This study provides multiple insights to consider for the application of bacterial and viral indicators in sewage to surface water quality monitoring across the contiguous United States, ranging from method selection considerations to future research directions. Systematic testing of a large collection of sewage samples confirmed that crAssphage genetic markers occur at a higher average concentration than key human-associated Bacteroides spp. on a national scale. Geospatial testing also suggested that some methods may be more suitable than others for widespread implementation. Nationwide characterization of indicator geospatial trends in untreated sewage represents an important step toward the validation of these newer methods for future water quality monitoring applications. In addition, the large paired-measurement data set reported here affords the opportunity to conduct a range of secondary analyses, such as the generation of new or updated quantitative microbial risk assessment models used to estimate public health risk.
KEYWORDS: bacteriophage, microbial source tracking, wastewater, geospatial, general fecal indicators, sewage
ABSTRACT
Cultivated fecal indicator bacteria such as Escherichia coli and enterococci are typically used to assess the sanitary quality of recreational waters. However, these indicators suffer from several limitations, such as the length of time needed to obtain results and the fact that they are commensal inhabitants of the gastrointestinal tract of many animals and have fate and transport characteristics dissimilar to pathogenic viruses. Numerous emerging technologies that offer same-day water quality results or pollution source information or that more closely mimic persistence patterns of disease-causing pathogens that may improve water quality management are now available, but data detailing geospatial trends in wastewater across the United States are sparse. We report geospatial trends of cultivated bacteriophage (somatic, F+, and total coliphages and GB-124 phage), as well as genetic markers targeting polyomavirus, enterococci, E. coli, Bacteroidetes, and human-associated Bacteroides spp. (HF183/BacR287 and HumM2) in 49 primary influent sewage samples collected from facilities across the contiguous United States. Samples were selected from rural and urban facilities spanning broad latitude, longitude, elevation, and air temperature gradients by using a geographic information system stratified random site selection procedure. Most indicators in sewage demonstrated a remarkable similarity in concentration regardless of location. However, some exhibited predictable shifts in concentration based on either facility elevation or local air temperature. Geospatial patterns identified in this study, or the absence of such patterns, may have several impacts on the direction of future water quality management research, as well as the selection of alternative metrics to estimate sewage pollution on a national scale.
IMPORTANCE This study provides multiple insights to consider for the application of bacterial and viral indicators in sewage to surface water quality monitoring across the contiguous United States, ranging from method selection considerations to future research directions. Systematic testing of a large collection of sewage samples confirmed that crAssphage genetic markers occur at a higher average concentration than key human-associated Bacteroides spp. on a national scale. Geospatial testing also suggested that some methods may be more suitable than others for widespread implementation. Nationwide characterization of indicator geospatial trends in untreated sewage represents an important step toward the validation of these newer methods for future water quality monitoring applications. In addition, the large paired-measurement data set reported here affords the opportunity to conduct a range of secondary analyses, such as the generation of new or updated quantitative microbial risk assessment models used to estimate public health risk.
INTRODUCTION
The presence of untreated sewage in surface waters can lead to public health, economic, and ecological impacts. Sewage wastewater is typically generated by a community of individuals and may contain a variety of pollutants, such as human pathogens (1), pharmaceuticals (2), antimicrobial-resistant bacteria (3), and toxic substances (4). It is estimated that the United States generates approximately 121 billion liters of sewage wastewater per day (4). To safely manage such large quantities of wastewater, the United States has built over 2.1 million kilometers of sewer lines transporting waste to an estimated 14,758 wastewater facilities (5). Even with this enormous sewage management infrastructure, researchers estimate that 1020 to 1023 bacteria enter U.S. water bodies on a daily basis from sewage infrastructure alone (6). Many water quality authorities rely on methods using general fecal indicator bacteria (FIB), such as culture-based Escherichia coli and enterococci, to determine if surface waters impacted by sewage are safe for swimming and other recreational activities. However, these cultivation-based procedures have several limitations, e.g., they typically require 18 h or more to yield results, making it challenging to ascertain water safety on the same day of use (7), the methods target bacteria when viral pathogens are thought to be the dominant public health risk in sewage pollution (8), and FIB are present in fecal waste across a broad range of animal groups (9), making it impossible to determine if sewage or another pollution source is the cause of water contamination.
In response to these limitations, rapid molecular biology-based technologies that can measure enterococci and E. coli in a matter of hours, offering the option for same-day water quality notification, have been developed (10–12). Other researchers are investigating the use of viral cultures for surface water quality monitoring that target somatic and F+ coliphages (13–16). Coliphage monitoring may offer a more public health-protective approach due to the increased similarities of coliphages to enteric viral pathogens in morphology, inactivation in the environment, and persistence during treatment (17–19). Human-associated methods targeting fecal bacteria (20–26) and viruses (27–30) have also been developed, allowing for the characterization of sewage pollution, even when surface waters are polluted by other animal wastes originating from agricultural, wildlife, and domestic pet fecal sources. The recent development of quantitative PCR (qPCR) methods targeting crAssphage bacteriophage (27, 30) are of particular interest, due to their extraordinarily high concentration in sewage and a strong association with human fecal waste (27, 31–36). Some of these alternative tools are now being adopted (37) or are under consideration by regulatory authorities such as the U.S. Environmental Protection Agency (U.S. EPA) for use in recreational water quality management (38). For example, the Entero1a qPCR method for rapid measurement of enterococci is formally recommended by the U.S. EPA as a tool for recreational water quality monitoring (39). In addition, the U.S. EPA just released nationally validated, standardized procedures for the characterization of human fecal waste in environmental surface waters by the use of two methods that target Bacteroides microorganisms (40, 41).
A reliable sewage fecal pollution target should be broadly distributed across U.S. populations and should occur in a predictable manner regardless of geographic location. It should also occur at a consistent and sufficiently high concentration such that once released and diluted into surface waters, it can still be routinely measured to identify sewage pollution. The contiguous United States covers over 8 × 106 km2, contains 48 states and the District of Columbia, and is home to more than 300 million individuals comprising 99.3% of the total U.S. population (42). Community populations range from less than 100 individuals to more than 8 million. U.S. wastewater treatment facilities are situated across a wide range of elevations, ranging from sea level to higher than 3,000 m. Local air temperature conditions are also diverse, presenting an enormous range that could influence the distribution of sewage fecal pollution indicators.
Numerous studies have shown that microbial communities can change in composition and function based on geospatial factors such as latitude, elevation, and air temperature gradients in natural (43–46) and built (47–49) environments. For example, significant links between Bacteroidetes microorganism population structure and elevation have been reported in multiple studies (47, 50). Many alternative sewage pollution metrics target members of this phylum, such as HF183/BacR287 (21), HumM2 (26), and GenBac3 (51), or serve as a host for bacteriophage infection, including crAssphage (52) and GB-124 (53). Little is known about the biogeography of many of these alternative bacterial and viral targets in sewage fecal pollution across the contiguous United States. This is, in part, because many studies focus on regional sewage samples collected near a respective research laboratory or use an insufficient number of samples to represent a broad geographic range. The few studies investigating sewage pollution metrics on a larger geographic scale (>10 disparate locations) do not include newer technologies such as crAssphage or HF183/BacR287 nor report paired measurements of both bacterial and viral targets (49, 54–56). Testing a larger sewage sample collection with the simultaneous measurement of multiple bacterial and viral fecal pollution indicators may help confirm the suitability of an alternative approach for widespread use across the United States and may help compare and contrast any geospatial variables potentially influencing occurrence.
The goal of this study was to characterize the concentrations of seven viral and five bacterial sewage pollution targets in wastewaters from treatment facilities situated across the contiguous United States We report geospatial trends in the concentration of cultivated bacteriophage (53, 57, 58) as well as genetic markers targeting crAssphage (27), polyomavirus (28), enterococci (11, 12), E. coli (10), Bacteroidetes (51), and human-associated Bacteroides spp. (21, 26) measured from 49 primary influent sewage samples. Sewage samples were collected from rural and urban facilities across broad latitude, longitude, elevation, and air temperature gradients by using a geographic information system (GIS) stratified random site selection procedure. Most indicator methods demonstrated a remarkable similarity in concentrations regardless of location. However, some exhibited predictable shifts in concentrations based on either facility elevation or local air temperature at the time of sampling. Geospatial patterns identified in this study may have several impacts on the direction of future water quality management research, as well as the selection of alternative sewage pollution metrics for widespread use.
RESULTS
Culture-based bacteriophage measurements.
