Abstract
Impairment of group I metabotropic glutamate receptors (mGluRs) results in altered glutamate signalling, which is associated with several neurological disorders including Huntington’s Disease (HD), an autosomal neurodegenerative disease. In this study, we assessed in vivo pathological changes in mGluR1 availability in the Q175DN mouse model of HD using longitudinal positron emission tomography (PET) imaging with the radioligand [11C]ITDM. Ninety-minute dynamic PET imaging scans were performed in 22 heterozygous (HET) Q175DN mice and 22 wild-type (WT) littermates longitudinally at 6, 12, and 16 months of age. Analyses of regional volume of distribution with an image-derived input function (VT (IDIF)) and voxel-wise parametric VT (IDIF) maps were performed to assess differences between genotypes. Post-mortem evaluation at 16 months was done to support in vivo findings. [11C]ITDM VT (IDIF) quantification revealed higher mGluR1 availability in the brain of HET mice compared to WT littermates (e.g. cerebellum: + 15.0%, + 17.9%, and + 17.6% at 6, 12, and 16 months, respectively; p < 0.001). In addition, an age-related decline in [11C]ITDM binding independent of genotype was observed between 6 and 12 months. Voxel-wise analysis of parametric maps and post-mortem quantifications confirmed the elevated mGluR1 availability in HET mice compared to WT littermates. In conclusion, in vivo measurement of mGluR1 availability using longitudinal [11C]ITDM PET imaging demonstrated higher [11C]ITDM binding in extra-striatal brain regions during the course of disease in the Q175DN mouse model.
Electronic supplementary material
The online version of this article (10.1007/s12035-019-01866-5) contains supplementary material, which is available to authorized users.
Keywords: [11C]ITDM, Glutamate, mGluR1, Brain imaging, Receptor
Introduction
Glutamate is the primary excitatory neurotransmitter in the central nervous system and exerts its action through both ionotropic and metabotropic receptors. Group I metabotropic glutamate receptors (mGluRs) are post-synaptic G protein-coupled receptors and include mGluR type 1 (mGluR1) and type 5 (mGluR5) [1]. These receptors mainly promote intracellular Ca2+ release and protein kinase C activation [2]. Impairment in their functionality can cause altered glutamate signalling and consequently excitotoxicity, a reason why both mGluR1 and mGluR5 have been associated with several neurological disorders, including Huntington’s Disease (HD) [3, 4].
HD is an inherited autosomal dominant neurodegenerative disorder caused by an expanded polyglutamine (CAG) repeat in the exon 1 of the HTT gene [5]. This mutated gene translates to mutant huntingtin (mHTT), which is the causative agent of the disease. As a consequence of mHTT accumulation, neuronal dysfunction and death occurs, leading to progressive motor, psychiatric, and cognitive impairments in individuals with HD [6, 7].
Of note, mGluR1 and mGluR5 display distinct cerebral expression patterns: mGluR5 is highly expressed in striatum, hippocampus, and cortex, whereas mGluR1 has a primarily thalamic and cerebellar distribution, with low levels in the other brain regions [8, 9].
Thus, likely due to the striatal and cortical distribution, previous studies mainly focused on mGluR5 and described altered receptor density in both HD mice [10] and human post-mortem tissue [11], although an understanding of the underlying mechanism is still a topic of debate. Knockout and pharmacological antagonism of mGluR5 have been shown to reduce formation of mHTT aggregates [12, 13], whilst mGluR5 positive allosteric modulation improved synaptic plasticity [14, 15]. Additionally, we recently characterized the longitudinal changes of mGluR5 density occurring during disease progression in the Q175 mouse model of HD [16] by means of positron emission tomography (PET) and found that mGluR5 levels were decreased in HD mice [17].
On the contrary, mGluR1 has received limited attention, and little is known on possible changes in mGluR1 availability during the progression of HD. However, the extra-striatal distribution of mGluR1 is exactly in regions implicated in movement disorders, such as the cerebellum and thalamus, and it is thus of high relevance, among others, to HD [3]. The aim of this study was to investigate whether changes in mGluR1 availability occur during disease progression in the same mouse model of HD we used to quantify mGluR5 [17] in order to provide a first evidence before clinical investigation. We performed longitudinal non-invasive PET imaging of mGluR1 at 6, 12, and 16 months of age using the selective radioligand [11C]ITDM (N-[4-[6-(isopropylamino)-pyrimidin-4-yl]-1,3-thiazol-2-yl]-N-methyl-4-[11C]-methylbenzamide) [18], for which we recently validated the pharmacokinetic methodology to perform accurate quantification of mGluR1 availability in the mouse brain [19]. By using the same Q175 mouse model, these findings will offer new insights into the characterization of both mGluR1 and mGluR5 during HD progression.
