Summary
Krüppel-associated box domain zinc finger proteins (KZFPs) are the largest family of transcriptional regulators encoded by higher vertebrates. Characterized by an N-terminal KRAB domain and a C-terminal array of DNA-binding zinc fingers, KZFPs are encoded in the hundreds by the human genome, and participate, together with their cofactor KAP1, in repressing sequences derived from transposable elements during the epigenetic reprogramming that takes place in the first few days of embryogenesis. TEs likely account for more than two thirds of the human genome, and this so-called endovirome is considered both as a genomic threat and an important motor of evolution. Until recently, the KZFP/KAP1-mediated repression of the endovirome was thought to lead to irreversible silencing, and the evolutionary selection of KZFPs was taken as the host component of an arms race against transposable elements. Recent advances partly invalidate this view, and indicate that KZFPs and their TE targets partner up to establish species-specific regulatory networks that influence multiple aspects of vertebrate development and physiology.
Keywords: Endovirome, transposable element, KRAB-ZFP, co-option, gene regulation, speciation
Introduction
All biological events are regulated by complex transcriptional networks, from the self-renewal of pluripotent embryonic stem cells to the making of a neuron, and from the activation of a lymphocyte meeting its antigen to the fine-tuning of glycemia on fat and lean days alike. Keys to this process are the interactions of transcription factors with cis-acting genomic sequences and their modulation by epigenetic modifications of the DNA and protein constituents of the chromatin. Almost 70 years ago, Barbara McClintock proposed that part of these regulatory DNA sequences lay in mobile genetic elements (McClintock, 1950), and twenty years later Roy Britten and Eric Davidson outlined that the repetitive nature of these genetic invaders might explain how multiple changes in gene activity can so remarkably result from a single initiatory event (Britten and Davidson, 1969). Nevertheless, transposable elements (TEs) were long considered mostly as genetic threats in need of the strictest silencing, and were otherwise dismissed as purely selfish or junk DNA (Doolittle and Sapienza, 1980). The sequencing of the human genome at the dawn of the century changed this view, and it is increasingly recognized that TEs are crucial components of transcriptional regulatory networks that play essential roles not only in the evolution but also the biology of most organisms.
Ancestral RNA interference mechanisms, based on various forms of small RNAs, play an important role in silencing TEs in the germ line (reviewed in Friedli and Trono, 2015)). However, while these can be subjected to some dynamic modulation, a protein-based system is much more amenable to all kinds of regulation, hence would seem better suited for exploiting fully the transcriptional modulatory power of the endovirome. Recent work indicates that Krüppel-associated box domain zinc finger proteins (KZFPs), which fulfill such requirement, control transposable elements in higher vertebrates, and as such exert key influences on the biology of these organisms, including humans.
KZFP genes first emerged more than 400 million years ago, and are now encoded in the hundreds by all examined modern tetrapods, with the notable exception of birds, where they are much fewer (Emerson and Thomas, 2009, Liu et al., 2014, Imbeault et al., 2017, Kauzlaric et al., 2017)}. Their protein products are characterized by an N-terminal Krüppel-associated box (KRAB) domain and a C-terminal array of C2H2 zinc fingers (ZNF) (Urrutia, 2003). In spite of their numerical abundance, the functions of KZFPs have long remained ill defined, although cumulated data implicated some of them in processes as diverse as imprinting, cell differentiation, metabolic control and sexual dimorphism (reviewed in Lupo et al., 2013)). The picture changed when their KRAB-binding cofactor KAP1/TRIM28 was demonstrated to be essential for the early embryonic repression of transposable elements in both mouse and human, and when a few individual KZFPs could be linked to this function as well (Wolf and Goff, 2007, Wolf and Goff, 2009, Wolf et al., 2015, Rowe et al., 2013a, Jacobs et al., 2014). It then became suspected that the primary role of KZFPs was to silence TEs, and that their evolutionary selection represented the host component of an arms race against these genetic invaders. Recent data, while partly validating this view, reveal that KZFPs fulfill a role that is far more elaborate, as the instruments of a massive enterprise of TE domestication for the benefit of the host (Ecco et al., 2016, Imbeault et al., 2017).
In this primer, after a brief presentation of the endovirome, we sum up our current understanding of the KZFP family. We discuss its evolution, its role in controlling TEs, and how the selection of both TEs and KZFPs represents a dynamic partnership generating largely species-specific transcriptional networks that likely influence most aspects of human biology.
Transposable elements and their genomic impact
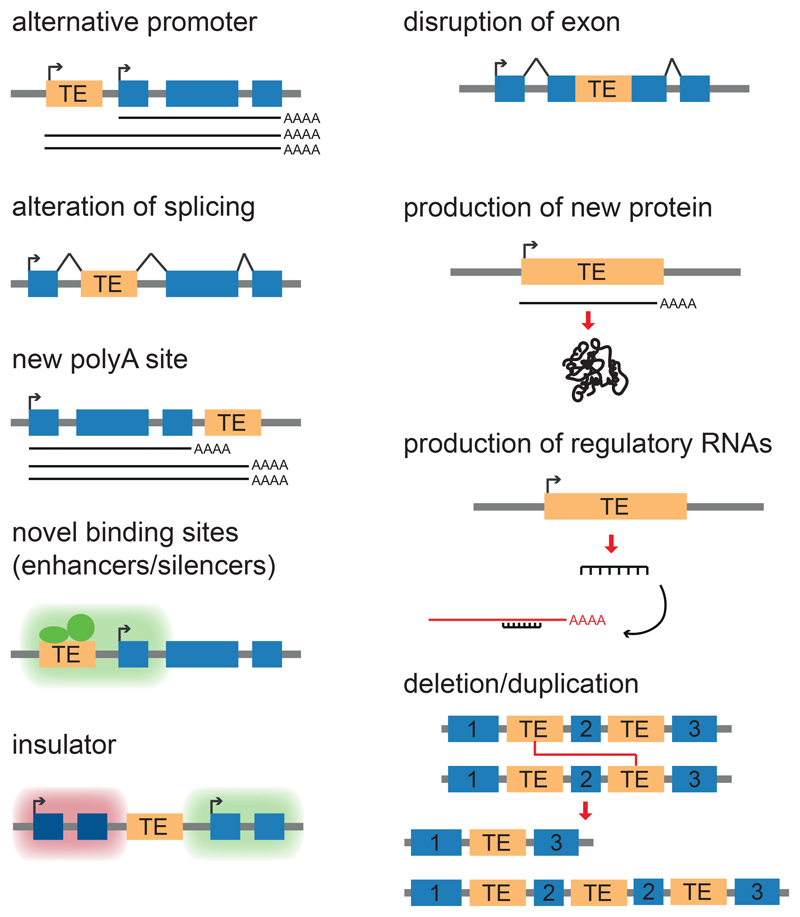
TEs can be classified according to their transposition mechanism, overall genetic structure and phylogenesis (Box 1). Most TEs present in the human genome are retroelements, whether endogenous retroviruses (HERV, or LTR- long terminal repeat- retrotransposons) or non-LTR-retrotransposons of the LINE, SINE (which include Alu) and SVA subgroups. All retroelements spread via a copy-and-paste mechanism leading to their amplification. Some 5 million sequences derived from TEs can be readily identified in the human genome. This endovirome accounts for a readily recognizable 50% of our DNA, but since TEs become unrecognizable over time due to mutational drifting, it is likely that this represents an underestimate of their contribution to our genetic make up. TEs fuel genetic diversity but can induce deleterious mutations responsible for disease, although less than 1 out of 10’000 human TE is still capable of transposition (about 100 LINEs, 1,000 Alus and a few tens of SVAs). Yet a far greater proportion can alter gene expression, as TEs can bear promoters, enhancers, repressors, insulators, splice sites or transcriptional stop signals (Figure 2). Accordingly, they can disrupt genes (via alternative splicing, truncation or insertion of new exons) or modify their expression (via promoter, enhancer or repressor effects). Owing to their highly repetitive nature, TEs also lay the ground for recombination events that can lead to deletions, duplications, rearrangements or translocations. Finally, they can alter genome architecture via insulator sequences or by nucleating short- and long-range chromatin interactions, or provide entirely novel open reading frames (reviewed in Friedli and Trono, 2015, Rebollo et al., 2012b, Warren et al., 2015). Consistent with their disruptive potential, TEs are counter-selected when inserted inside genes, notably in the sense orientation (Zhang et al., 2011, Medstrand et al., 2002).
Box 1. TE classification.