The concentrations of select bacteriophage were determined in primary influent sewage from 49 wastewater treatment facilities (Fig. 1). A measurable level was observed for nearly all samples tested, regardless of bacteriophage type, with only one site yielding no plaques with the GB-124 method. The average log10 PFU/10 ml concentration was highest for somatic coliphage (3.61 ± 0.91), followed by F+ (3.42 ± 0.64) and CB-390 (3.38 ± 0.86) coliphages. The GB-124 Bacteroides phage yielded the lowest average concentration (1.76 ± 0.66 log10 PFU/10 ml).
FIG 1.
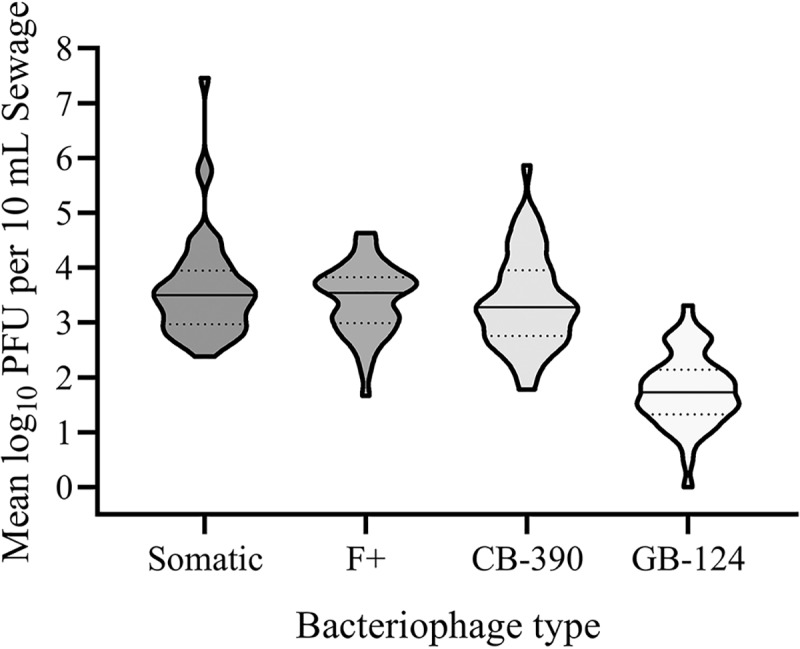
Frequency distribution plots of culturable bacteriophage in mean log10 PFU per 10 ml of sewage in primary wastewater samples. Solid lines represent median values; dotted lines represent quartiles.
Genetic measurements with qPCR.
qPCR calibration model performance metrics are summarized in Table 1. Amplification inhibition was not identified in any samples with multiplex HF183/BacR287 or HumM2 experiments (data not shown). Internal amplification control (IAC) acceptance thresholds ranged from 33.2 quantification cycle (Cq) to 34.1 Cq (HF183/BacR287) and 34.0 Cq to 35.7 Cq (HumM2). Competition thresholds ranged from 27.9 Cq to 28.0 Cq for HF183/BacR287 and 26.7 Cq to 27.1 Cq for HumM2. Extraction blank and no-template controls indicated the absence of contamination in the range of quantification for 100% of control reactions (n = 1,174). The mean log10 copies per 10 ml of primary influent sewage are depicted in Fig. 2. Genetic markers were detected at concentrations above the respective lower limit of quantification (LLOQ) threshold in all samples, except for Entero1a (n = 10 markers detected) and HPyV (n = 19 markers detected; n = 1 nondetected). Because of the high proportions of results below the LLOQ for Entero1a (20.4%) and HPyV (40.8%), these data sets were excluded from additional analyses. The average concentration in log10 copies per 10 ml of primary influent sewage was highest for GenBac3 (7.77 ± 0.43), followed by CPQ_064 (7.43 ± 0.53), CPQ_056 (7.35 ± 0.54), HF183/BacR287 (6.83 ± 0.49), EC23S857 (6.01 ± 0.40), and HumM2 (5.71 ± 0.49), Entero1a (4.93 ± 0.49; detections > LLOQ only), and HPyV (4.65 ± 0.45; detections > LLOQ only).
TABLE 1.
Summary of qPCR assay calibration model parameters
| Assay | Master slopea | Eb |
y intercepta
|
LLOQc
|
||
|---|---|---|---|---|---|---|
| Min | Max | Min | Max | |||
| CPQ_056 | −3.31 ± 0.02 | 1.00 | 40.12 ± 0.13 | 40.52 ± 0.13 | 37.06 | 37.80 |
| CPQ_064 | −3.34 ± 0.02 | 0.99 | 41.7 ± 0.16 | 42.09 ± 0.22 | 38.65 | 39.18 |
| HF183/BacR287 | −3.26 ± 0.03 | 1.03 | 37.68 ± 0.22 | 37.82 ± 0.23 | 34.72 | 35.01 |
| HPyV | −3.29 ± 0.03 | 1.02 | 36.84 ± 0.15 | 37.21 ± 0.21 | 33.76 | 34.34 |
| HumM2 | −3.24 ± 0.02 | 1.03 | 39.69 ± 0.16 | 40.11 ± 0.11 | 36.76 | 37.08 |
| Entero1a | −3.55 ± 0.04 | 0.91 | 36.87 ± 0.16 | 37.06 ± 0.14 | 33.32 | 33.48 |
| EC23S857 | −3.56 ± 0.04 | 0.91 | 37.48 ± 0.16 | 37.75 ± 0.09 | 33.82 | 34.02 |
| GenBac3 | −3.57 ± 0.03 | 0.91 | 37.73 ± 0.12 | 37.92 ± 0.14 | 34.03 | 34.32 |
Values are reported as the mean ± standard error. Min, miminum; max, maximum.
E, amplification efficiency (E = 10(−1/slope) − 1).
LLOQ, lower limit of quantification Cq value.
FIG 2.
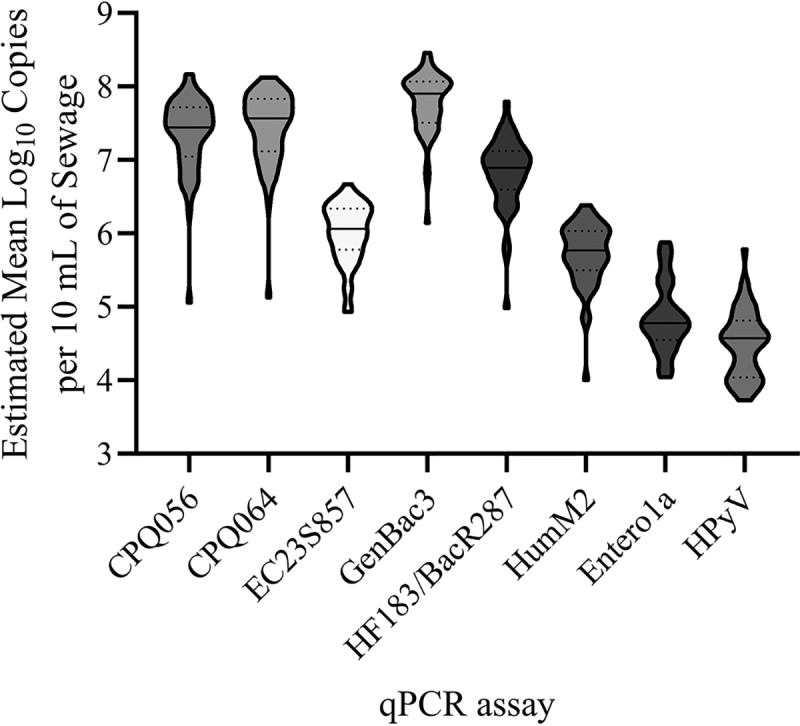
Frequency distribution plots of bacterial and viral indicators in estimated mean log10 copies per 10 ml of sewage enumerated by qPCR in primary wastewater samples. Solid lines represent median values; dotted lines represent quartiles.
Comparative analyses.
Viral and bacterial fecal pollution metric paired-measurement combinations were compared to identify potential correlations and identify any significant differences in mean concentrations. Correlation coefficients (r) for all data combinations are reported in Table 2 and ranged from 0.991 (CPQ_056 versus CPQ_064) to −0.006 (HF183/BacR287 versus somatic coliphage). One-way analysis of variance (ANOVA) was used to compare genetic marker mean concentrations (in log10 copies/10 ml of primary influent sewage) for all eligible data combinations. All genetic marker data combinations were significantly different in concentration (P ≤ 0.022), except for the combination of CPQ_056 and CPQ_064 (P = 0.940). The concentration for GenBac3 was significantly higher than those for all other genetic markers (P < 0.001), followed by crAssphage (CPQ_056 and CPQ_064). The concentrations for CrAssphage genetic markers were significantly higher than those for HF183/BacR287, HumM2, and EC23S857 (P < 0.001). The concentration for HF183/BacR287 was significantly higher than those for HumM2 and EC23S857 (P < 0.001). The concentration for EC23S857 was significantly higher than that for HumM2 (P = 0.022). The concentration for HumM2 was significantly lower than those for all other eligible (Entero1a and HPyV not considered) genetic markers (P ≤ 0.022). One-way ANOVA was also used to compare log10 PFU/100 ml cultivated bacteriophage concentrations. Results for coliphage indicators (somatic, F+, and CB-390) were not significantly different from each other (P ≥ 0.469), but they were all higher than the results for Bacteroides fragilis bacteriophage GB-124 (P < 0.001).