Materials and Methods
Animals
Thirty-seven 6 months old male heterozygous (HET) Q175DN mice (C57BL/6J background) and 37 age-matched wild-type (WT) littermates were obtained from the Jackson Laboratories (Bar Harbour, Maine, USA). Since C57BL/6J mice present sporadic congenital portosystemic shunt [20], animals were screened before inclusion in the study to avoid this variable as confounding factor. This mouse model of HD exhibits motor, cognitive, molecular, and electrophysiological abnormalities, including in vivo decrease in several striatal markers and HD hallmarks similarly to patients with HD [16, 17, 21–24]. Only HET mice were included in the study to better resemble the clinical condition as homozygousity is rare in patients with HD.
Twenty-two mice per genotype were allocated to the longitudinal study, while the remaining ones (n = 15/genotype) were used for radiometabolite measurements. Animals were single-housed in individually ventilated cages with temperature- and humidity-controlled environment on a 12 h light/dark cycle with access to food and water ad libitum. Animals were given at least 1 week to habituate to the facility before the start of the procedures.
Radiotracer Synthesis
[11C]ITDM synthesis was performed on an automated synthesis module (Carbosynthon I, Comecer, The Netherlands) optimizing the reported procedure [18] to our system as recently described [19]. Average molar activity was 90.1 ± 27.6 GBq/μmol with radiochemical purity greater than 99%. [3H]ITDM was synthetized by Pharmaron, UK, with a molar radioactivity of 2.66 GBq/μmol and purity above 99.9%.
[11C]ITDM Dynamic PET Imaging
Image Acquisition
Dynamic microPET/computed tomography (CT) imaging was performed on two Siemens Inveon PET/CT scanners (Siemens Preclinical Solution, Knoxville, USA). Following isoflurane anaesthesia (induction 5%, maintenance 1.5–2% supplemented in oxygen) (Forene, Belgium), animals were catheterized in the tail vein for intravenous (i.v.) bolus injection of the tracer and were placed on the scanner bed. The whole body was in the PET scanner’s field of view to allow the extraction of the image-derived input function (IDIF) from the lumen of the left ventricle of the heart as previously described and validated [19, 25]. A monitoring acquisition module (Minerve, France) was used to constantly monitor the respiration of the animal during the entire preparation and scanning period. The core body temperature of the animals was maintained stable at 37 ± 1 °C using a heating pad during the entire preparation phase and a feedback-controlled warm air flow (Minerve, France) during the scanning period.
[11C]ITDM image acquisition was performed as previously validated [19]. At the onset of the 90 min dynamic microPET scan, animals were injected with a bolus of [11C]ITDM over a 12 s interval (1 ml/min) using an automated pump (Pump 11 Elite, Harvard Apparatus, USA). Activity was injected in a trace dose, keeping the cold mass similar across time points, and below a maximum limit (2.0 μg/kg). PET scans were acquired in list-mode format and followed by a 10 min 80 kV/500 μA CT scan for attenuation correction and co-registration.
Animals (n = 44) were imaged longitudinally at 6, 12, and 16 months of age. A total of 2 WT and 4 HET Q175DN mice died during longitudinal imaging (in part due to recovery related to anaesthesia). Details regarding body weight, molar radioactivity, injected radioactivity and mass, and number of animals for each time point are provided in Supplementary Table 1.
Metabolite Analysis
In order to assess possible genotypic difference in peripheral metabolism, a population-based metabolite analysis was performed in a cohort of WT (n = 15) and HET (n = 15) mice at 0, 5, 15, and 30 min post injection (p.i.). The procedure was as previously described [19]. Briefly, mice were injected with the radioligand via the lateral tail vein, and blood was collected via cardiac puncture. Next, samples were centrifuged at 2377 × rcf for 5 min, and both plasma and residual fractions were counted in a gamma counter (Wizard2, PerkinElmer). Following addition of equal amounts of ice-cold acetonitrile to the plasma samples and centrifugation at 2377 × rcf for 5 min, supernatant was separated from the precipitate, and both fractions were counted in the gamma counter to calculate the plasma extraction efficiency (WT: 96.4 ± 2.3%; HET: 97.3 ± 0.8%). Finally, 100 μl of supernatant were loaded onto a preconditioned reverse-phase (RP)-HPLC system (Kinetex, 150 × 4.6 mm, 5 μm HPLC column + Phenomenex security guard precolumn), fractions were collected, and radioactivity was measured in the gamma counter to determine the unchanged fraction. Blood spiked in vitro with 37 kBq of radiotracer indicated no degradation occurred during the workup (unchanged radioligand = 99.3 ± 0.4%).