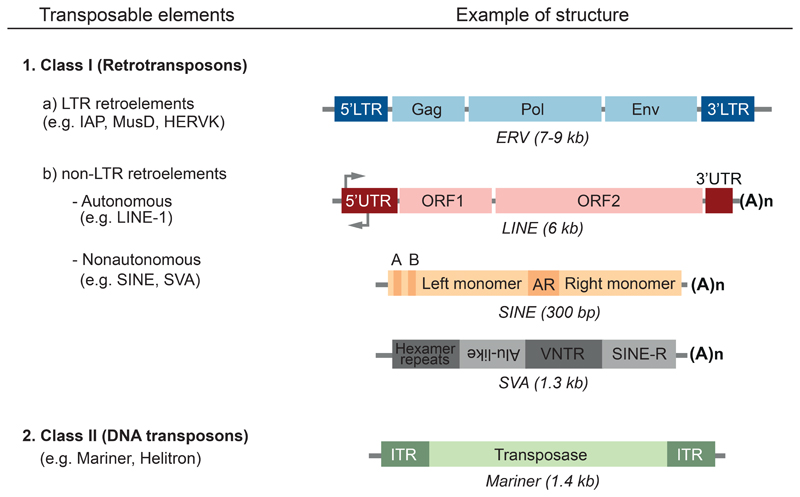
TEs are classified into two classes according to their transposition mechanism (Figure 1) (Rebollo et al., 2012b, Warren et al., 2015, Feschotte and Pritham, 2007). Class I elements, or retrotransposons, replicate via an RNA intermediate, using a copy-and-paste mechanism. They are further divided into long terminal repeat (LTR)-containing and non-LTR retroelements. The main representatives of LTR retroelements are endogenous retroviruses (ERVs), elements reminiscent of ancient retroviral infections. Several hundreds murine ERVs are active and capable of retrotransposition, accounting for up to 10% of spontaneous mutations observed in inbred mice (Maksakova et al., 2006). In contrast, all humans ERVs are transposition-defective. Non-LTR retrotransposons are further divided into autonomous, or long interspersed elements (LINE), and non-autonomous elements, comprising the short interspersed elements (SINE) and, in hominoid primates, the SINE-R, VNTR, Alu (SVA) elements. SINEs and SVAs depend on trans acting functions encoded by LINEs for their retrotransposition (Raiz et al., 2012, Hancks and Kazazian, 2010). A few hundred LINEs, about a thousand SINEs (of Alu subtype) and a few tens of SVAs are still transposition-competent in humans, accounting for 0.1% of de novo mutations in humans (Maksakova et al., 2006, Antony et al., 2011).
Class II elements, or DNA transposons, replicate via a DNA intermediate, either by a cut-and-paste mechanism (classic DNA transposons harboring transposases), by rolling-circle DNA replication (Helitrons), or by mechanisms not yet fully understood (Mavericks). DNA transposons are not active in humans, with the last transposition-competent element in primates dating back to 37 million years (Padeken et al., 2015, Pace and Feschotte, 2007).
Figure 2. How transposable elements can impact on host genomes.
Pathologies associated with new TE insertions or other types of deregulation include cancers, hemophilia, muscular dystrophy and other congenital or acquired human diseases (reviewed in Ayarpadikannan et al., 2015, Hancks and Kazazian, 2012, Mager and Stoye, 2015). Most TE-associated human disorders are related to non-LTR retrotransposons. A known cause of breast cancer is the insertion of a primate-specific Alu SINE into the BRCA1/2 genes (Miki et al., 1996, Puget et al., 1999), and cases of hemophilia A and B are associated with insertional mutations of LINE-1 or Alu elements into genes coding for the corresponding coagulation factors (Kazazian et al., 1988, Li et al., 2001). LTR retrotransposons have also been associated with some diseases, especially cancer, as ERV transcripts are upregulated in some tumors and there are reports of LTRs driving oncogene expression in human lymphomas (Lamprecht et al., 2010, Romanish et al., 2010). In mouse, many LTR elements are transcriptionally proficient and ERVs related to MMTV (mouse mammary tumor virus) and MLV (mouse leukemia virus) can cause cancer via activation of proto-oncogenes (Rosenberg and Jolicoeur, 1997). Finally, expression of ERV proteins can be detrimental to the host and might be associated with autoimmune diseases such as systemic lupus erythematosus in mice and multiple sclerosis in humans (Baudino et al., 2010, Antony et al., 2011).
However, it is on an evolutionary scale that the impact of transposable elements is best appreciated. TEs sprinkle the genomes of their host species with binding sites for transcription factors, which can then contribute to species-restricted phenotypes (reviewed in Thompson et al., 2016)). For instance mammals generally produce amylase in the pancreas, yet primates release this enzyme in saliva too, owing to the insertion upstream the amylase coding sequence of a HERV-E LTR driving expression in salivary glands (Samuelson et al., 1996, Ting et al., 1992). Many other cases of LTR promoter exaptation have been documented, generally resulting in new or altered tissue-specificity for cellular genes (Cohen et al., 2009, Stavenhagen and Robins, 1988, Rebollo et al., 2012a). Examples of TE-based species-specific enhancers also abound, and in mammals include MER130 elements acting as neocortex-specific units, a SINE integrant functioning as a distal enhancer of Fgf8 in the diencephalon, RLTR13D5 ERVs co-opted as placenta-specific enhancers, and the MER41-mediated dispersion of interferon-responsive elements in primates (Notwell et al., 2015, Nakanishi et al., 2012, Chuong et al., 2013). Retroelements also contribute to embryonic stem cells (ESC) regulatory networks, as many binding sites for pluripotency factors (such as Oct4 and Nanog) reside within primate- or human-specific ERVs in the human genome, and LTR elements are implicated in the regulation of specific genes in early embryogenesis (Bourque et al., 2008, Fort et al., 2014b, Macfarlan et al., 2012, Peaston et al., 2004). TEs further correlate with p53 recruitment, with more than one third of the genomic targets of this tumor suppressor overlapping with primate-specific ERVs (Wang et al., 2007).
Finally, ERV-derived proteins can themselves be sources of genetic diversity, as illustrated in placental mammals where formation of the syncytiotrophoblast, a placenta layer with extensive cellular fusion, is mediated by the ERV envelope-derived syncytins (Mi et al., 2000, Dupressoir et al., 2009, Dupressoir et al., 2011). Interestingly, across mammals, these proteins derive from the env gene of distinct groups of ERVs, indicating convergent evolution with multiple and independent events of ERV co-option (Lavialle et al., 2013).
The impact of the endovirome on the evolution and biology of complex organisms is thus enormous, yet it would be likely far more limited were it not for host-encoded activities capable of modulating its influences. In higher vertebrates, this is a function notably accomplished by KRAB zinc finger proteins.
The make up of a KRAB zinc finger protein
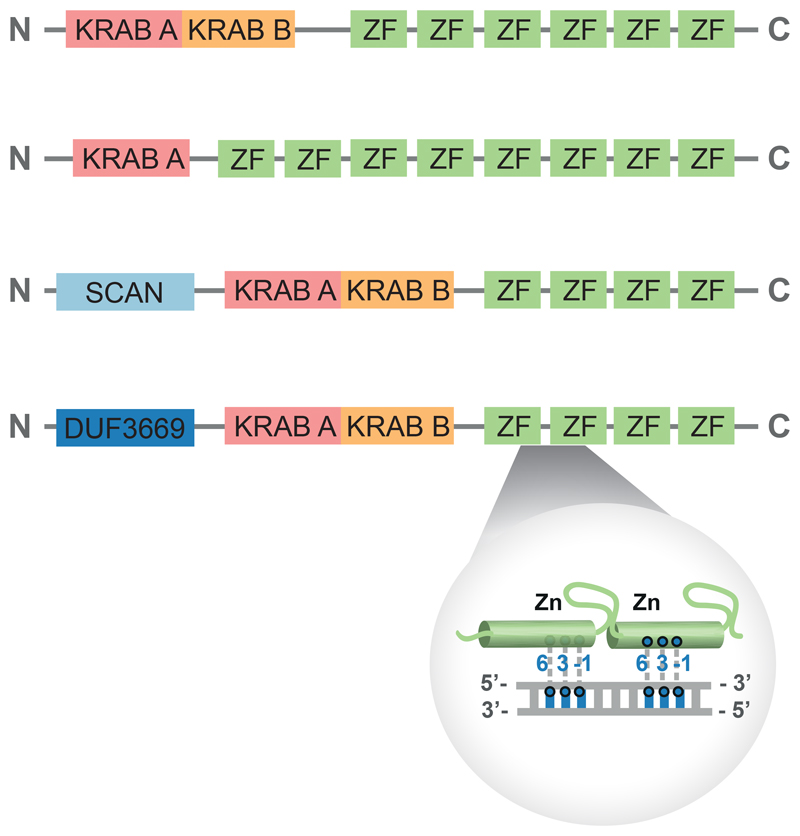
KZFPs are characterized by the presence of a KRAB domain and an array of C2H2 zinc fingers (Figure 3). The KRAB domain encompasses approximately 75 amino acids often split in two modules, the A-box – primarily responsible for repressive activity - and the B-box – thought to potentiate KRAB-A effectiveness (Bellefroid et al., 1991, Mannini et al., 2006, Witzgall et al., 1994). The repressor activity of KZFPs stems from the KRAB-mediated recruitment of KAP1 (KRAB-associated protein 1, also known as TRIM28 –tripartite motif protein 28-, Tif1β or KRIP-1) (Friedman et al., 1996), a scaffold protein that recruits mediators of heterochromatin formation (Iyengar and Farnham, 2011).