TABLE 2.
Pearson product momentum correlation analysis results comparing paired water quality measurements for indicatorsa
| Indicator | Somatic coliphage | F+ coliphage | CB-390 coliphage | GB-124 | CPQ_056 | CPQ_064 | EC23S857 | GenBac3 | HF183 | HumM2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Somatic coliphage (n = 49) | 0.455 | 0.690 | 0.239 | 0.012 | 0.040 | 0.208 | 0.127 | −0.006 | 0.008 | |
| F+ coliphage (n = 49) | 0.001 | 0.484 | −0.021 | 0.266 | 0.282 | 0.412 | 0.352 | 0.255 | 0.319 | |
| CB-390 coliphage (n = 49) | <0.001 | <0.001 | 0.372 | 0.077 | 0.113 | 0.219 | 0.205 | −0.019 | 0.044 | |
| GB-124 (n = 49) | 0.098 | 0.883 | 0.008 | 0.097 | 0.075 | 0.0001 | 0.072 | −0.086 | 0.022 | |
| CPQ_056 (n = 49) | 0.932 | 0.065 | 0.599 | 0.508 | 0.991 | 0.796 | 0.920 | 0.855 | 0.882 | |
| CPQ_064 (n = 49) | 0.786 | 0.049 | 0.441 | 0.609 | <0.001 | 0.801 | 0.926 | 0.855 | 0.873 | |
| EC23S857 (n = 49) | 0.151 | 0.003 | 0.131 | 0.997 | <0.001 | <0.001 | 0.870 | 0.809 | 0.799 | |
| GenBac3 (n = 49) | 0.385 | 0.013 | 0.158 | 0.624 | <0.001 | <0.001 | <0.001 | 0.917 | 0.919 | |
| HF183 (n = 49) | 0.966 | 0.077 | 0.895 | 0.556 | <0.001 | <0.001 | <0.001 | <0.001 | 0.923 | |
| HumM2 (n = 49) | 0.955 | 0.025 | 0.762 | 0.883 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
Paired water quality measurements with r values (negative values indicate a negative correlation) are shown in the upper right portion (shaded), and P values are shown in the bottom left portion (statistically significant [α = 0.05] values are in bold). HF183 = HF183/BacR287.
Geospatial analysis.
A mixed model approach was used to identify potential significant relationships between eligible fecal pollution metric data sets and average air temperature prior to sampling, facility elevation, and urban/rural designation (Table 3). The average air temperature 7 days prior to sampling across all sampling sites was 6.0 ± 6.6°C and ranged from −5.1°C (North Dakota) to 21.2°C (Florida). The average elevation was 250.5 ± 327.2 m and ranged from 1 m to 1,746 m. Air temperature was significantly correlated with latitude (r = −0.77, P < 0.001) but not elevation (r = 0.24, P = 0.10). No fecal pollution metrics exhibited a significant correlation between urban/rural wastewater treatment designation (P > 0.05). In contrast, average air temperature showed a positive correlation with somatic coliphage (P = 0.046), CB-390 total coliphage (P = 0.001), GenBac3 (P = 0.008), and EC23S857 (P = 0.023). Elevation was not significantly correlated with any fecal pollution metric (P > 0.05), except GB-124 (P = 0.016).
TABLE 3.
Mixed model geospatial analysis comparing each fecal pollution metric with average 7-day air temperature prior to sampling (°C), facility elevation (m), and urban/rural designation
| Method | Metric | Microbe type | na | Geospatial variableb |
||
|---|---|---|---|---|---|---|
| Urban/rural | Air temp | Elevation | ||||
| Culture based | Somatic coliphage | Virus | 48 | 0.051 | (+) 0.046 | 0.740 |
| F+ coliphage | Virus | 49 | 0.414 | 0.320 | 0.059 | |
| CB-390 coliphage | Virus | 49 | 0.564 | (+) 0.001 | 0.840 | |
| GB-124 | Virus | 49 | 0.549 | 0.136 | (−) 0.016 | |
| qPCR | CPQ056 | Virus | 48 | 0.084 | 0.132 | 0.374 |
| CPQ064 | Virus | 48 | 0.107 | 0.083 | 0.412 | |
| HF183/BacR287 | Bacterium | 49 | 0.110 | 0.187 | 0.748 | |
| HumM2 | Bacterium | 49 | 0.275 | 0.109 | 0.472 | |
| GenBac3 | Bacterium | 49 | 0.208 | (+) 0.008 | 0.642 | |
| EC23S857 | Bacterium | 49 | 0.683 | (+) 0.023 | 0.300 | |
n, number of samples used in the mixed model.
Symbols in parentheses denote the direction of correlation. Bold values indicate significantly different parameters (α = 0.05).
DISCUSSION
Indicator concentrations in wastewater across the contiguous United States.
This study reports the simultaneous measurement of bacterial and viral alternative sewage pollution measuring technologies from wastewater facilities selected by a GIS stratified random procedure across the contiguous United States. Observed concentrations provide novel information and confirm many trends reported by other researchers.
(i) Human-associated viruses.
The use of virus-based methods for sewage identification is an attractive approach due to the potential for a high degree of host specificity (28) and a closer similarity to disease-causing enteric pathogens (17–19). Four viral sewage pollution metrics targeting crAssphage (CPQ_056 and CPQ_064), polyomavirus (HPyV), and a B. fragilis bacteriophage (GB-124) were tested. This study confirms previous reports that crAssphage is highly abundant in untreated sewage, with estimated concentrations ranging from 5.06 to 8.17 (CPQ_056) and 5.13 to 8.13 (CPQ_064) log10 copies/10 ml. Similar crAssphage concentrations have been reported in the United Kingdom (3.3 to 7.3 log10 copies/10 ml) (35), Australia (6.91 to 7.56 log10 copies/10 ml) (33, 34), Florida (USA) (7.08 to 7.98 log10 copies/10 ml) (32), and Southeast Asia (4.28 to 6.38 log10 copies/10 ml) (36). HPyV concentrations ranged from a nondetectable level to 5.79 log10 copies/10 ml in this study and were approximately 870 times lower than the crAssphage concentration on average. These observed measurements parallel raw sewage concentrations reported in Australia (59, 60), New Zealand (61), French Polynesia (62), and Florida (USA) (28). Interestingly, HPyV concentrations reported in Argentina are markedly higher than those in other locations (5.86 to 7.41 log10 copies/10 ml) (63), suggesting the potential for geographic variability in polyomavirus occurrence in some populations. Additional research is warranted to explore potential geospatial trends in polyomavirus on an international scale. Average B. fragilis bacteriophage (GB-124) concentrations (1.76 ± 0.66 log10 PFU/10 ml) were remarkably similar to those reported in primary influent wastewater samples collected from eight treatment facilities in the United States (1.61 log10 PFU/10 ml) (55), suggesting that GB-124 bacteriophage occurrence may be uniform across the contiguous United States, albeit at a much lower concentration than coliphage.
(ii) Human-associated Bacteroides spp.
The recent public release of U.S. EPA draft methods for HF183/BacR287 and HumM2 (17–19, 40, 41) has led to an increased interest in the use of these technologies to characterize sewage pollution in environmental waters polluted by sewage. The average HumM2 concentration observed in this study (5.71 ± 0.49 log10 copies/10 ml) was similar to a previously reported value (5.98 ± 0.48 log10 copies/10 ml) from another large-scale study (54 wastewater samples) (56). The same study also reported the average concentration (6.21 ± 0.57 log10 copies/10 ml) of the HF183/BFDrev genetic marker (22), using an earlier version of the HF183 qPCR assay shown to routinely form nonspecific amplification products leading to reduced sensitivity and precision (21). The updated HF183/BacR287 average concentration observed in this study was slightly higher (6.83 ± 0.49 log10 copies/10 ml), mirroring a similar pattern reported in HF183/BFDrev and HF183/BacR287 head-to-head experiments comparing 58 raw sewage samples, where HF183/BacR287 levels were approximately 5-fold higher on average (21). Similar average concentrations of human-associated Bacteroides DNA targets suggest two important conclusions. First, the combination of the current (n = 49) and past (n = 54) studies demonstrates consistent recovery from more than 100 wastewater facility locations, making these genetics-based fecal source identification technologies perhaps the most intensively characterized methodologies for sewage pollution characterization to date in the United States. Second, samples were collected more than 10 years apart, suggesting a high level of temporal consistency in the shedding of these human-associated genetic markers by U.S. populations.