Image Processing and Data Analysis
Acquired 90 min PET data were histogrammed and reconstructed into 39 frames of increasing length (12 × 10 s, 3 × 20 s, 3 × 30 s, 3 × 60 s, 3 × 150 s, and 15 × 300 s). Images were reconstructed using a list-mode iterative reconstruction with proprietary spatially variant resolution modelling with 8 iterations and 16 subsets of the 3D ordered subset expectation maximization (OSEM 3D) algorithm. Normalization, dead time, and CT-based attenuation corrections were applied. PET image frames were reconstructed on a 128 × 128 × 159 grid with 0.776 × 0.776 × 0.776 mm3 voxels. Processing and analysis of the PET images was performed using PMOD 3.6 software (Pmod Technologies, Zurich, Switzerland).
Spatial normalization of the PET images was achieved by normalization of the images to a [11C]ITDM PET template as previously described [19, 24]. Time activity curves (TACs) were extracted from the images using a volume of interest brain atlas adapted from the Waxholm one [26]. mGluR1 is primarily expressed in cerebellum and thalamus. Nonetheless, it can be measured at lower levels also in other brain regions, so we considered striatum, motor cortex, and hippocampus given their relevance to HD. Finally, pons was included in order to assess the application of reference region-based analysis.
Kinetic modelling was performed by fitting the TACs using a two-tissue compartment model (2TCM) with a blood volume fraction (VB) fixed at 3.6% [27] and using the Logan model [28] with the start of the linear phase (t*) calculated according to the maximum error criterion (10%; t* ranging 12.5–30 min depending on the brain region). The total volume of distribution based on image-derived input function (VT (IDIF)) was calculated as a non-invasive surrogate of the VT given the high linear correlation when compared to the invasive approach (r = 0.977, r2 = 0.954, p < 0.0001) as we recently validated [19].
The IDIF was obtained as recently described [19]. Briefly, using the CT image for anatomical information, a sphere (3.5 mm in diameter) was generated with centre in the lumen of the left ventricle of the heart. Next, by selecting the early PET frame exhibiting maximal activity, a threshold set to 50% of max was applied in order to obtain a stable volume across subjects and standardize the procedure. The volume was then used to extract the activity directly from the PET image. The IDIF measured for each genotype and time point are shown in Supplementary Fig. 1. No radiometabolite correction was applied to the IDIF as no difference in metabolism between genotypes was measured (Supplementary Fig. 2).
Also, since an excellent agreement exists between values obtained with 2TCM and Logan plot (r = 0.993, r2 = 0.986, p < 0.0001) [19], only VT (IDIF) values determined with Logan are reported. Finally, to investigate the consequence of applying a reference region-based analysis on mGluR1 quantification, we estimated the binding potential (BPND) using pons as reference region as it was previously reported [29]. In addition to the distribution volume ratio minus 1 (DVR-1) [30], calculated as the ratio of VT (IDIF) of a target region over the VT (IDIF) in the reference region minus 1, we used the simplified reference tissue model 2 (SRTM2) [31]. Given the agreement between the two approaches (r = 0.992, r2 = 0.984, p < 0.0001), only BPND values based on SRTM2 are reported. The simplified reference tissue model (SRTM) [32] was not included as it failed to fit the data of few subjects. For this reason, the k2’ values for SRTM2 were based on the average value of each group across different regions.
Parametric VT (IDIF) maps were generated in PXMOD (Pmod Technologies, Zurich, Switzerland) through voxel-wise Logan graphical analysis using the IDIF as input function with the t* calculated according to the maximum error criterion (10%). The averages for each genotype were overlaid to a 3D mouse brain template for anatomical reference. Finally, PET images were smoothed using an isotropic Gaussian filter (FWHM = 0.5 mm), and a voxel-wise analysis with statistical parametric mapping (SPM) was performed using SPM12 (Wellcome Department of Imaging Neuroscience, London, UK). For voxel-wise analysis, data obtained from WT and HET Q175DN mice at each time point were compared, considering both contrasts (WT > HET and HET > WT). Statistical T-maps were calculated for a peak voxel threshold of p = 0.05 (uncorrected) and cluster threshold of 100 voxels (0.8 mm3). Only significant clusters with p < 0.05 were considered and reported.
