854.1 for P. stutzeri SLG510A3-8. The mass spectrometry proteomics data reported in this paper have been deposited in the open-access database ProteomeXchange Consortium via the iProX partner repository (50) with the data set identifier PXD016754.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Program of China (2018YFA0902100 and 2018YFA0902103), the National Natural Science Foundation of China (grant no. 31770118, 31770120, and 31761133006), and the Beijing Institute of Technology Research Fund Program for Young Scholars.
Footnotes
Supplemental material is available online only.
[This article was published on 1 April 2020 with a standard copyright line (“© 2020 American Society for Microbiology. All Rights Reserved.”). The authors elected to pay for open access for the article after publication, necessitating replacement of the original copyright line with the one above, and this change was made on 6 April 2020.]
REFERENCES
- 1.Head IM, Gray ND. 2016. Microbial biotechnology 2020: microbiology of fossil fuel resources. Microb Biotechnol 9:626–634. doi: 10.1111/1751-7915.12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Safdari MS, Kariminia HR, Rahmati M, Fazlollahi F, Polasko A, Mahendra S, Wilding WV, Fletcher TH. 2018. Development of bioreactors for comparative study of natural attenuation, biostimulation, and bioaugmentation of petroleum-hydrocarbon contaminated soil. J Hazard Mater 342:270–278. doi: 10.1016/j.jhazmat.2017.08.044. [DOI] [PubMed] [Google Scholar]
- 3.Liu H, Xu J, Liang R, Liu J. 2014. Characterization of the medium- and long-chain n-alkanes degrading Pseudomonas aeruginosa strain SJTD-1 and its alkane hydroxylase genes. PLoS One 9:e105506. doi: 10.1371/journal.pone.0105506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dvořák P, Nikel PI, Damborský J, de Lorenzo V. 2017. Bioremediation 3.0: engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol Adv 35:845–866. doi: 10.1016/j.biotechadv.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Fuentes S, Mendez V, Aguila P, Seeger M. 2014. Bioremediation of petroleum hydrocarbons: catabolic genes, microbial communities, and applications. Appl Microbiol Biotechnol 98:4781–4794. doi: 10.1007/s00253-014-5684-9. [DOI] [PubMed] [Google Scholar]
- 6.Nyer EK, Payne F, Suthersan S. 2003. Treatment technology/environment vs. bacteria or let’s play ‘name that bacteria’. Groundwater Monit Remediat 23:36–45. doi: 10.1111/j.1745-6592.2003.tb00780.x. [DOI] [Google Scholar]
- 7.Marecik R, Chrzanowski Ł, Piotrowska-Cyplik A, Juzwa W, Biegańska-Marecik R. 2015. Rhizosphere as a tool to introduce a soil-isolated hydrocarbon-degrading bacterial consortium into a wetland environment. Int Biodeterior Biodegrad 97:135–142. doi: 10.1016/j.ibiod.2014.11.006. [DOI] [Google Scholar]
- 8.Dombrowski N, Donaho JA, Gutierrez T, Seitz KW, Teske AP, Baker BJ. 2016. Reconstructing metabolic pathways of hydrocarbon-degrading bacteria from the Deepwater Horizon oil spill. Nat Microbiol 1:16057. doi: 10.1038/nmicrobiol.2016.57. [DOI] [PubMed] [Google Scholar]
- 9.Gurav R, Lyu H, Ma J, Tang J, Liu Q, Zhang H. 2017. Degradation of n-alkanes and PAHs from the heavy crude oil using salt-tolerant bacterial consortia and analysis of their catabolic genes. Environ Sci Pollut Res Int 24:11392–11403. doi: 10.1007/s11356-017-8446-2. [DOI] [PubMed] [Google Scholar]