(iii) Coliphage.
Several literature reviews have examined somatic and F+ coliphage concentrations in primary effluents worldwide and reported average concentrations for somatic coliphage of 4.26 ± 0.96 (13) and 4.0 ± 1.3 (16) log10 PFU/10 ml, with moderately lower levels observed for F+ coliphage (4.24 ± 0.92 and 3.8 ± 1.0 log10 PFU/10 ml, respectively). These values are slightly higher than average somatic (3.61 ± 0.91 log10 PFU/10 ml) and F+ (3.42 ± 0.64 log10 PFU/10 ml) coliphage concentrations observed in this study but still within an overlapping range, suggesting that the two groups are uniformly distributed in untreated wastewater. The CB-390 total coliphage procedure is a recently developed dual-coliphage host assay that enumerates both somatic and F+ groups simultaneously. To date, this protocol has been extensively tested in wastewaters from Europe (64, 65) and South America (66) but has undergone limited screening in the United States (67). Reported total coliphage concentrations range from 4 to 5 log10 PFU/10 ml (65, 67). For CB-390 total coliphage concentrations in untreated sewage across the contiguous United States, we observed an average concentration of 3.38 ± 0.86 log10 PFU/10 ml, up to 41 times lower than previously reported average concentrations. There are several possible explanations for these slightly lower total coliphage levels, such as potential differences in sample collection, cultivation conditions, and/or wastewater facility geospatial factors. Additional research is needed to investigate potential elements influencing interlaboratory variability in coliphage occurrence from untreated sewage.
(iv) Rapid general FIB.
The application of qPCR-based procedures to measure concentrations of general fecal indicator bacteria (FIB) such as enterococci (Entero1a), E. coli (EC23S857), and Bacteroidetes (GenBac3) in surface waters can allow for same-day public health notification at recreational beaches. However, little is known about the distribution of these genetic targets in untreated sewage across the United States. The most comprehensive study to date measured enterococci (Entero1a; average, 5.08 ± 0.58 log10 copies/10 ml), E. coli (uidA450 [68]; average, 5.18 ± 0.31 log10 copies/10 ml), and Bacteroidetes (GenBac3; average, 7.46 ± 0.40 log10 copies/10 ml) in untreated wastewater from 54 facilities (54). Average concentrations observed in this study for enterococci (4.93 ± 0.49 log10 copies per 10 ml; detections > LLOQ only) and Bacteroidetes (7.77 ± 0.43 log10 copies per 10 ml) are similar, suggesting that these general FIB genetic markers are ubiquitous in U.S. sewage. It is important to note that the previously reported E. coli concentrations were measured with a different qPCR assay targeting a single-copy gene (uidA), resulting in a 6.8-fold-lower average concentration than the EC23S857 results from this study. The EC23S857 assay targets the 23S rRNA gene, which is reported to have an average of seven copies per genome (10), suggesting that E. coli uidA450 and EC23S857 average concentrations are almost indistinguishable across studies after accounting for differences in genomic copy number.
Comparative analysis of indicators.
Comparative analysis of indicator paired measurements reveals several interesting trends (Table 2). The highest correlation was observed between CPQ_056 and CPQ_064 crAssphage genetic markers (r = 0.991), strongly suggesting that these two assays target the same virus group. In addition, paired measurements indicate that crAssphage (CPQ_056 and CPQ_064) occurs at a significantly higher mean concentration (log10 copies/10 ml) than HF183/BacR287 (P < 0.001) in untreated sewage from across the contiguous United States. It is also interesting to note the strong correlation between crAssphage and human-associated Bacteroides genetic markers (r range, 0.854 to 0.881). These correlations may be due, in part, to the predicted bacterial host specificity of crAssphage for Bacteroides microorganisms (52). It is also possible that crAssphage can infect some of the same Bacteroides subpopulations that harbor the HF183/BacR287 and HumM2 genetic markers, resulting in a high degree of correlation. The notion that crAssphage infects Bacteroides spp. is further supported by the strong correlations between crAssphage and Bacteroidetes (GenBac3) genetic markers (r range, 0.912 to 0.926). Additional research is warranted to further investigate potential links between crAssphage and Bacteroides human-associated bacterial targets.
Although mean log10 PFU/10 ml concentrations of coliphage (somatic, F+, and total coliphage) were not significantly different from each other (P ≥ 0.469), they exhibited a markedly different pattern than other indicators. Somatic and total coliphage were not correlated with any human-associated genetic markers (P ≥ 0.441), while F+ coliphage was correlated with most human-associated and general FIB genetic markers (P ≤ 0.049), suggesting that geospatial factors may differentially influence these two groups of coliphage (somatic versus F+). In contrast, GB-124 was significantly correlated only with total coliphage (P = 0.008). This discrepancy also suggests that geospatial factors may influence GB-124 occurrence in a different manner than other indicators tested in this study.
Geospatial trends in bacterial and viral indicators.
All indicators tested in this study, regardless of whether the target was viral or bacterial, exhibited no geographic variability based on urban or rural facility designation (Table 3). This is notable given recent studies reporting that sewage microbial communities can significantly vary by city (48, 49). The stability observed in this study is ideal for any indicator under consideration for routine water quality monitoring across the contiguous United States. In addition, all crAssphage (CPQ_056 and CPQ_064), human-associated Bacteroides spp. (HF183/BacR287 and HumM2), and F+ coliphage methods yielded consistent concentrations across broad air temperature (−5.1°C to 21.2°C) and elevation (0 to 1,746 m) gradients, further supporting potential implementation on a national scale.
B. fragilis bacteriophage (GB-124) demonstrated a significant negative correlation with elevation (P = 0.016), where concentrations were higher at facilities situated at sea level and predictably decreased at higher elevations. The notion that microbial communities can shift in function (44) and composition (43, 69) due to elevation gradients is not new. A study investigating the influence of elevation on wastewater bacterial structures reported that richness and evenness significantly decreased with increased elevation (47) and was most pronounced in facilities at elevations greater than 1,200 m above sea level. Interestingly, one facility in this study is located above 1,200 m (1,756 m), and it yielded the lowest concentration of GB-124. It is possible that the host bacterium (B. fragilis) for GB-124 is not readily amenable to infection due to a decrease in abundance and/or increased stress levels resulting from a higher-altitude environment.
Finally, somatic coliphage, total coliphage (CB-390), Bacteroidetes (GenBac3), and E. coli (EC23S857) concentrations showed a significant correlation with average air temperature prior to sampling (Table 3). A recent review suggests that ambient water temperature is a key factor influencing the decay of many indicators (70), especially coliphage (71, 72). In addition, meta-analyses of somatic and F+ coliphage decay rates confirmed the high sensitivity of these viral groups to water temperature across multiple studies (73), potentially explaining this geospatial trend in somatic coliphage assuming there is a predictable link between air and sewage temperatures in this study. F+ coliphage did not vary by air temperature in our study, potentially contradicting previous reports. This disparity between somatic and F+ coliphage groups could be due to different data measurement distributions impacting the geospatial mixed model outcomes (Fig. 1); however, a comparison of mean log10 PFU/10 ml concentrations did not show a significant difference (P = 0.618). Instead, this difference may be due to variability in coliphage subpopulations between somatic and F+ groups. It is possible that some subpopulations of somatic coliphage occurring in untreated sewage are more sensitive to the surrounding air temperature, resulting in the observed differences in this study. This subpopulation hypothesis is also potentially supported by the GenBac3 assay results. Some Bacteroidetes subpopulations are reported to differ by latitude in untreated sewage (74). This could be important, because the average air temperature prior to sampling was significantly correlated with latitude (r = −0.77, P < 0.0001) in this study, suggesting that a similar scenario is possible for somatic and total coliphages. Regardless of explanation, susceptibility to air temperature may result in seasonal and geography-driven fluctuations in these indicator concentrations, making application to future water quality criteria more challenging. Additional research is necessary to characterize coliphage and Bacteroidetes subpopulations and confirm geospatial trends observed in this study.
Conclusions.
Nationwide characterization of viral and bacterial indicator geospatial trends in untreated sewage represents an important step toward the validation of these newer methods for future water quality monitoring applications. In addition, the large paired-measurement data set reported here also affords the opportunity to conduct a range of secondary analyses, such as the generation of new or updated quantitative microbial risk assessment models used to estimate public health risk. However, it is important to note that even though this research provides novel information, more extensive studies are necessary to confirm geospatial trends and address study limitations. For example, this study relied on single grab samples collected over a 3-month period during the winter. Future studies should investigate whether the observed geospatial trends vary over time due to seasonal changes or fluctuations in untreated sewage composition.