Figure 3. Domain structure of KZFPs.
All KZFPs contain a KRAB domain and an array of zinc fingers. Those are responsible for interacting with the DNA, and in each three amino acids (at positions 6, 3, and -1) are engaged in mediating contacts with DNA. KRAB, Krüppel-associated box; ZF, zinc finger; SCAN, SREZBP, CTfin51, AW-1, and Number 18 cDNA; DUF3669, domain of unknown function 3669.
The C-terminal C2H2 ZNF arrays of KZFPs are tandem repeats of the CX2-4CX12HX2–6H motif (where X is any amino acid) interspaced by seven residue-long linkers (Iuchi, 2001). Human KZFPs harbor from 2 to more than forty ZNFs, for an average of 12 (Urrutia, 2003). Each zinc finger can theoretically interact with 3 nucleotides of the primary DNA strand via amino acids at positions -1, 3, and 6 of the C2H2 helix, with some contacts established with the secondary strand via amino acid 2. KZFP genes display signs of strong positive selection at codons encoding for these positions, consistent with interactions of their products with DNA targets themselves capable of rapid evolution, such as transposable elements or viruses (Emerson and Thomas, 2009). While the length of many KZFP ZNF arrays should allow for a very high degree of specificity in the recognition of long DNA targets, KZFP binding motifs are usually shorter than predicted, suggesting that some ZNFs rather engage in other types of interactions, for instance with RNA or proteins (Najafabadi et al., 2015, Imbeault et al., 2017, Schmitges et al., 2016).
Some highly conserved KZFPs contain additional elements in their N-terminus, such as SCAN or DUF3669 domains. The vertebrate-specific SCAN can mediate oligomerization notably with other SCAN-containing proteins (Honer et al., 2001), whereas the function of the DUF3669 domain remains largely unknown, as indicated by its acronym (Domain of Unknown Function).
Genomic targets of human KZFPs
The genomic targets of a large fraction of human KZFPs were cumulatively characterized through three recent studies using chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) and tagged proteins overexpressed in 293T cells as baits (Najafabadi et al., 2015, Imbeault et al., 2017, Schmitges et al., 2016). Our lab succeeded in identifying the genomic targets of 222 human KZFPs (Imbeault et al., 2017). In line with the other two studies, we found that a great majority of human KZFPs associate with at least one subfamily of transposable elements, most of them retrotransposons. Some KZFPs bound sequences in different TE families (e.g. HERVs and LINEs), although usually with variable affinities. Conversely, many TE subfamilies were recognized by several KZFPs, which most often targeted clearly distinct regions of their integrants. Illustrative of this situation, HERVH-int sequences could recruit, from 5’ to 3’, ZNF90, ZNF534, ZFP69B and ZNF257. Interestingly ZNF90 and ZNF534, which bound in very close proximity around the PBS (primer binding site)-coding sequence of HERVH, were often found on the same proviruses, whereas ZFP69B and ZNF257, which recognized sequences located further downstream, were largely mutually exclusive. As well, primate-specific L1PA integrants could be bound, from 5’ to 3’, by various combinations of ZNF141, ZNF649, ZNF765, ZNF93, ZNF382, ZNF17, ZNF425 and ZNF248. Interestingly, the age of these elements influenced their pattern of KZFP recruitment. L1PA4s, which are approximately 20 million year old (myo), were recognized by many of these KZFPs. In contrast, most human-specific L1Hs were devoid of binding sites for factors recruited near the L1 promoter, such as ZNF93 (~20 myo), ZNF649 (~105 myo), ZNF765 (~7 myo) and ZNF141 (~43 myo). However, 3’ binders such as ZNF382, ZNF84 (both ~105 myo) and ZNF429 (~29 myo) bound a significant fraction of all L1PA integrants, from the ~40 myo L1PA16 to the youngest L1Hs (Imbeault et al., 2017).
About a third of the tested human KZFPs did not significantly associate with TEs, and were instead recruited to other types of genomic targets such as promoters, simple repeats and poly-zinc finger protein genes. Many of the promoter-binding KZFPs are evolutionary conserved and contain SCAN or DUF3669 domains; most do not recruit KAP1, and are of yet unknown function. Some associated with wide arrays of promoters, for instance ZNF202, which was found binding in the vicinity of several thousand TSS. Others bound to some promoters in a combinatorial fashion, ranging from the overlapping binding sites of the evolutionary conserved DUF3669-containing ZNF282 and ZNF398 to KZFP couples binding at variable distances from each other (ZNF282 and ZNF777, ZNF282 and ZNF202, ZNF534 and ZNF202, ZKSCAN2 and ZNF263) (Imbeault et al., 2017).
ZNF75D and ZNF274 both associated with the 3’ region of poly-zinc finger protein genes, where they recognized conserved and partly overlapping motifs within the proximal part of ZNF-encoding sequences. This resulted in their spatially rhythmic recruitment over these gene targets, 75 of them for ZNF274, which was previously identified as responsible for tethering KAP1 to the 3’ end of a subset of zinc finger genes (Frietze et al., 2010), and around 300 for ZNF75D, which recognizes a shorter binding motif and was also found at thousands of other genomic locations. Interestingly, ZNF75D has a paralogue in human and many other mammals, ZNF75A, which harbors the same zinc finger signature and is therefore expected to bind the same sequences.
Biological impact of KZFPs
KAP1 binding has been confirmed for a sizeable fraction of human and murine KZFPs (Schmitges et al., 2016) (and our unpublished data). KAP1 acts as as a scaffold for a silencing complex that comprises the histone methyltransferase SETDB1 (also known as ESET) (Schultz et al., 2002), the nucleosome remodeling and deacetylation (NuRD) complex (Schultz et al., 2001), heterochromatin protein 1 (HP-1) (Nielsen et al., 1999, Sripathy et al., 2006) and DNA methyltransferases (Quenneville et al., 2012) (Figure 4). Accordingly, many KZFPs act as transcriptional repressors via the KAP1-nucleated induction of heterochromatin and, in early embryonic cells, DNA methylation (Wolf and Goff, 2009, Quenneville et al., 2012, Rowe et al., 2013a, Jacobs et al., 2014, Najafabadi et al., 2015, Ecco et al., 2016, Schmitges et al., 2016, Imbeault et al., 2017). However, not all KZFPs bind KAP1, and the interactome of more ancient human family members, notably those endowed with SCAN or DUF3669 domains, reveals associations with other types of proteins, including transcriptional activators (Schmitges et al., 2016) (and our unpublished data).
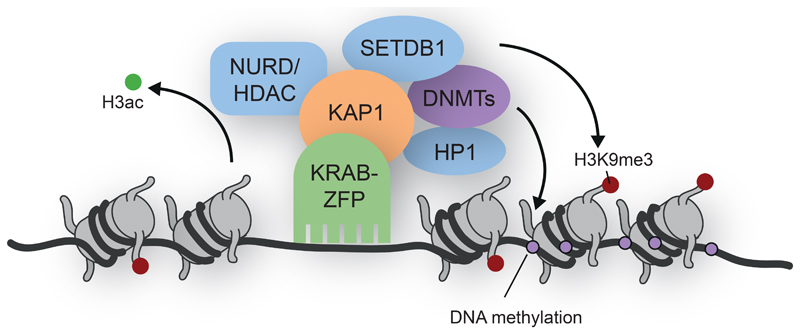
Figure 4. The KRAB/KAP1 repressive complex.
KZFPs bind to DNA via their zinc fingers and recruit KAP1 via their KRAB domain. KAP1 then recruits components of a repressor complex, which leads to heterochromatin formation and transcriptional silencing. H3ac, acetylated histone H3; H3K9me3, histone H3 trimethylated at Lys9; NURD, nucleosome remodeling deacetylase complex; HDAC, histone deacetylase; KAP1, Krüppel-associated box (KRAB)-associated protein 1; KZFP, KRAB-zinc finger protein; SETDB1, SET domain bifurcated 1; HP1, heterochromatin protein 1; DNMT, DNA methyltransferase.