- 10.McGenity TJ, Folwell BD, McKew BA, Sanni GO. 2012. Marine crude-oil biodegradation: a central role for interspecies interactions. Aquat Biosyst 8:10. doi: 10.1186/2046-9063-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gieg LM, Fowler SJ, Berdugo-Clavijo C. 2014. Syntrophic biodegradation of hydrocarbon contaminants. Curr Opin Biotechnol 17:21–29. doi: 10.1016/j.copbio.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Cardoso D, Sandionigi A, Cretoiu MS, Casiraghi M, Stal L, Bolhuis H. 2017. Comparison of the active and resident community of a coastal microbial mat. Sci Rep 7:2969. doi: 10.1038/s41598-017-03095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pande S, Kost C. 2017. Bacterial unculturability and the formation of intercellular metabolic networks. Trends Microbiol 25:349–361. doi: 10.1016/j.tim.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Worm P, Koehorst JJ, Visser M, Sedano-Núñez VT, Schaap PJ, Plugge CM, Sousa DZ, Stams A. 2014. A genomic view on syntrophic versus non-syntrophic lifestyle in anaerobic fatty acid degrading communities. Biochim Biophys Acta 1837:2004–2016. doi: 10.1016/j.bbabio.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Ghazali FM, Rahman R, Salleh AB, Basri M. 2004. Biodegradation of hydrocarbons in soil by microbial consortium. Int Biodeter Biodegr 54:61–67. doi: 10.1016/j.ibiod.2004.02.002. [DOI] [Google Scholar]
- 17.Zanaroli G, Di Toro S, Todaro D, Varese GC, Bertolotto A, Fava F. 2010. Characterization of two diesel fuel degrading microbial consortia enriched from a non acclimated, complex source of microorganisms. Microb Cell Fact 9:10. doi: 10.1186/1475-2859-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, Morgan XC, Huttenhower C. 2015. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol 13:360–372. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiele I, Palsson BØ. 2010. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc 5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun W, Li J, Jiang L, Sun Z, Fu M, Peng X. 2015. Profiling microbial community structures across six large oilfields in China and the potential role of dominant microorganisms in bioremediation. Appl Microbiol Biotechnol 99:8751–8764. doi: 10.1007/s00253-015-6748-1. [DOI] [PubMed] [Google Scholar]
- 21.Song W, Wang J, Yan Y, An L, Zhang F, Wang L, Xu Y, Tian M, Nie Y, Wu X. 2018. Shifts of the indigenous microbial communities from reservoir production water in crude oil- and asphaltene-degrading microcosms. Int Biodeter Biodeg 132:18–29. doi: 10.1016/j.ibiod.2018.04.015. [DOI] [Google Scholar]
- 22.Go P, Tian H, Wang Y, Li Y, Xie J, Li Y, Xie J, Zeng B, Zhou J, Li G, Ma T. 2016. Spatial isolation and environmental factors drive distinct bacterial and archaeal communities in different types of petroleum reservoirs in China. Sci Rep 6:20174. doi: 10.1038/srep20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gharibzahedi SMT, Razavi SH, Mousavi M. 2014. Potential applications and emerging trends of species of the genus Dietzia: a review. Ann Microbiol 64:421–429. doi: 10.1007/s13213-013-0699-5. [DOI] [Google Scholar]
- 24.Wang X, Chi C, Nie Y, Tang Y, Tan Y, Wu G, Wu X. 2011. Degradation of petroleum hydrocarbons (C6-C40) and crude oil by a novel Dietzia strain. Bioresour Technol 102:7755–7761. doi: 10.1016/j.biortech.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Nie Y, Liang J, Fang H, Tang Y, Wu X. 2014. Characterization of a CYP153 alkane hydroxylase gene in a Gram-positive Dietzia sp. DQ12-45-1b and its “team role” with alkW1 in alkane degradation. Appl Microbiol Biotechnol 98:163–173. doi: 10.1007/s00253-013-4821-1. [DOI] [PubMed] [Google Scholar]