MATERIALS AND METHODS
Sewage facility selection.
GIS mapping with ArcGIS ArcMap (version 10.3; ESRI, Redlands, CA) was used to select 50 sewage facilities from urban (n = 25) and rural (n = 25) locations in the contiguous United States using a stepwise process. First, candidate facilities were selected from the U.S. EPA Facility Register Service geospatial database (https://www.epa.gov/frs/geospatial-data-download-service), with only “major” wastewater treatment plant facilities selected. Next, candidate urban and rural facilities were classified with the Spatial Analysis Zonal Statistics tool using U.S. Census Bureau 2016 shape files for urban areas (https://www.census.gov/geographies/mapping-files/time-series/geo/carto-boundary-file.html) combined with human population data from the EnviroAtlas Dasymetric toolbox (75). An urban facility was defined as a location in an urban area with ≥1,000,000 estimated population. In contrast, a rural facility was defined as an area with ≤3,000 estimated population that was at least 50 km or more from an urban area. A stratified random selection process was then used to select candidate urban and rural facilities across the contiguous United States for sewage sample collection using the National Oceanic and Atmospheric Administration Sampling Design Tool (https://coastalscience.noaa.gov/project/sampling-design-tool-arcgis/). Study participation was voluntary, with no facility treatment or service information requested during sampling.
Sewage sample collection.
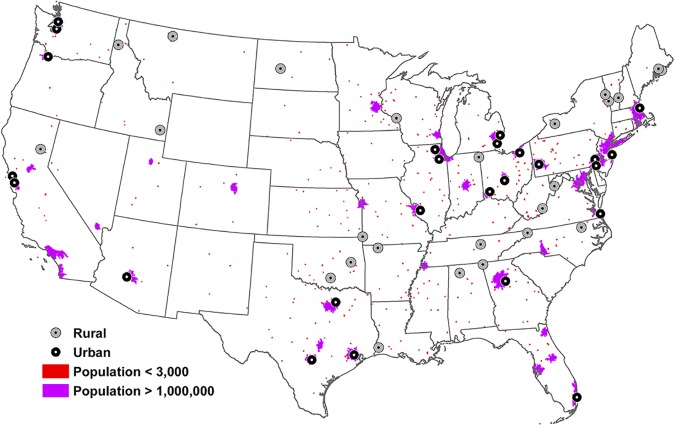
Single grab primary influent sewage was collected at each selected facility (n = 49; one facility elected to not participate in the study) across the contiguous United States over an 85-day period of time (29 January 2018 to 23 April 2018) (Fig. 3), as previously described (54). Briefly, 1 liter of untreated primary influent sewage was collected from each facility and immediately stored on ice. Samples were then packed and shipped on ice overnight to Cincinnati, OH, for laboratory testing (maximum holding time, 24 h).
FIG 3.
Geospatial information system (GIS) map of selected wastewater treatment facilities. Gray circles with a black center represent rural facilities. Black circles with a gray center represent urban facilities. Purple-shaded areas indicate locales with a reported population of greater than 1,000,000 individuals (https://www.census.gov/geographies/mapping-files/time-series/geo/carto-boundary-file.html), and red-shaded areas depict regions with a population of less than 3,000 (75). The map was generated with ArcGIS ArcMap (version 10.3; ESRI, Redlands, CA) using public domain data layers (U.S. Census Bureau, Washington, DC).
Culture-based bacteriophage enumeration.
The single agar layer (SAL) method was used to enumerate somatic, F+, and CB-390 coliphages as well as B. fragilis GB-124 bacteriophage from 100 ml of primary influent sewage samples. The somatic and F+ coliphage SAL method was performed as previously described (57, 58). For CB-390 and GB-124 bacteriophages, existing double agar layer methods (53, 64) were modified to the SAL format to accommodate processing of 100-ml samples. Modifications included a proportional increase of all reagents and the amount of bacterial host culture and utilization of 100 ml of 2× bottom agar formulations. During each sampling event, a positive control and two negative controls were used for each method. The positive control for coliphages consisted of adding either Phi X174 (somatic coliphage; ATCC 13706-B1) or MS2 (F+ coliphage; ATCC 15597-B1) to 100 ml of sterile 0.01 M phosphate-buffered saline (PBS), pH 7.4 (Sigma-Aldrich, St. Louis, MO), followed by sample processing as described above. For Bacteroides bacteriophage, GB-124 previously isolated from sewage acted as a positive control. Negative controls consisted of method blanks in which sample was replaced with 0.01 M PBS and medium sterility checks in which plates containing only agar were incubated. For the duration of the study, positive controls yielded expected results (i.e., plaques characteristic of each coliphage type) and no plaques were observed on any of the negative controls, indicating absence of contamination. All data were log10 transformed and expressed as log10 PFU per 10 ml.
Total DNA extraction and quantification.
A large-scale genomic and viral DNA purification procedure was performed with the QIAamp DNA blood maxi kit spin protocol as described by the manufacturer (Qiagen, Valencia, CA). Total DNA extractions were performed on 10-ml primary influent sewage sample volumes and eluted in 600 μl AE buffer. Total DNA extraction yields were determined with a NanoDrop ND-2000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). All DNA extracts were diluted to a concentration of 2.5 ng/μl and stored at –20°C in 50-μl aliquots in GeneMate Slick low-adhesion microtubes (ISC BioExpress) until the time of analysis (<6 months). DNA extracts were normalized to a fixed test concentration (5 ng per reaction) to standardize test conditions and eliminate any potential differences in DNA concentration between samples that could impact amplification chemistry. Extraction controls, with purified water substituted for sewage DNA extract, were used each day samples were extracted to monitor for potential extraneous DNA contamination.
Primers and probes.
Primer and probe sequences for eight qPCR assays are reported in Table 4. The panel of human-associated DNA markers target both bacteria (HF183/BacR287 and HumM2) and viruses (CPQ_056, CPQ_064, and HPyV) (21, 26–28, 40, 41). The remaining three qPCR assays target FIB (Entero1a, EC23S857, and GenBac3) (10–12, 51).
TABLE 4.
Primer and probe sequences of qPCR assay human-associated DNA marker and general fecal sewage metrics
| Assay | Primer/probe | Sequence (5′ to 3′)a | Reference(s) |
|---|---|---|---|
| CPQ_056 | crAss056_F1 | CAGAAGTACAAACTCCTAAAAAACGTAGAG | 27 |
| crAss056_R2 | GATGACCAATAAACAAGCCATTAGC | ||
| crAss056_P1 | FAM-AATAACGATTTACGTGATGTAAC-MGB | ||
| CPQ_064 | crAss064_F1 | TGTATAGATGCTGCTGCAACTGTACTC | |
| crAss064_R1 | CGTTGTTTTCATCTTTATCTTGTCCAT | ||
| crAss064_P1 | FAM-CTGAAATTGTTCATAAGCAA-MGB | ||
| HPyV | SM2 | AGTCTTTAGGGTCTTCTACCTTT | 21, 41 |
| P6 | GGTGCCAACCTATGGAACAG | ||
| KGj3 | FAM-TCATCACTGGCAAACAT-MGB | ||
| HF183/BacR287 | HF183 | ATCATGAGTTCACATGTCCG | 28 |
| BacR287 | CTTCCTCTCAGAACCCCTATCC | ||
| BacP234 | FAM-CTAATGGAACGCATCCC-MGB | ||
| Bac234IAC | VIC-AACACGCCGTTGCTACA-MGB | ||
| HumM2 | HumM2F | CGTCAGGTTTGTTTCGGTATTG | 26, 40 |
| HumM2R | TCATCACGTAACTTATTTATATGCATTAGC | ||
| HumM2P | FAM-TATCGAAAATCTCACGGATTAACTCTTGTGTACGC-TAMRA | ||
| UC1P1 | VIC-CCTGCCGTCTCGTGCTCCTCA-TAMRA | ||
| Entero1a | EnteroF1A | GAGAAATTCCAAACGAACTTG | 11, 12 |
| EnteroR1 | CAGTGCTCTACCTCCATCATT | ||
| GPL813TQ | FAM-TGGTTCTCTCCGAAATAGCTTTAGGGCTA-TAMRA | ||
| EC23S857 | EC23SF2-1 | GGTAGAGCACTGTTTTGGCA: | 10 |
| EC23SR2-1 | TGTCTCCCGTGATAACTTTCTC | ||
| EC23SP2b | FAM-TCATCCCGACTTACCAACCCG-TAMRA | ||
| GenBac3 | GenBactF3 | GGGGTTCTGAGAGGAAGGT | 51 |
| GenBactR4 | CCGTCATCCTTCACGCTACT | ||
| GenBactP2 | FAM-CAATATTCCTCACTGCTGCCTCCCGTA-TAMRA |
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
Reference DNA preparation.