KZFPs have roles in different biological contexts (Box 2), but the best-characterized function of these proteins is the locus-specific induction of heterochromatin during early embryogenesis via the KRAB-mediated recruitment of KAP1. At imprinting control regions, where a methylated hexanucleotide is recognized in mouse and human by ZFP57, this results in the trans-generational preservation of imprinting (Quenneville et al., 2011, Li et al., 2008a, Strogantsev et al., 2015). At sequences derived from transposable elements, this allows for the taming of transcriptional influences that would otherwise hamper early development, from zygotic genome activation to the establishment and normal differentiation of pluripotent stem cells (Rowe et al., 2010, Matsui et al., 2010, Rowe et al., 2013b, Turelli et al., 2014, Macfarlan et al., 2012). KZFPs display exquisitely regulated patterns of expression during the first few days of embryogenesis in both human and mouse, mirroring the tightly orchestrated transcription of TE-containing loci during this period (Corsinotti et al., 2013, Theunissen et al., 2016, Macfarlan et al., 2012, Fort et al., 2014a, Gifford et al., 2013, Goke et al., 2015, Grow et al., 2015, Kunarso et al., 2010, Xue et al., 2013, Yan et al., 2013), and removal of KAP1 or its partner histone methyltransferase SETDB1 in murine or human embryonic stem cells activates the expression of multiple TEs (Matsui et al., 2010, Rowe et al., 2010, Turelli et al., 2014). ZFP809, a murine-specific KZFP, was demonstrated early on to silence exogenous MLV in embryonic carcinoma cells through recognition of the provirus primer binding site-coding sequence (Wolf and Goff, 2007, Wolf and Goff, 2009). Curiously, depletion of ZFP809 in the mouse leads to de-repression of MLV-related ERVs in adult tissues, but not in ES cells (Wolf et al., 2015). While functional data on the role of individual human KZFPs during this period are still missing, it is noteworthy that HERVH integrants, which appear to play an important role in human ES cells pluripotency, are recognized by several KZFPs, the levels of which change during the switch of these cells from naïve to primed state (Theunissen et al., 2016). Other KZFPs controlling TEs in ES cells are ZNF91 and ZNF93, which respectively repress SVAs and LINE-1 (Jacobs et al., 2014), and the murine paralogs ZFP932 and Gm15446, which regulate ERVKs (Ecco et al., 2016). It is now established that, by controlling TEs, the KZFP/KAP1 complex ensures the transcriptional homeostasis and normal differentiation of embryonic stem cells. Upon KAP1 or KZFPs depletion in ES cells, repressive chromatin marks at TEs are replaced by active histone modifications typically found on enhancers, and nearby genes can become activated (Rowe et al., 2013b, Jacobs et al., 2014, Turelli et al., 2014, Ecco et al., 2016).
Box 2. KZFPs influence a variety of biological processes.
While TE-based sequences appear to represent an important fraction of KZFPs targets, these regulators have been implicated in various biological events, although in many cases the underlying mechanism was not completely elucidated.
Metabolism
ZFP69 was reported to mediate liver fat accumulation and mild insulin resistance in mice (Chung et al., 2015). ZNF224, in humans, is associated with glycolysis and oxidative metabolism (Iacobazzi et al., 2009, Lupo et al., 2011).
ZNF74 binds RNA and is tightly associated with the nuclear matrix, suggesting a role for this protein in RNA metabolism (Grondin et al., 1996). ZNF255, an isoform of ZNF224, interacts with a Wilms’ tumour 1 (WT1) protein isoform that has affinity for RNA and has been implicated in transcript processing, suggesting a role for this KZFP in RNA maturation and post-transcriptional control (Florio et al., 2010).
Differentiation and development
Many KZFPs are expressed in embryonic stem cells and early progenitors (Corsinotti et al., 2013), where many are engaged together with their cofactor KAP1 in repressing transposable elements. At this or later stages, they influence other aspects of development. In the mouse, ZFP689, ZFP13 and KAP1 play an important role in erythropoiesis by regulating an miRNA cascade that governs mitophagy in red cell precursors (Barde et al., 2013b). In humans, there is evidence that ZNF589, ZNF268, and ZNF300 influence hematopoietic differentiation (Venturini et al., 2016, Zeng et al., 2012, Xu et al., 2010). Other events influenced by KZFPs include osteogenesis, (Jheon et al., 2001), mammary gland development (Oliver et al., 2012), and formation of extra-embryonic tissues (Shibata and Garcia-Garcia, 2011, Shibata et al., 2011).
Imprinting
The well-studied ZFP57 is implicated in the maintenance of imprinting marks during early embryogenesis through binding to imprinting control regions (ICR) followed by recruitment of KAP1 and other chromatin inducing proteins, which drive subsequent methylation (Zuo et al., 2012, Quenneville et al., 2011). Loss of ZFP57 in mouse embryos and ES cells leads to loss of DNA methylation at multiple imprinted regions (Li et al., 2008b, Zuo et al., 2012).
Sexual dimorphism
In mouse, the KZFPs RSL1 and RSL2 are involved in sexually dimorphic gene expression in liver, repressing male-specific hepatic genes, such as members of the cytochrome P450 (Cyp) families (Krebs et al., 2003). These dimorphic cytochrome P450 genes are also upregulated in KAP1 KO liver (Bojkowska et al., 2012). Interestingly, it has been reported that the control of one of RSL1 targets, the gene encoding for the sex-limited protein (SLP), seems to occur via binding to an ancient endogenous retrovirus (Stavenhagen and Robins, 1988, Krebs et al., 2012).
Until recently, it was generally believed that most TEs are irreversibly silenced during this period, alleviating the need for subsequent sequence-specific control including by the KZFP/KAP1 system (Maksakova et al., 2008, Walsh et al., 1998). However, recent evidence proves otherwise. First, deep transcriptome analyses indicate that some TE loci can be transcriptionally active in adult tissues, providing alternative promoters or fulfilling other regulatory functions (Faulkner et al., 2009, Belancio et al., 2010). Second, a significant fraction of TEs bound by KAP1 in human ES cells still carries the co-repressor in mature T lymphocytes (Turelli et al., 2014). Third, KAP1 deletion in neuronal progenitors activates some EREs (Fasching et al., 2015), and selected ERVs are similarly induced in murine B-lymphocytes or mouse embryonic fibroblasts (MEFs) depleted for SETDB1 (Collins et al., 2015, Wolf et al., 2015). Correspondingly, human KZFPs display extensive and cell-specific patterns of expression in all adult tissues examined (Imbeault et al., 2017). Furthermore, the mouse-specific paralogs ZFP932 and Gm15446 are also involved in controlling their TE targets in somatic tissues, where they modulate the TE-mediated regulation of neighboring genes, including in vivo (Ecco et al., 2016). More broadly, by comparing KZFP binding sites with the ENCODE database, we observed significant overlap between the TE targets of a number of human KZFPs and the binding regions of other transcription factors such as YY1, CEBPZ, GATA3, FOXA1, and STAT1 (Imbeault et al., 2017). Finally, by examining the chromatin state of KZFP-bound TEs in a subset of these tissues, we found that a significant fraction could display a cell-specific enrichment of activation marks instead of those associated with repressive heterochromatin. Moreover, in these cases, nearby genes were on average expressed at higher levels, consistent with KZFP-controlled, TE-based enhancer effects on these genes (Imbeault et al., 2017). Considering the limited scope of this type of analysis, which can detect neither long range effects nor trans acting influences by TE-derived regulatory RNAs, and the fact that chromatin data were available only for a few cell types, it is very likely that the KZFP-mediated control of TEs impacts on the physiology of an extremely broad range of developing and adult tissues. This is consistent with data obtained through the conditional knockout of Kap1 in the mouse, which revealed that the master regulator partakes in processes as diverse as the management of behavioral stress, the differentiation of erythroid precursors, the maturation and activation of B- and T-lymphocytes and the metabolism of hormones and xenobiotics in the liver (Jakobsson et al., 2008, Barde et al., 2013a, Santoni de Sio et al., 2012a, Santoni de Sio et al., 2012b, Bojkowska et al., 2012, Chikuma et al., 2012).
The evolutionary path towards human KZFPs
A survey of more than 200 vertebrate genomes reveals that KZFP genes first appeared in an early Devonian common Sarcopterygian precursor of African coelacanth, lungfish and tetrapods, some 420 million years ago (Imbeault et al., 2017). The genomes of all analyzed modern species derived from this ancestor contain several hundreds of KZFP genes, except for birds in which they are scarce. All 300 or so KZFP genes found in coelacanth seem mono-exonic, whereas in all other species the KRAB and zinc finger domains are most often encoded by separate exons. The switch to multi-exonic genes may have facilitated the reshuffling of zinc finger arrays and the independent evolution of the KRAB domain, paving the way to its coupling in some proteins to SCAN or DUF3669 domains, and to the emergence of non-canonical KRAB units not functionally limited to KAP1 recruitment.