- 26.Liang J, Nie Y, Wang M, Xiong G, Wang Y, Maser E, Wu X. 2016. Regulation of alkane degradation pathway by a TetR family repressor via an autoregulation positive feedback mechanism in a Gram-positive Dietzia bacterium. Mol Microbiol 99:338–359. doi: 10.1111/mmi.13232. [DOI] [PubMed] [Google Scholar]
- 27.Hu B, Nie Y, Geng S, Wu X. 2015. Complete genome sequence of the petroleum-emulsifying bacterium Pseudomonas stutzeri SLG510A3-8. J Biotechnol 211:1–2. doi: 10.1016/j.jbiotec.2015.06.421. [DOI] [PubMed] [Google Scholar]
- 28.Mulet M, David Z, Nogales B, Bosch R, Lalucat J, García-Valdés E. 2011. Pseudomonas diversity in crude-oil-contaminated intertidal sand samples obtained after the prestige oil spill. Appl Environ Microbiol 77:1076–1085. doi: 10.1128/AEM.01741-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joshi MN, Dhebar SV, Dhebar SV, Bhargava P, Pandit A, Patel RP, Saxena A, Bagatharia SB. 2014. Metagenomics of petroleum muck: revealing microbial diversity and depicting microbial syntrophy. Arch Microbiol 196:531–544. doi: 10.1007/s00203-014-0992-0. [DOI] [PubMed] [Google Scholar]
- 30.Kaczorek E, Sałek K, Guzik U, Jesionowski T, Cybulski Z. 2013. Biodegradation of alkyl derivatives of aromatic hydrocarbons and cell surface properties of a strain of Pseudomonas stutzeri. Chemosphere 90:471–478. doi: 10.1016/j.chemosphere.2012.07.065. [DOI] [PubMed] [Google Scholar]
- 31.Pierce KM, Schale SP. 2011. Predicting percent composition of blends of biodiesel and conventional diesel using gas chromatography-mass spectrometry, comprehensive two-dimensional gas chromatography-mass spectrometry, and partial least squares analysis. Talanta 83:1254–1259. doi: 10.1016/j.talanta.2010.07.084. [DOI] [PubMed] [Google Scholar]
- 32.Aislabie J, Saul D, Foght J. 2006. Bioremediation of hydrocarbon-contaminated polar soil. Extremophiles 10:171–179. doi: 10.1007/s00792-005-0498-4. [DOI] [PubMed] [Google Scholar]
- 33.Atlas RM. 2007. Microbial hydrocarbon degradation-bioremediation of oil spills. J Chem Technol Biotechnol 52:149–156. doi: 10.1002/jctb.280520202. [DOI] [Google Scholar]
- 34.Furuno S, Päzolt K, Rabe C, Neu TR, Harms H, Wick LY. 2010. Fungal mycelia allow chemotactic dispersal of polycyclic aromatic hydrocarbon-degrading bacteria in water-unsaturated systems. Environ Microbiol 12:1391–1398. doi: 10.1111/j.1462-2920.2009.02022.x. [DOI] [PubMed] [Google Scholar]
- 35.Ellegaard-Jensen L, Knudsen BE, Johansen A, Albers CN, Aamand J, Rosendahl S. 2014. Fungal-bacterial consortia increase diuron degradation in water-unsaturated systems. Sci Total Environ 466–467:699–705. doi: 10.1016/j.scitotenv.2013.07.095. [DOI] [PubMed] [Google Scholar]
- 36.Gill AJ. 2018. Succinate dehydrogenase (SDH)-deficient neoplasia. Histopathology 72:106–116. doi: 10.1111/his.13277. [DOI] [PubMed] [Google Scholar]
- 37.Hartman T, Weinrick B, Vilchèze C, Berney M, Tufariello J, Cook GM, Jacobs WR Jr. 2014. Succinate dehydrogenase is the regulator of respiration in Mycobacterium tuberculosis. PLoS Pathog 10:e1004510. doi: 10.1371/journal.ppat.1004510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiefler I, Bringer S, Bott M. 2017. Metabolic engineering of Gluconobacter oxydans 621H for increased biomass yield. Appl Microbiol Biotechnol 101:5453–5467. doi: 10.1007/s00253-017-8308-3. [DOI] [PubMed] [Google Scholar]