Reference DNA sources for human-associated genetic markers consisted of a gBLOCK and plasmid construct (Integrated DNA Technologies, Coralville, IA). The calibration model standard consisted of a single gBLOCK preparation (all DNA targets on same construct), while the plasmid construct served as an IAC target. The IAC plasmid construct was linearized by NotI restriction digest (New England BioLabs, Beverly, MA) and purified by use of a QIAquick PCR purification kit (Qiagen, Valencia, CA). The gBLOCK and IAC plasmid constructs were then quantified with a NanoDrop ND-2000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE) and diluted in 10 mM Tris–0.1 mM EDTA (pH 8.0) to generate 10, 102, 103, 104, and 105 copies/2 μl for calibration standards and 102 copies/2 μl for IAC reference material. All reference DNA material preparations were stored in GeneMate Slick low-adhesion microcentrifuge tubes (ISC BioExpress, Kaysville, UT) at –20°C. A previously reported plasmid DNA standard (76) was used for FIB genetic markers (Entero1a, EC23S857, and GenBac3).
qPCR amplification.
All qPCR assays were used as previously described (Table 4). Briefly, reaction mixtures contained 1× TaqMan environmental master mix (version 2.0; Thermo Fisher Scientific, Grand Island, NY), 0.2 mg/ml bovine serum albumin (Sigma-Aldrich, St. Louis, MO), 1 μM each primer, 80 nM 6-carboxyfluorescein (FAM)-labeled probe, and 80 nM VIC-labeled probe (HF183/BacR287 and HumM2 only). All reaction mixtures contained either a reference DNA standard dilution ranging between 10 and 1 × 105 copies/2 μl or 2 μl of DNA sample extract (5 ng of total DNA) in a total reaction volume of 25 μl. Multiplex reaction mixtures with HF183/BacR287 and HumM2 also contained 102 copies of IAC template. All reactions were performed in triplicate in MicroAmp optical 96-well reaction plates with MicroAmp 96-well optical adhesive film (Thermo Fisher Scientific, Grand Island, NY). The thermal cycling profile for all assays was 2 min at 95°C, followed by 40 cycles of 5 s at 95°C and 30 s at 60°C (except for EC23S857, which uses a 56°C annealing temperature). The threshold was manually set to either 0.03 (HF183/BacR287, CPQ_056, CPQ_064, HPyV, Entero1a EC23S857, and GenBac3) or 0.08 (HumM2), and Cq values were exported to Microsoft Excel for further data analysis. Six no-template controls with purified water substituted for template DNA were performed with each instrument run to identify potential qPCR amplification contamination. HF183/BacR287 and HumM2 multiplex IAC procedures were used to monitor for amplification inhibition, as previously reported (77). Any DNA extract indicating evidence of amplification inhibition was discarded.
Data analysis.
“Mixed” calibration models (generated from a master slope derived from six independent standard curves and instrument run-specific y-intercept control data) (78), LLOQ, and concentration estimates of qPCR genetic markers (mean log10 copy number per 10 ml of sewage) were calculated using a Bayesian Markov Chain Monte Carlo approach on the publicly available software WinBUGS, version 1.4.1 1 (www.mrc-bsu.cam.ac.uk/software/bugs/the-bugs-project-winbugs/). LLOQ was defined as the upper bound of the 95% credible interval from repeated measurements of the lowest standard dilution tested. Amplification efficiency (E) was calculated as follows: E = 10 (−1/slope) − 1. One-way ANOVA with Tukey’s multiple-comparison test was performed using GraphPad Prism version 8.0.1 (GraphPad Software, La Jolla, CA) to compare mean log10 concentration per 10 ml of sewage values between each sewage pollution metric. Pearson product momentum correlation analysis was performed between eligible bacterial and viral sewage pollution metric paired measurements (α = 0.05). For geospatial analysis, a multiple linear mixed model (Proc Mixed) was used to regress each fecal pollution metric against two continuous predictors (average air temperature [°C] and elevation [m]) and one categorical (urban or rural) predictor. Average air temperature (°C) prior to sampling indicates the 7-day average from Weather Underground (https://www.wunderground.com/) historic data sets (5 days used for two sites with incomplete historic records). Elevation was reported as the respective urban or rural wastewater facility height above mean sea level (in meters). A repeated variance spatial power [SP(…) (Lat Long)] covariance structure was used for urban and rural categorical error correlation across space. Covariance structure models (…) included exponential (exp), anisotropy exponential (expa), and power linear (lin). Model fit was assessed with q-q residual plots (qqplot). Outliers were defined as samples that deviated from the 1:1 qqplot residual. Two outliers were identified, including (i) site 9 for somatic coliphage and (ii) site 25 for CPQ_056. Residual serial correlation analysis with ARIMA (Proc ARIMA) indicated no residual serial correlation among all models (P > 0.06). All statistics were calculated with SAS software (version 9.4; SAS, Cary, NC) unless noted otherwise.
ACKNOWLEDGMENTS
The U.S. Environmental Protection Agency, through its Office of Research and Development, funded and managed the research described herein.
Information has been subjected to U.S. EPA peer and administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the U.S. EPA. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
REFERENCES
- 1.Strubbia S, Phan MVT, Schaeffer J, Koopmans M, Cotten M, Le Guyader FS. 2019. Characterization of norovirus and other human enteric viruses in sewage and stool samples through next-generation sequencing. Food Environ Virol 11:400–409. doi: 10.1007/s12560-019-09402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomez-Canela C, Sala-Comorera T, Pueyo V, Barata C, Lacorte S. 2019. Analysis of 44 pharmadeuticals consumed by elderly using liquid chromatography coupled to tandem mass spectrometry. J Pharm Biomed Anal 168:55–63. doi: 10.1016/j.jpba.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Hendriksen RS, Global Sewage Surveillance Project Consortium, Munk P, Njage P, van Bunnik B, McNally L, Lukjancenko O, Röder T, Nieuwenhuijse D, Pedersen SK, Kjeldgaard J, Kaas RS, Clausen PTLC, Vogt JK, Leekitcharoenphon P, van de Schans MGM, Zuidema T, de Roda Husman AM, Rasmussen S, Petersen B, Amid C, Cochrane G, Sicheritz-Ponten T, Schmitt H, Alvarez JRM, Aidara-Kane A, Pamp SJ, Lund O, Hald T, Woolhouse M, Koopmans MP, Vigre H, Petersen TN, Aarestrup FM. 2019. Global monitoring of antimicrobial resistance based on metagenomic analyses of urban sewage. Nat Commun 10:1124. doi: 10.1038/s41467-019-08853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.U.S. EPA. 2004. Report to Congress: impacts and control of CSOs and SSOs. EPA 833-R-04-001 U.S. EPA, Washington, DC. [Google Scholar]
- 5.American Society of Civil Engineers. 2017. Report card for America’s infrastructure. ASCE, Reston, VA: https://www.infrastructurereportcard.org/. Accessed September 2019. [Google Scholar]
- 6.Newton RJ, McClary JS. 2019. The flux and impact of wastewater infrastructure microorganisms on human ecosystem health. Curr Opin Biotechnol 57:145–150. doi: 10.1016/j.copbio.2019.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorevitch S, Shrestha A, DeFlorio-Barker S, Breitenbach C, Heimler I. 2017. Monitoring urban beaches with qPCR vs. culture measures of fecal indicator bacteria: implications for public notification. Environ Health 16:45. doi: 10.1186/s12940-017-0256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ. 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res 44:4674–4691. doi: 10.1016/j.watres.2010.06.049. [DOI] [PubMed] [Google Scholar]