To trace putative DNA binding orthologues, we and others compared the zinc fingerprints of KZFPs, that is, the series of amino acid triplets within their ZNF arrays predicted to dictate their DNA binding specificity (Liu et al., 2014, Imbeault et al., 2017). This delineated clusters specific for most taxonomic orders and the identification of KZFPs restricted to each species (Imbeault et al., 2017). Interestingly, no zinc fingerprint orthologue of coelacanth KZFPs is found in any other species, suggesting that the genomic targets of these proteins are unique to this organism and possibly untested close relatives, consistent with species-restricted transposable elements. Many species- and class-specific KZFPs can similarly be detected in most analyzed genomes, indicating ongoing amplification and turnover of the family with regular addition of new members (Imbeault et al., 2017). A closer examination of the mouse genome identifies about twice more elements than previously annotated as either KZFP genes or pseudogenes, notably by assigning to this family an entity formerly considered as a large group of Satellite repeats (Kauzlaric et al., 2017). It also outlines an organization in clusters distributed throughout the genome, with signs of recombination, translocation, duplication and seeding of new sites by retrotransduction of KZFP genes. Finally, it provides evidence that closely related paralogs have evolved through both drifting and shifting of sequences encoding for zinc finger arrays (Kauzlaric et al., 2017).
Invasion by new families of endogenous retroviruses coincided with the appearance of novel KZFP duplicates in primates (Thomas and Schneider, 2011). The guinea pig, opossum and to a lesser extent mouse genomes display an unusually high number of species-specific paralogues. The mouse genome is known to harbor a significant fraction of retrotransposition-competent TEs, including ERVs and LINEs (Kazazian, 2004, DeBerardinis et al., 1998), supporting a model whereby new TE variants contribute to fix recently emerged KZFP paralogues. Conversely, a few KZFPs highly conserved in other mammals were lost in primates, and some human KZFP pseudogenes have functional orthologues in closely related species, indicating divergence in the selective pressures responsible for their maintenance (Imbeault et al., 2017).
It is remarkable that all examined bird genomes stand out for their very low content in KZFP genes. Interestingly, avian genomes are significantly smaller than those of other amniotes (Organ et al., 2007, Wallis et al., 2004), and a much smaller fraction of the chicken and zebra finch genomes can be readily attributed to TEs, compared with most other tetrapods (15% vs. 40-50% on average) (Chalopin et al., 2015). It further suggests that TE burden and activity contribute to maintaining a pool of functional KZFPs. However, it may also be that birds, when they emerged from theropod dinosaurs, evolved another TE control system with many of the same functional properties as KZFPs, rendering the latter dispensable.
An arms race turned into a massive enterprise of domestication
Several lines of evidence indicate that transposable elements have served as an important motor for the evolution of KZFP genes. During evolution, KZFP genes underwent strong positive selection at positions encoding for amino acids predicted to determine the DNA binding specificity of their products (Emerson and Thomas, 2009, Liu et al., 2014). Furthermore, KZFP paralogs exhibit not only significant differences in zinc fingerprints, but also differential expression and splicing patterns across tissues, consistent with the acquisition of new functions following gene duplication events (Nowick et al., 2010, Kauzlaric et al., 2017). An analysis of data from the 1,000 Genomes Project revealed that human KZFP genes harboring non-synonymous SNPs in sequences encoding their predictive DNA-contacting residues are generally expressed at lower levels, are evolutionarily younger, and seem to be less evolutionarily constrained than those without such polymorphism, suggesting that they are on their way to become pseudogenes (Kapopoulou et al., 2016).
Most importantly, both KZFPs and TEs underwent parallel waves of expansion in the genomes of tetrapods (Thomas and Schneider, 2011). Also, in human embryonic stem cells, a dynamic regulation model of LINE elements by KZFP/KAP1 can be documented, whereby the expression of newly emerged LINE-1 families is initially repressed by small-RNA-induced DNA methylation, before KAP1-mediated repression takes over through the selection of KZFPs sequentially capable of recognizing these TEs, until these are ultimately deprived of any activity by mutations (Castro-Diaz et al., 2014).
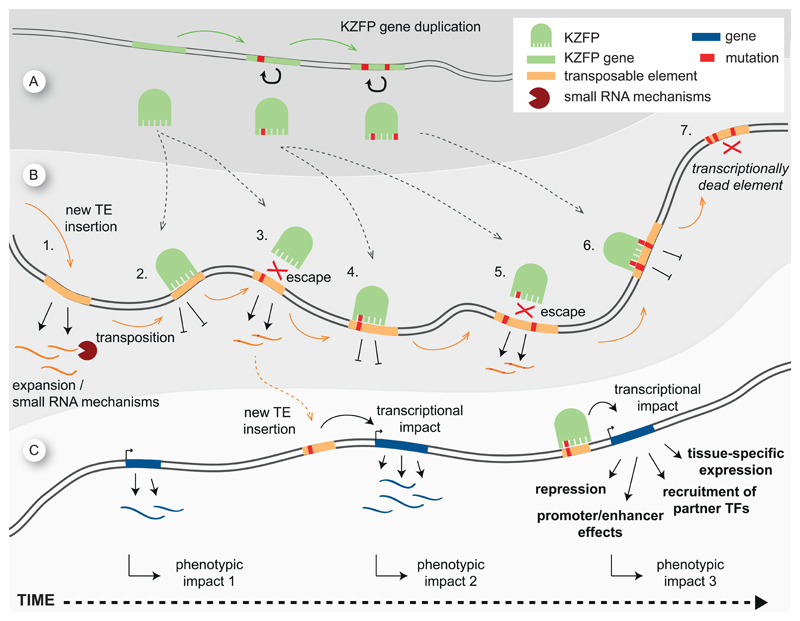
The arms race model (Figure 5 A-B) asserts that a dynamic competition between transposable elements and KZFPs is driving their co-evolution, with TEs controlled by a KZFP mutating away to escape repression while the pool of KZFP genes evolves proteins with novel zinc finger arrays, which get fixed once they can recognize the renegade TE (Imbeault and Trono, 2014). It is best studied within the context of the primate-specific L1PA subfamily of LINE elements, as these TEs devoid of extracellular phase display a linear evolutionary path, each new subfamily deriving from the one previously expanded in the genome of its host species and ancestors. With ERVs, the situation is more complicated, since these TEs are endogenized following waves of genomic invasion originating from external sources, with potential iterations precluding firm dating. Compelling evidence for the arms race model stems from the characterization of ZNF93 and its binding to L1PA elements – in particular the loss via deletion of the ZNF93 recognition site in newer L1PA subfamilies (Jacobs et al., 2014). Additional support comes from the recent identification of the TE targets of a large set of human KZFPs, as it reveals the sequential recruitment at the 5’ ends of primate-specific L1 elements not only of ZNF93 but also ZNF141, ZNF649, and ZNF765, with zinc finger mutations accumulating coincidentally with the appearance of new L1PA subfamilies, and loss of binding sites for all of these KZFPs in the newest human-specific LINE-1. In addition, it retraces specific mutation events in the binding motifs of KZFPs that correlated with loss of binding in the youngest elements, generally subtler than the 129-bp deletion event that led to escape from ZNF93 (Imbeault et al., 2017). In the mouse, the KZFP paralogs ZFP932 and Gm15446 regulate overlapping but distinct sets of ERVKs (Ecco et al., 2016). Both proteins bind to the 3’end of members from the same families of retroelements, but with different preferences. While ZFP932 and Gm15446 are similarly enriched at RLTR44-int, IAP-d-int, and MMERVK10D3_I-int elements, Gm15446 is more frequently found at MMERVK10C-int, IAPEy-int, and IAPEY3-int (Ecco et al., 2016). Further analyses suggest that ZFP932 appeared first and that Gm15446 arose secondarily by imperfect duplication, with subsequent accumulation of mutations leading to a partial shift in target range (Kauzlaric et al., 2017).
Figure 5. The dual evolutionary drive of KZFPs: arms race and TE domestication.