- 39.Johns NI, Blazejewski T, Gomes AL, Wang H. 2016. Principles for designing synthetic microbial communities. Curr Opin Microbiol 31:146–153. doi: 10.1016/j.mib.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cortes-Tolalpa L, Salles JF, van Elsas JD. 2017. Bacterial synergism in lignocellulose biomass degradation—complementary roles of degraders as influenced by complexity of the carbon source. Front Microbiol 8:1628. doi: 10.3389/fmicb.2017.01628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones JA, Vernacchio VR, Sinkoe AL, Collins SM, Ibrahim MHA, Lachance DM, Hahn J, Koffas M. 2016. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoid. Metab Eng 35:55–63. doi: 10.1016/j.ymben.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 42.Jones JA, Vernacchio VR, Collins SM, Shirke AN, Xiu Y, Englaender JA, Cress BF, McCutcheon CC, Linhardt RJ, Gross RA, Koffas M. 2017. Complete biosynthesis of anthocyanins using E. coli polycultures. mBio 8:e00621-17. doi: 10.1128/mBio.00621-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones JA, Wang X. 2018. Use of bacterial co-cultures for the efficient production of chemicals. Curr Opin Biotechnol 53:33–38. doi: 10.1016/j.copbio.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 44.Bihari Z, Szvetnik A, Szabó Z, Blastyák A, Zombori Z, Balázs M, Kiss I. 2011. Functional analysis of long-chain n-alkane degradation by Dietzia spp. FEMS Microbiol Lett 316:100–107. doi: 10.1111/j.1574-6968.2010.02198.x. [DOI] [PubMed] [Google Scholar]
- 45.Klitgord N, Segrè D. 2010. Environments that induce synthetic microbial ecosystems. PLoS Comput Biol 6:e1001002. doi: 10.1371/journal.pcbi.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinken A, Haraldsdóttir HS, Wachowiak J, Keating SM, Vlasov V, Magnusdóttir S, Ng CY, Preciat G, Žagare A, Chan SHJ, Aurich MK, Clancy CM, Modamio J, Sauls JT, Noronha A, Bordbar A, Cousins B, Assal DCE, Valcarcel LV, Apaolaza I, Ghaderi S, Ahookhosh M, Guebila MB, Kostromins A, Sompairac N, Le HM, Ma D, Sun Y, Wang L, Yurkovich JT, Oliveira MAP, Vuong PT, Assal LPE, Kuperstein I, Zinovyev A, Hinton HS, Bryant WA, Artacho FJA, Planes FJ, Stalidzans E, Maass A, Vempala S, Hucka M, Saunders MA, Maranas CD, Lewis NE, Sauter T, Palsson BØ, Thiele I, Fleming R. 2019. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat Protoc 14:639–702. doi: 10.1038/s41596-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Zhang Q, Hu M, Yu K, Fu J, Zhou F, Liu X. 2015. Proteomic analyses of intracellular Salmonella enterica serovar Typhimurium reveal extensive bacterial adaptations to infected host epithelial cells. Infect Immun 83:2897–2906. doi: 10.1128/IAI.02882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calderón-Celis F, Encinar JR, Sanz-Medel A. 2018. Standardization apporaches in absolute quantitative proteomics with mass spectrometry. Mass Spectrom Rev 37:715–737. doi: 10.1002/mas.21542. [DOI] [PubMed] [Google Scholar]
- 49.Mujahid M, Prasuna ML, Sasikala C, Ramana CV. 2015. Integrated metabolomic and proteomic analysis reveals systemic responses of Rubrivivax benzoatilyticus JA2 to aniline stress. J Proteome Res 14:711–727. doi: 10.1021/pr500725b. [DOI] [PubMed] [Google Scholar]
- 50.Ma J, Chen T, Wu S, Yang C, Bai M, Shu K, Li K, Zhang G, Jin Z, He F, Hermjakob H, Zhu Y. 2019. iProX: an integrated proteome resource. Nucleic Acids Res 47:1211–1217. 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA sequencing data are deposited at GenBank under accession numbers CP046567.1, CP046568.1, and CP046569.1 for Dietzia sp. DQ12-45-1b and CP011854.1 for P. stutzeri SLG510A3-8. The mass spectrometry proteomics data reported in this paper have been deposited in the open-access database ProteomeXchange Consortium via the iProX partner repository (50) with the data set identifier PXD016754.