- 9.Frick C, Vierheilig J, Linke R, Savio D, Zornig H, Antensteiner R, Baumgartner C, Bucher C, Blaschke AP, Derx J, Kirschner AKT, Ryzinska-Paier G, Mayer R, Seidl D, Nadiotis-Tsaka T, Sommer R, Farnleitner A. 2018. Poikilothermic animals as a previously unrecognized source of fecal indicator bacteria in a backwater ecosystem of a large river. Appl Environ Microbiol 84:e00715-18. doi: 10.1128/AEM.00715-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chern EC, Siefring S, Paar J, Doolittle M, Haugland R. 2011. Comparison of quantitative PCR assays for Escherichia coli targeting ribosomal RNA and single copy genes. Lett Appl Microbiol 52:298–306. doi: 10.1111/j.1472-765X.2010.03001.x. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig W, Schleifer KH. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst Appl Microbiol 23:556–562. doi: 10.1016/S0723-2020(00)80030-2. [DOI] [PubMed] [Google Scholar]
- 12.Siefring SC, Varma M, Atikovic E, Wymer LJ, Haugland RA. 2008. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. J Water Health 6:225–237. doi: 10.2166/wh.2008.022. [DOI] [PubMed] [Google Scholar]
- 13.McMinn BR, Ashbolt NJ, Korajkic A. 2017. Bacteriophages as indicators of faecal pollution and enteric virus removal. Lett Appl Microbiol 65:11–26. doi: 10.1111/lam.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMinn BR, Huff EM, Rhodes ER, Korajkic A. 2017. Concentration and quantification of somatic and F+ coliphages from recreational waters. J Virol Methods 249:58–65. doi: 10.1016/j.jviromet.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wanjugi P, Sivaganesan M, Korajkic A, McMinn B, Kelty CA, Rhodes E, Cyterski M, Zepp R, Oshima K, Stachler E, Kinzelman J, Kurdas SR, Citriglia M, Hsu F-C, Acrey B, Shanks OC. 2018. Incidence of somatic and F+ coliphage in Great Lake Basin recreational waters. Water Res 140:200–210. doi: 10.1016/j.watres.2018.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nappier SP, Hong T, Ichida A, Goldstone A, Eftim SE. 2019. Occurrence of coliphage in raw wastewater and in ambient water: a meta-analysis. Water Res 153:263–273. doi: 10.1016/j.watres.2018.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gantzer C, Maul A, Audic JM, Schwartzbrod L. 1998. Detection of infectious enteroviruses, enterovirus genomes, somatic coliphages, and Bacteroides fragilis phages in treated wastewater. Appl Environ Microbiol 64:4307–4312. doi: 10.1128/AEM.64.11.4307-4312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Havelaar A, van Olphen M, Drost YC. 1993. F-specific RNA bacteriophages are adequate model organisms for enteric viruses in freshwater. Appl Environ Microbiol 59:2956–2962. doi: 10.1128/AEM.59.9.2956-2962.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pouillot R, Van Doren JM, Woods J, Plante D, Smith M, Goblick G, Roberts C, Locas A, Hajen W, Stobo J, White J, Holtzman J, Buenaventura E, Burkhardt W, Catford A, Edwards R, DePaola A, Calci KR. 2015. Meta-analysis of the reduction of norovirus and male-specific coliphage concentrations in wastewater treatment plants. Appl Environ Microbiol 81:4669–4681. doi: 10.1128/AEM.00509-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol 66:4571–4574. doi: 10.1128/aem.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green HC, Haugland RA, Varma M, Millen HT, Borchardt MA, Field KG, Walters WA, Knight R, Sivaganesan M, Kelty CA, Shanks OC. 2014. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl Environ Microbiol 80:3086–3094. doi: 10.1128/AEM.04137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haugland RA, Varma M, Kelty CA, Peed L, Sivaganesan M, Shanks OC. 2010. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by real-time PCR. Syst Appl Microbiol 33:348–357. doi: 10.1016/j.syapm.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res 41:3701–3715. doi: 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 24.Lee CS, Lee J. 2010. Evaluation of new gyrB-based real-time PCR system for the detection of B. fragilis as an indicator of human-specific fecal contamination. J Microbiol Methods 82:311–318. doi: 10.1016/j.mimet.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Reischer GH, Kasper DC, Steinborn R, Farnleitner AH, Mach RL. 2007. A quantitative real-time PCR assay for the highly sensitive and specific detection of human faecal influence in spring water from a large alpine catchment area. Lett Appl Microbiol 44:351–356. doi: 10.1111/j.1472-765X.2006.02094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shanks OC, Kelty CA, Sivaganesan M, Varma M, Haugland RA. 2009. Quantitative PCR for genetic markers of human fecal pollution. Appl Environ Microbiol 75:5507–5513. doi: 10.1128/AEM.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stachler E, Kelty CA, Sivaganesan M, Li X, Bibby K, Shanks OC. 2017. Development of crAssphage quantitative real-time PCR assays for human fecal pollution measurement. Environ Sci Technol 51:9146–9154. doi: 10.1021/acs.est.7b02703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McQuaig SM, Scott TM, Lukasik JO, Paul JH, Harwood VJ. 2009. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl Environ Microbiol 75:3379–3388. doi: 10.1128/AEM.02302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosario K, Symonds EM, Sinigalliano C, Stewart J, Breitbart M. 2009. Pepper mild mottle virus as an indicator of fecal pollution. Appl Environ Microbiol 75:7261–7267. doi: 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Aljaro C, Balleste E, Muniesa M, Jofre J. 2017. Determination of crAssphage in water samples and applicability for tracking human faecal pollution. Microb Biotechnol 10:1775–1780. doi: 10.1111/1751-7915.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed W, Gyawali P, Feng S, McLellan SL. 2019. Host specificity and sensitivity of established and novel sewage-associated marker genes in human and nonhuman fecal samples. Appl Environ Microbiol 85:e00641-19. doi: 10.1128/AEM.00641-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed W, Lobos A, Senkbeil J, Peraud J, Gallard J, Harwood VJ. 2018. Evaluation of the novel crAssphage marker for sewage pollution tracking in storm drain outfalls in Tampa, Florida. Water Res 131:142–150. doi: 10.1016/j.watres.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 33.Ahmed W, Payyappat S, Cassidy M, Besley C. 2019. A duplex PCR assay for the simultaneous quantification of Bacteroides HF183 and crAssphage CPQ_056 marker genes in untreated sewage and stormwater. Environ Int 126:252–259. doi: 10.1016/j.envint.2019.01.035. [DOI] [PubMed] [Google Scholar]
- 34.Ahmed W, Payyappat S, Cassidy M, Besley C, Power K. 2018. Novel crAssphage marker genes ascertain sewage pollution in a recreational lake receiving urban stormwater runoff. Water Res 145:769–778. doi: 10.1016/j.watres.2018.08.049. [DOI] [PubMed] [Google Scholar]
- 35.Farkas K, Adriaenssens EM, Walker DI, McDonald JE, Malham SK, Jones DL. 2019. Critical evaluation of crAssphage as a molecular marker for human-derived wastewater contamination in the aquatic environment. Food Environ Virol 11:113–119. doi: 10.1007/s12560-019-09369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kongprajug A, Mongkolsuk S, Sirikanchana K. 2019. CrAssphage as a potential human sewage marker for microbial source tracking in Southeast Asia. Environ Sci Technol Lett 6:159–164. doi: 10.1021/acs.estlett.9b00041. [DOI] [Google Scholar]
- 37.Griffith JF, Layton BA, Boehm AB, Holden P, Jay J, Hagedorn C, McGee C, Weisberg SB. 2013. The California microbial source identification manual: a tiered approach to identifying fecal pollution sources to beaches. Southern California Coastal Water Research Project, Costa Mesa, CA. [Google Scholar]
- 38.U.S. EPA. 2015. Review of coliphages as possible indicators of fecal contamination from ambient water quality. U.S. EPA, Washington, DC. [Google Scholar]
- 39.U.S. EPA. 2012. Recreational water quality criteria. Office of Water, U.S. EPA, Washington, DC. [Google Scholar]
- 40.U.S. EPA. 2019. Method 1697: characterization of human fecal pollution in water by HumM2 TaqMan quantitative polymerase chain reaction (qPCR) assay. U.S. EPA, Washington, DC. [Google Scholar]
- 41.U.S. EPA. 2019. Method 1696: characterization of human fecal pollution in water by HF183/BacR287 TaqMan quantitative polymerase chain reaction (qPCR) assay. U.S. EPA, Washington, DC. [Google Scholar]
- 42.U.S. Census Bureau. 2010. Resident population data. https://web.archive.org/web/20111028061117/http://2010.census.gov/2010census/data/apportionment-dens-text.php.