(A) KZFPs gene expansion over time. Duplication events and accumulation of mutations are depicted. (B) An evolutionary arms race between KZFPs and TEs. When a novel TE enters the host, it starts to be expressed and to transpose, albeit partly controlled by a first, small RNA-based, line of defense (1). Over time, KZFPs genes duplicate and paralogs emerge that can bind these TEs (2). In parallel, transposons accumulate mutations and escape repression (3 and 5), yet new KZFPs appear that can suppress the expression of these escapees (4 and 6). Eventually some of these TEs accumulate mutations and are rendered inactive (7) (C) From deleterious mutations to co-option. Left, a hypothetical gene would initially have a certain phenotypic impact (phenotypic impact 1). Middle, transposon insertions near genes can lead to transcriptional effects with phenotypic consequences (causing a phenotypic impact 2). Right, some of these could be beneficial for the host, notably if modulated by KZFP-mediated control, which can incur a variety of transcriptional and phenotypic effects. The TE/KZFP pair then can become fixed in evolution, completing the co-option process. KZFPs, KRAB-zinc finger proteins; TEs, transposable elements; TFs, transcription factors
However, cumulated evidence also demonstrates that a host-invader arms race cannot have been the sole motor of the evolutionary selection of KZFP genes. First, the recognition of many TEs by several KZFPs would constitute, analogous to antiviral combination therapies, a major obstacle to mutational escape if these factors were all simultaneously engaged in their repression. Second, LINE1 integrants controlled by KAP1 in human ES cells are between ~7 and 25 million years of age, and have all long lost all transposition potential (Castro-Diaz et al., 2014), as do all HERVs, including the tens of thousands of integrants still controlled by KAP1; therefore, the conservation of KZFP binding sites in these elements does not arise from the need to suppress their replication. Third, numerous TEs have kept spreading or even started invading the human ancestral genome long after KZFPs capable of recognizing their sequence had emerged. For instance ZNF649, which as ZNF93 binds the L1PA promoter and exhibits a very similar expression pattern, dates back to the time of mammalian radiation, some 60 million years before either ZNF93 or any of its target L1PA subfamilies appeared. Finally, our data suggest that enrichment for certain KZFPs is positively selected on some TEs, as for ZNF382 and ZNF84 on L1Hs (Imbeault et al., 2017).
Collectively, the phylogenetic study of the KZFP gene family, the characterization of the genomic targets of its human products, preliminary analyses of their protein interactome, as well as functional data indicating that KZFPs partner up with TEs to create regulator hubs strongly suggest that these proteins, rather than just engaged in blocking the transposition potential of TEs, participate in their domestication, along a general model (Figure 5 A-C) that can be described as follows: when a new TE enters the genome, whether from an exogenous source (for ERVs) or by mutation of an endogenous predecessor, it is initially silenced via ancestral RNA-based mechanisms, such as mediated by piRNAs.
Over time, its integrants accumulate mutations that progressively hamper their transposition potential. Meanwhile, KZFP paralogs with novel DNA binding specificities are generated, some of which recognize these TEs and get fixed, either because they contribute to preventing the further spread of these elements or because they partake in their co-option for the benefit of the host. Based on the observed evolutionary dynamic of KAP1-control of human LINE1 in human ES cells, it seems that, at least in recent time and for this class of retroelements, matching between a newly appeared TE and an inhibitory KZFP can take more than 7 million years, since KAP1 does not repress any human-specific LINEs (Castro-Diaz et al., 2014). Over time, KZFP/KAP1-controlled TE integrants continue to undergo a mutational drift, so that in some cases only their KZFP-recruiting region remains to serve as a transcription regulatory platform, which maybe explains why we frequently find the oldest human KZFPs at promoters, without identifiable TE signature. KZFPs themselves might evolve to become capable of recruiting activities distinct from KAP1-nucleated repression. Ultimately, all that might be left from the TE/repressor pair is a DNA target motif and its sequence-specific polypeptidic ligand, with no recognizable trace of their source elements.
Concluding remarks: of the species-specificity of human biology
An important implication is that TEs and their KZFP controllers confer a high degree of species-specificity to the conduct of many biological processes relevant to human development and physiology. Indeed, a large fraction of the human endovirome is unique to our species and its close relatives, both in the sequence and genomic distribution of its individual components and, correspondingly, many human KZFPs are relatively recent products of our evolution (Nowick et al., 2010, Liu et al., 2014, Imbeault et al., 2017). Therefore, while animal models may be valuable for delineating general principles and studying canonical aspects of gene regulation, many cis- and trans-regulatory features likely essential to human biology can only be studied in the human system, whether in tissue culture or in vivo.
Considering that species-restricted KZFPs and TEs likely shape regulatory networks in all mammals, the high degree of similarity in the physiology of these organisms might seem surprising. However, while the dynamic partnership between TEs and their KZFPs provides plenty of ground for divergence, for most organ systems functional evolution is limited by prior decisions and environmental constraints. For instance, even though early embryogenesis is regulated by different sets of TEs and KZFPs in mouse and human, major deviations are difficult to introduce in this highly orchestrated process. Similarly, gene expression in murine and human hepatocytes or blood cells is influenced by species-distinct TEs and KZFPs, but the functions to be fulfilled by cells from either system are grossly similar between the two species. One organ that likely escapes at last partly this type of restriction is the central nervous system, since at least in humans a very wide range of cognitive and psychological phenotypes are compatible with normal life expectancy and efficient reproduction. It is thus interesting to note that remarkably elevated levels of TE activity have been recorded in the brain (Erwin et al., 2014), that the hominoid-restricted SVAs are vastly overrepresented in neuron-specific enhancers (our unpublished results), that a higher range of KZFPs is expressed in the brain than in most other adult human tissues (http://fantom.gsc.riken.jp/) and that KZFPs disproportionately contribute to differences between the brain gene networks of chimpanzees and humans (Nowick et al., 2009). It suggests that the endovirome and its KZFP controllers have played an important role in the expansion of higher brain functions that was key to the emergence of modern humans.
Figure 1. Classification of transposable elements.
The different classes of transposable elements and examples of their structures are depicted. LTR, long terminal repeat; Gag, group-specific antigen; Pol, polymerase; Env, envelope protein; UTR, untranslated region; ORF, open reading frame; (A)n, poly(A) tail; A and B, component sequences of the RNA polymerase III promoter; AR, the adenosine-rich segment separating the 7SL monomers; VNTR, variable number target repeats; ITR, inverted terminal repeat.
Acknowledgments
We thank the Swiss National Foundation and the European Research Council for financial support, and all former and current members of the Trono Lab for discussions. We regret being unable to cite all work relevant to this primer due to space constraints.
References
- Antony JM, Deslauriers AM, Bhat RK, Ellestad KK, Power C. Human endogenous retroviruses and multiple sclerosis: innocent bystanders or disease determinants? Biochim Biophys Acta. 2011;1812:162–76. doi: 10.1016/j.bbadis.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayarpadikannan S, Lee H-E, Han K, Kim H-S. Transposable element-driven transcript diversification and its relevance to genetic disorders. Gene. 2015;558:187–194. doi: 10.1016/j.gene.2015.01.039. [DOI] [PubMed] [Google Scholar]
- Barde I, Rauwel B, Marin-Florez RM, Corsinotti A, Laurenti E, Verp S, Offner S, Marquis J, Kapopoulou A, Vanicek J, Trono D. A KRAB/KAP1-MiRNA Cascade Regulates Erythropoiesis Through Stage-Specific Control of Mitophagy. Science. 2013a doi: 10.1126/science.1232398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barde I, Rauwel B, Marin-Florez RM, Corsinotti A, Laurenti E, Verp S, Offner S, Marquis J, Kapopoulou A, Vanicek J, Trono D. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science. 2013b;340:350–3. doi: 10.1126/science.1232398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudino L, Yoshinobu K, Morito N, Santiago-Raber ML, Izui S. Role of endogenous retroviruses in murine SLE. Autoimmun Rev. 2010;10:27–34. doi: 10.1016/j.autrev.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010;38:3909–22. doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid EJ, Poncelet DA, Lecocq PJ, Revelant O, Martial JA. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc Natl Acad Sci U S A. 1991;88:3608–12. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkowska K, Aloisio F, Cassano M, Kapopoulou A, Santoni de Sio F, Zangger N, Offner S, Cartoni C, Thomas C, Quenneville S, Johnsson K, et al. Liver-specific ablation of Kruppel-associated box-associated protein 1 in mice leads to male-predominant hepatosteatosis and development of liver adenoma. Hepatology. 2012;56:1279–90. doi: 10.1002/hep.25767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, Liu ET. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–62. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–57. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 2014;28:1397–409. doi: 10.1101/gad.241661.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalopin D, Naville M, Plard F, Galiana D, Volff JN. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol. 2015;7:567–80. doi: 10.1093/gbe/evv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikuma S, Suita N, Okazaki IM, Shibayama S, Honjo T. TRIM28 prevents autoinflammatory T cell development in vivo. Nat Immunol. 2012;13:596–603. doi: 10.1038/ni.2293. [DOI] [PubMed] [Google Scholar]
- Chung B, Stadion M, Schulz N, Jain D, Scherneck S, Joost HG, Schurmann A. The diabetes gene Zfp69 modulates hepatic insulin sensitivity in mice. Diabetologia. 2015;58:2403–13. doi: 10.1007/s00125-015-3703-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MA, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 2013;45:325–9. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–14. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Collins PL, Kyle KE, Egawa T, Shinkai Y, Oltz EM. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc Natl Acad Sci U S A. 2015;112:8367–72. doi: 10.1073/pnas.1422187112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsinotti A, Kapopoulou A, Gubelmann C, Imbeault M, Santoni De Sio FR, Rowe HM, Mouscaz Y, Deplancke B, Trono D. Global and stage specific patterns of Kruppel-associated-box zinc finger protein gene expression in murine early embryonic cells. PLoS One. 2013;8:e56721. doi: 10.1371/journal.pone.0056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis RJ, Goodier JL, Ostertag EM, Kazazian HH., Jr Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nat Genet. 1998;20:288–90. doi: 10.1038/3104. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–3. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci U S A. 2009;106:12127–32. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Harper F, Guegan J, Dessen P, Pierron G, Heidmann T. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci U S A. 2011;108:E1164–73. doi: 10.1073/pnas.1112304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D. Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Dev Cell. 2016;36:611–23. doi: 10.1016/j.devcel.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci. 2014;15:497–506. doi: 10.1038/nrn3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching L, Kapopoulou A, Sachdeva R, Petri R, Jonsson ME, Manne C, Turelli P, Jern P, Cammas F, Trono D, Jakobsson J. TRIM28 represses transcription of endogenous retroviruses in neural progenitor cells. Cell Rep. 2015;10:20–8. doi: 10.1016/j.celrep.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, Waki K, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–68. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio F, Cesaro E, Montano G, Izzo P, Miles C, Costanzo P. Biochemical and functional interaction between ZNF224 and ZNF255, two members of the Kruppel-like zinc-finger protein family and WT1 protein isoforms. Hum Mol Genet. 2010;19:3544–56. doi: 10.1093/hmg/ddq270. [DOI] [PubMed] [Google Scholar]
- Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, Noro Y, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014a;46:558–66. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, Noro Y, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014b;46:558–66. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- Friedli M, Trono D. The developmental control of transposable elements and the evolution of higher species. Annual Review of Cellular and Developmental Biology. 2015;31:13.1–13.23. doi: 10.1146/annurev-cellbio-100814-125514. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–78. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Frietze S, O'Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3' ends of ZNF genes. PLoS One. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford WD, Pfaff SL, Macfarlan TS. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol. 2013;23:218–26. doi: 10.1016/j.tcb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goke J, Lu X, Chan YS, Ng HH, Ly LH, Sachs F, Szczerbinska I. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16:135–41. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Grondin B, Bazinet M, Aubry M. The KRAB zinc finger gene ZNF74 encodes an RNA-binding protein tightly associated with the nuclear matrix. J Biol Chem. 1996;271:15458–67. doi: 10.1074/jbc.271.26.15458. [DOI] [PubMed] [Google Scholar]
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, Pera RA, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–5. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH., Jr SVA retrotransposons: Evolution and genetic instability. Semin Cancer Biol. 2010;20:234–45. doi: 10.1016/j.semcancer.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honer C, Chen P, Toth MJ, Schumacher C. Identification of SCAN dimerization domains in four gene families. Biochim Biophys Acta. 2001;1517:441–8. doi: 10.1016/s0167-4781(00)00274-8. [DOI] [PubMed] [Google Scholar]
- Iacobazzi V, Infantino V, Convertini P, Vozza A, Agrimi G, Palmieri F. Transcription of the mitochondrial citrate carrier gene: identification of a silencer and its binding protein ZNF224. Biochem Biophys Res Commun. 2009;386:186–91. doi: 10.1016/j.bbrc.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Imbeault M, Helleboid P-Y, Trono D. KRAB zinc finger proteins contribute to the evolution of gene regualtory networks. Nature. 2017 doi: 10.1038/nature21683. [DOI] [PubMed] [Google Scholar]
- Imbeault M, Trono D. As time goes by: KRABs evolve to KAP endogenous retroelements. Dev Cell. 2014;31:257–8. doi: 10.1016/j.devcel.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58:625–35. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem. 2011;286:26267–76. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516:242–5. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson J, Cordero MI, Bisaz R, Groner AC, Busskamp V, Bensadoun JC, Cammas F, Losson R, MansuyI IM, Sandi C, Trono D. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60:818–31. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Jheon AH, Ganss B, Cheifetz S, Sodek J. Characterization of a novel KRAB/C2H2 zinc finger transcription factor involved in bone development. J Biol Chem. 2001;276:18282–9. doi: 10.1074/jbc.M010885200. [DOI] [PubMed] [Google Scholar]
- Kapopoulou A, Mathew L, Wong A, Trono D, Jensen JD. The evolution of gene expression and binding specificity of the largest transcription factor family in primates. Evolution. 2016;70:167–80. doi: 10.1111/evo.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauzlaric A, Ecco G, Cassano M, Duc J, Imbeault M, Trono D. The mouse genome displays highjly dynamicx populations of KRAB-zinc finger protein genes and related genetic units. PLOS ONE. 2017 doi: 10.1371/journal.pone.0173746. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–32. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–6. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Krebs CJ, Larkins LK, Price R, Tullis KM, Miller RD, Robins DM. Regulator of sex-limitation (Rsl) encodes a pair of KRAB zinc-finger genes that control sexually dimorphic liver gene expression. Genes Dev. 2003;17:2664–74. doi: 10.1101/gad.1135703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs CJ, Schultz DC, Robins DM. The KRAB zinc finger protein RSL1 regulates sex- and tissue-specific promoter methylation and dynamic hormone-responsive chromatin configuration. Mol Cell Biol. 2012;32:3732–42. doi: 10.1128/MCB.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–4. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, Kochert K, Bouhlel MA, Richter J, Soler E, Stadhouders R, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med. 2010;16:571–9. doi: 10.1038/nm.2129. 1p following 579. [DOI] [PubMed] [Google Scholar]
- Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T. Paleovirology of 'syncytins', retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci. 2013;368 doi: 10.1098/rstb.2012.0507. 20120507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Developmental cell. 2008a;15:547–57. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008b;15:547–57. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Scaringe WA, Hill KA, Roberts S, Mengos A, Careri D, Pinto MT, Kasper CK, Sommer SS. Frequency of recent retrotransposition events in the human factor IX gene. Hum Mutat. 2001;17:511–9. doi: 10.1002/humu.1134. [DOI] [PubMed] [Google Scholar]
- Liu H, Chang LH, Sun Y, Lu X, Stubbs L. Deep vertebrate roots for mammalian zinc finger transcription factor subfamilies. Genome Biol Evol. 2014;6:510–25. doi: 10.1093/gbe/evu030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo A, Cesaro E, Montano G, Izzo P, Costanzo P. ZNF224: Structure and role of a multifunctional KRAB-ZFP protein. The International Journal of Biochemistry & Cell Biology. 2011;43:470–473. doi: 10.1016/j.biocel.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Lupo A, Cesaro E, Montano G, Zurlo D, Izzo P, Costanzo P. KRAB-Zinc Finger Proteins: A Repressor Family Displaying Multiple Biological Functions. Curr Genomics. 2013;14:268–78. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Stoye JP. Mammalian Endogenous Retroviruses. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0009-2014. MDNA3-0009-2014. [DOI] [PubMed] [Google Scholar]
- Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol Life Sci. 2008;65:3329–47. doi: 10.1007/s00018-008-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, Van De Lagemaat LN, Mager DL. Retroviral Elements and Their Hosts: Insertional Mutagenesis in the Mouse Germ Line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannini R, Rivieccio V, D'Auria S, Tanfani F, Ausili A, Facchiano A, Pedone C, Grimaldi G. Structure/function of KRAB repression domains: structural properties of KRAB modules inferred from hydrodynamic, circular dichroism, and FTIR spectroscopic analyses. Proteins. 2006;62:604–16. doi: 10.1002/prot.20792. [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–31. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Mcclintock B. The Origin and Behavior of Mutable Loci in Maize. Proceedings of the National Academy of Sciences of the United States of America. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand P, Van De Lagemaat LN, Mager DL. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 2002;12:1483–95. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, Lavallie E, Tang XY, Edouard P, Howes S, Keith JC, Jr, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–9. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Miki Y, Katagiri T, Kasumi F, Yoshimoto T, Nakamura Y. Mutation analysis in the BRCA2 gene in primary breast cancers. Nat Genet. 1996;13:245–7. doi: 10.1038/ng0696-245. [DOI] [PubMed] [Google Scholar]
- Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, Greenblatt J, et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol. 2015;33:555–62. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- Nakanishi A, Kobayashi N, Suzuki-hirano A, Nishihara H, Sasaki T, Hirakawa M, Sumiyama K, Shimogori T, Okada N. A SINE-derived element constitutes a unique modular enhancer for mammalian diencephalic Fgf8. PLoS One. 2012;7:e43785. doi: 10.1371/journal.pone.0043785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–95. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notwell JH, Chung T, Heavner W, Bejerano G. A family of transposable elements co-opted into developmental enhancers in the mouse neocortex. Nat Commun. 2015;6 doi: 10.1038/ncomms7644. 6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, Gernat T, Almaas E, Stubbs L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci U S A. 2009;106:22358–63. doi: 10.1073/pnas.0911376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, Hamilton AT, Zhang H, Stubbs L. Rapid sequence and expression divergence suggest selection for novel function in primate-specific KRAB-ZNF genes. Mol Biol Evol. 2010;27:2606–17. doi: 10.1093/molbev/msq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver CH, Khaled WT, Frend H, Nichols J, Watson CJ. The Stat6-regulated KRAB domain zinc finger protein Zfp157 regulates the balance of lineages in mammary glands and compensates for loss of Gata-3. Genes Dev. 2012;26:1086–97. doi: 10.1101/gad.184051.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ CL, Shedlock AM, Meade A, Pagel M, Edwards SV. Origin of avian genome size and structure in non-avian dinosaurs. Nature. 2007;446:180–4. doi: 10.1038/nature05621. [DOI] [PubMed] [Google Scholar]
- Pace JK, 2nd, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–32. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeken J, Zeller P, Gasser SM. Repeat DNA in genome organization and stability. Curr Opin Genet Dev. 2015;31:12–9. doi: 10.1016/j.gde.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, De Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Puget N, Sinilnikova OM, Stoppa-Lyonnet D, Audoynaud C, Pages S, Lynch HT, Goldgar D, Lenoir GM, Mazoyer S. An Alu-mediated 6-kb duplication in the BRCA1 gene: a new founder mutation? Am J Hum Genet. 1999;64:300–2. doi: 10.1086/302211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep. 2012;2:766–73. doi: 10.1016/j.celrep.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, Trono D. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–72. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, Lower J, Stratling WH, Lower R, Schumann GG. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2012;40:1666–83. doi: 10.1093/nar/gkr863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Farivar S, Mager DL. C-GATE - catalogue of genes affected by transposable elements. Mob DNA. 2012a;3:9. doi: 10.1186/1759-8753-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012b;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- Romanish MT, Cohen CJ, Mager DL. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin Cancer Biol. 2010;20:246–53. doi: 10.1016/j.semcancer.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Rosenberg N, Jolicoeur P. Retroviral Pathogenesis. In: Coffin JM, H SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 2013a;140:519–29. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-liesching H, Verp S, Marquis J, Spitz F, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–40. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013b;23:452–61. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson LC, Phillips RS, Swanberg LJ. Amylase gene structures in primates: retroposon insertions and promoter evolution. Mol Biol Evol. 1996;13:767–79. doi: 10.1093/oxfordjournals.molbev.a025637. [DOI] [PubMed] [Google Scholar]
- Santoni de Sio FR, Barde I, Offner S, Kapopoulou A, Corsinotti A, Bojkowska K, Genolet R, Thomas JH, Luescher IF, Pinschewer D, Harris N, et al. KAP1 regulates gene networks controlling T-cell development and responsiveness. FASEB J. 2012a;26:4561–75. doi: 10.1096/fj.12-206177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni de Sio FR, Massacand J, Barde I, Offner S, Corsinotti A, Kapopoulou A, Bojkowska K, Dagklis A, Fernandez M, Ghia P, Thomas JH, et al. KAP1 regulates gene networks controlling mouse B-lymphoid cell differentiation and function. Blood. 2012b;119:4675–85. doi: 10.1182/blood-2011-12-401117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Radovani E, Najafabadi HS, Barazandeh M, Campitelli LF, Yin Y, Jolma A, Zhong G, Guo H, Kanagalingam T, Dai WF, et al. Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res. 2016;26:1742–1752. doi: 10.1101/gr.209643.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–32. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ., 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–43. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Blauvelt KE, Liem KF, Jr, Garcia-Garcia MJ. TRIM28 is required by the mouse KRAB domain protein ZFP568 to control convergent extension and morphogenesis of extra-embryonic tissues. Development. 2011;138:5333–43. doi: 10.1242/dev.072546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Garcia-Garcia MJ. The mouse KRAB zinc-finger protein CHATO is required in embryonic-derived tissues to control yolk sac and placenta morphogenesis. Dev Biol. 2011;349:331–41. doi: 10.1016/j.ydbio.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26:8623–38. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen JB, Robins DM. An ancient provirus has imposed androgen regulation on the adjacent mouse sex-limited protein gene. Cell. 1988;55:247–54. doi: 10.1016/0092-8674(88)90047-5. [DOI] [PubMed] [Google Scholar]
- Strogantsev R, Krueger F, Yamazawa K, Shi H, Gould P, Goldman-Roberts M, Mcewen K, Sun B, Pedersen R, Ferguson-Smith AC. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol. 2015;16:112. doi: 10.1186/s13059-015-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Friedli M, He Y, Planet E, O'neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, Imbeault M, Duc J, et al. Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 2011;21:1800–12. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PJ, Macfarlan TS, Lorincz MC. Long Terminal Repeats: From Parasitic Elements to Building Blocks of the Transcriptional Regulatory Repertoire. Mol Cell. 2016;62:766–76. doi: 10.1016/j.molcel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CN, Rosenberg MP, Snow CM, Samuelson LC, Meisler MH. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 1992;6:1457–65. doi: 10.1101/gad.6.8.1457. [DOI] [PubMed] [Google Scholar]
- Turelli P, Castro-Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, Tieng V, Quenneville S, Trono D. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 2014;24:1260–70. doi: 10.1101/gr.172833.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini L, Stadler M, Manukjan G, Scherr M, Schlegelberger B, Steinemann D, Ganser A. The stem cell zinc finger 1 (SZF1)/ZNF589 protein has a human-specific evolutionary nucleotide DNA change and acts as a regulator of cell viability in the hematopoietic system. Exp Hematol. 2016;44:257–68. doi: 10.1016/j.exphem.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Wallis JW, Aerts J, Groenen MA, Crooijmans RP, Layman D, Graves TA, Scheer DE, Kremitzki C, Fedele MJ, Mudd NK, Cardenas M, et al. A physical map of the chicken genome. Nature. 2004;432:761–4. doi: 10.1038/nature03030. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–7. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A. 2007;104:18613–8. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren IA, Naville M, Chalopin D, Levin P, Berger CS, Galiana D, Volff JN. Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates. Chromosome Res. 2015;23:505–31. doi: 10.1007/s10577-015-9493-5. [DOI] [PubMed] [Google Scholar]
- Witzgall R, O'leary E, Leaf A, Onaldi D, Bonventre JV. The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc Natl Acad Sci U S A. 1994;91:4514–8. doi: 10.1073/pnas.91.10.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–4. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Yang P, Fuchtbauer AC, Fuchtbauer EM, Silva AM, Park C, Wu W, Nielsen AL, Pedersen FS, Macfarlan TS. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015;29:538–54. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JH, Wang T, Wang XG, Wu XP, Zhao ZZ, Zhu CG, Qiu HL, Xue L, Shao HJ, Guo MX, Li WX. PU.1 can regulate the ZNF300 promoter in APL-derived promyelocytes HL-60. Leuk Res. 2010;34:1636–46. doi: 10.1016/j.leukres.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, Liu JY, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–7. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, Huang J, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–9. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wang W, Ma J, Wang X, Guo M, Li W. Knockdown of ZNF268, which is transcriptionally downregulated by GATA-1, promotes proliferation of K562 cells. PLoS One. 2012;7:e29518. doi: 10.1371/journal.pone.0029518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Romanish MT, Mager DL. Distributions of Transposable Elements Reveal Hazardous Zones in Mammalian Introns. PLoS Comput Biol. 2011;7:e1002046. doi: 10.1371/journal.pcbi.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Sheng J, Lau HT, Mcdonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, Li X. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem. 2012;287:2107–18. doi: 10.1074/jbc.M111.322644. [DOI] [PMC free article] [PubMed] [Google Scholar]