- 43.Fierer N, McCain CM, Meir P, Zimmerman M, Rapp JM, Silman MR, Knight R. 2011. Microbes do not follow the elevational diversity patterns of plants and animals. Ecology 92:797–804. doi: 10.1890/10-1170.1. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y, Gao Y, Wang S, Xu D, Yu H, Wu L, Lin Q, Hu Y, Li X, He Z, Deng Y, Zhou J. 2014. The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J 8:430–440. doi: 10.1038/ismej.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bryant JA, Lamanna C, Morlon H, Kerkhoff AJ, Enquist BJ, Green JL. 2008. Colloquium paper: microbes on mountain sides: contrasting elevational patterns of bacterial and plant diversity. Proc Natl Acad Sci U S A 105:11505–11511. doi: 10.1073/pnas.0801920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki TA, Worobey M. 2014. Geographical variation of human gut microbial composition. Biol Lett 10:20131037. doi: 10.1098/rsbl.2013.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu L, Li Y, Wang P, Zhang W, Wang C, Wang Q. 2015. Understanding the linkage between elevation and the activated-sludge bacterial community along a 3,600-meter elevation gradient in China. Appl Environ Microbiol 81:6567–6576. doi: 10.1128/AEM.01842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shanks OC, Newton RJ, Kelty CA, Huse SM, Sogin ML, McLellan SL. 2013. Comparison of the microbial community structures of untreated wastewaters from different geographic locales. Appl Environ Microbiol 79:2906–2913. doi: 10.1128/AEM.03448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newton RJ, McLellan SL, Dila DK, Vineis JH, Morrison HG, Eren AM, Sogin ML. 2015. Sewage reflects the microbiomes of human populations. mBio 6:e02574-14. doi: 10.1128/mBio.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Soininen J, He J, Shen J. 2012. Phylogenetic clustering increases with elevation in microbes. Environ Microbiol Rep 4:217–226. doi: 10.1111/j.1758-2229.2011.00324.x. [DOI] [PubMed] [Google Scholar]
- 51.Dick LK, Field KG. 2004. Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes. Appl Environ Microbiol 70:5695–5697. doi: 10.1128/AEM.70.9.5695-5697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GZ, Boling L, Barr JJ, Speth DR, Seguritan V, Aziz RK, Felts B, Dinsdale EA, Mokili JL, Edwards RE. 2014. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebdon JE, Sellwood J, Shore J, Taylor HD. 2012. Phages of Bacteroidales (GB-124): a novel tool for viral waterborne disease control? Environ Sci Technol 46:1163–1169. doi: 10.1021/es202874p. [DOI] [PubMed] [Google Scholar]
- 54.Kelty CA, Varma M, Sivaganesan M, Haugland RA, Shanks OC. 2012. Distribution of genetic marker concentrations for fecal indicator bacteria in sewage and animal feces. Appl Environ Microbiol 78:4225–4232. doi: 10.1128/AEM.07819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McMinn BR, Korajkic A, Ashbolt NJ. 2014. Evaluation of Bacteroides fragilis GB-124 bacteriophages as novel human-associated faecal indicators in the United States. Lett Appl Microbiol 59:115–121. doi: 10.1111/lam.12252. [DOI] [PubMed] [Google Scholar]
- 56.Shanks OC, White K, Kelty CA, Sivaganesan M, Blannon J, Meckes M, Varma M, Haugland RA. 2010. Performance of PCR-based assays targeting Bacteroidales genetic markers of human fecal pollution in sewage and fecal samples. Environ Sci Technol 44:6281–6288. doi: 10.1021/es100311n. [DOI] [PubMed] [Google Scholar]
- 57.U.S. EPA. 2001. Method 1602: male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure. U.S. EPA, Washington, DC. [Google Scholar]
- 58.U.S. EPA. 2018. Method 1643: male-specific (F+) and somatic coliphage in secondary (no disinfection) wastewater by the single agar layer (SAL) procedure. U.S. EPA Office of Water, Washington, DC. [Google Scholar]
- 59.Ahmed W, Sidhu JPS, Smith K, Beale DJ, Gyawali P, Toze S. 2016. Distributions of fecal markers in wastewater from different climatic zones for human fecal pollution tracking in Australian surface waters. Appl Environ Microbiol 82:1316–1323. doi: 10.1128/AEM.03765-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sidu JPS, Sena K, Hodgers L, Palmer A, Toze S. 2018. Comparative enteric viruses and coliphage removal during wastewater treatment processes in a sub-tropical environment. Sci Total Environ 616-617:669–677. doi: 10.1016/j.scitotenv.2017.10.265. [DOI] [PubMed] [Google Scholar]
- 61.Hewitt J, Greening GE, Leonard M, Lewis GD. 2013. Evaluation of human adenovirus and human polyomavirus as indicators of human sewage contamination in the aquatic environment. Water Res 47:6750–6761. doi: 10.1016/j.watres.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 62.Kaas L, Ogorzaly L, Lecellier G, Berteaux-Lecellier V, Cauchie H, Langlet J. 2019. Detection of human enteric viruses in French Polynesian wastewaters, environmental waters and giant clams. Food Environ Virol 11:52–64. doi: 10.1007/s12560-018-9358-0. [DOI] [PubMed] [Google Scholar]
- 63.Barrios ME, Fernandez MDB, Cammarata RV, Torres C, Mbayed VA. 2018. Viral tools for detection of fecal contamination and microbial source tracking in wastewater from food industries and domestic sewage. J Virol Methods 262:79–88. doi: 10.1016/j.jviromet.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 64.Guzman C, Moce-Llivina L, Lucena F, Jofre J. 2008. Evaluation of Escherichia coli host strain CB390 for simultaneous detection of somatic and F-specific coliphages. Appl Environ Microbiol 74:531–534. doi: 10.1128/AEM.01710-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aqullo-Barcelo M, Galofre B, Sala L, Garcia-Aljaro C, Lucena F, Jofre J. 2016. Simultaneous detection of somatic and F-specific coliphage in different settings by Escherichia coli strain CB390. FEMS Microbiol Lett 363:17. doi: 10.1093/femsle/fnw180. [DOI] [PubMed] [Google Scholar]
- 66.Campos C, Mendez J, Venegas C, Riano LF, Castano P, Leiton N, Riano E. 2019. Aptness of Escherichia coli host strain CB390 to detect total coliphage in Colombia. Sci Rep 9:9246. doi: 10.1038/s41598-019-45775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bailey ES, Price M, Casanova LM, Sobsey MD. 2017. E. coli CB390: an alternative E. coli host for simultaneous detection of somatic and F+ coliphage viruses in reclaimed and other waters. J Virol Methods 250:25–28. doi: 10.1016/j.jviromet.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 68.Chern EC, Brenner KP, Wymer L, Haugland R. 2009. Comparison fecal indicator bacteria densities in marine recreational waters by qPCR. Water Qual Expo Health 1:203–214. doi: 10.1007/s12403-009-0019-2. [DOI] [Google Scholar]
- 69.Wang X, Hu M, Xia Y, Wen X, Ding K. 2012. Pyrosequencing analysis of bacterial diversity in 14 wastewater treatment systems in China. Appl Environ Microbiol 78:7042–7047. doi: 10.1128/AEM.01617-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Korajkic A, Wanjugi P, Brooks L, Cao Y, Harwood VJ. 2019. Persistence and decay of fecal microbiota in aquatic habitats. Microbiol Mol Biol Rev 83:e00005-19. doi: 10.1128/MMBR.00005-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu J, Cao Y, Young B, Yuen Y, Jiang S, Melendez D, Griffith JF, Stewart JR. 2016. Decay of coliphages in sewage-contaminated freshwater: uncertainty and seasonal effects. Environ Sci Technol 50:11593–11601. doi: 10.1021/acs.est.6b03916. [DOI] [PubMed] [Google Scholar]
- 72.Bailey ES, Casanova LM, Sobsey MD. 2019. Effects of environmental storage conditions on survival of indicator organisms in a blend of surface and dual disinfected reclaimed water. J Appl Microbiol 126:985–994. doi: 10.1111/jam.14186. [DOI] [PubMed] [Google Scholar]
- 73.Boehm AB, Silverman AI, Schriewer A, Goodwin KD. 2019. Systematic review and meta-analysis of decay rates of waterborne mammalian viruses and coliphages in surface waters. Water Res 164:114898. doi: 10.1016/j.watres.2019.114898. [DOI] [PubMed] [Google Scholar]
- 74.Shanks OC, Kelty CA, Archibeque SL, Jenkins M, Newton RJ, McLellan SL, Huse SM, Sogin ML. 2011. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microbiol 77:2992–3001. doi: 10.1128/AEM.02988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pickard BR, Daniel J, Mehaffey M, Jackson LE, Neale A. 2015. EnviroAtlas: a new geospatial tool to foster ecosystem services science and resource management. Ecosystems Services 14:45–55. doi: 10.1016/j.ecoser.2015.04.005. [DOI] [Google Scholar]
- 76.Sivaganesan M, Varma M, Siefring S, Haugland RA. 2018. Quantification of plasmid DNA standards for U.S. EPA fecal indicator bacteria qPCR methods by droplet digital PCR analysis. J Microbiol Methods 152:135–142. doi: 10.1016/j.mimet.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shanks OC, Kelty CA, Oshiro R, Haugland RA, Madi T, Brooks L, Field KG, Sivaganesan M. 2016. Data acceptance criteria for standardized human-associated fecal source identification quantitative real-time PCR methods. Appl Environ Microbiol 82:2773–2782. doi: 10.1128/AEM.03661-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sivaganesan M, Haugland RA, Chern EC, Shanks OC. 2010. Improved strategies and optimization of calibration models for real-time PCR absolute quantification. Water Res 44:4726–4735. doi: 10.1016/j.watres.2010.07.066. [DOI] [PubMed] [Google Scholar]