Brain Tissue Collection
One week following the last PET scan, 16-month old animals (WT: n = 20; HET: n = 18) were euthanized by decapitation while under anaesthesia, and brains were snap-frozen in 2-metylbuthane at − 35 °C for 2 min and further preserved at − 80 °C until use. Sagittal sections (20 μm of thickness) were collected serially starting at 0.96 mm lateral according to Paxinos and Franklin [33] in triplicate on Superfrost Plus slides (Thermo Fisher Scientific, USA), using a cryostat (Leica, Germany).
In Vitro [3H]ITDM Autoradiography
Autoradiography was performed as recently validated [19]. Briefly, sections were pre-incubated for 20 min with binding buffer (50 mM Tris-HCl buffer, pH 7.4, containing 2 mM CaCl2 and 1.2 mM MgCl2). Next, 1 h incubation with total binding solution (TB, 0.5 nM of [3H]ITDM) or nonspecific-binding solution (NB, 0.5 nM of [3H]ITDM +1 μM of cold ITDM) at room temperature. Sections were washed in 50 mM Tris-HCl buffer on ice, followed by distilled water, and dried for 2 h at room temperature. Sections were exposed on imaging plates (BAS-TR2025, Fujifilm, Japan) for 90 h and imaged with a phosphor imager (Fuji FLA-700 image reader). The measured grey intensity was transformed into radioactivity based on intensity values obtained using tritium standards (American Radiolabeled Chemicals Inc., USA). Given the known decay-corrected molar activity of [3H]ITDM and tritium standards, the calculated radioactivity was transformed into picomoles.
Regional quantification was performed using ImageJ software (National Institute of Health, USA). Binding was measured in the same regions of interest manually drawn on each section as measured on the PET images. Regional specific binding was measured in triplicate (3 slices) for each region. The average of each animal was used for statistical analysis.
mGluR1 Immunohistochemistry
In vitro mGluR1 levels were also visualized by immunohistochemistry. First, sections were air dried for 5 min and incubated with 4% paraformaldehyde (PFA) for 15 min as tissue postfixation. Following rinsing with phosphate-buffered saline (PBS, pH 7.4), nonspecific binding sites were blocked using 5% normal goat serum (NGS) and 0.3% Triton X-100 in PBS for 1 h. Next, sections were incubated with the monoclonal primary antibody anti-mGluR1 (rabbit; 1:200; #12551, Cell Signalling Technologies) in antibody diluent containing 1% bovine serum albumin (BSA) and 0.3% Triton X-100 in PBS overnight at room temperature. The next day, sections were rinsed with PBS prior to a 1 h incubation with the secondary antibody (goat anti-rabbit conjugated with horseradish peroxidase; 1:1000; Jackson ImmunoResearch) in PBS. Following three washes with PBS, sections were exposed to the colorimetric diaminobenzidine reaction (DAB reagent, Dako) for 10 min and stopped with distilled water for 1 min. Finally, sections were dehydrated and mounted with DPX mounting medium (Sigma).
Images covering the entire slices were acquired at 10 × magnification for quantification with a high-throughput microscope (Nikon, Japan) with NIS elements software. Quantification of the mGluR1 immunoreactivity was performed using ImageJ software (National Institute of Health, USA). Images were converted into 8-bit grayscale, and an intensity threshold was applied to all images (threshold of 170 of 255). Regions-of-interest (ROIs) were manually drawn on each image, and the percentage of positive area after thresholding was assessed. Quantification was done in triplicate (3 slices) for each region, and the average was used for statistical analysis.
Statistical Analysis
The Shapiro-Wilk test confirmed the normal distribution of the data. Longitudinal analysis of the PET data was performed using a linear mixed-model, given its robustness and the possibility to include subjects with missing observations. The linear mixed-model analysis was performed using VT (IDIF) values as the dependent variable for each region separately, with genotype (WT and HET), time (6, 12, and 16 months), and the interaction between genotype and time (genotype*time) as fixed effects, and with subjects included as random effect, followed by multiple comparison correction for differences within and between genotypes performed using the Tukey-Kramer test. Regular two-way ANOVA with post hoc Bonferroni correction for multiple comparisons was applied to investigate differences between genotypes for post-mortem variables and reference region-based quantification. Linear mixed-model analysis was performed in JMP Pro 13 (SAS), while all the other analyses were performed with GraphPad Prism (v 6.0) statistical software. Data are represented as mean ± standard deviation (SD) unless specified otherwise; all tests were two-tailed, except for the voxel-based analyses. Statistical significance was set at p < 0.05.
Results
In Vivo mGluR1 Availability Was Higher in HD Mice
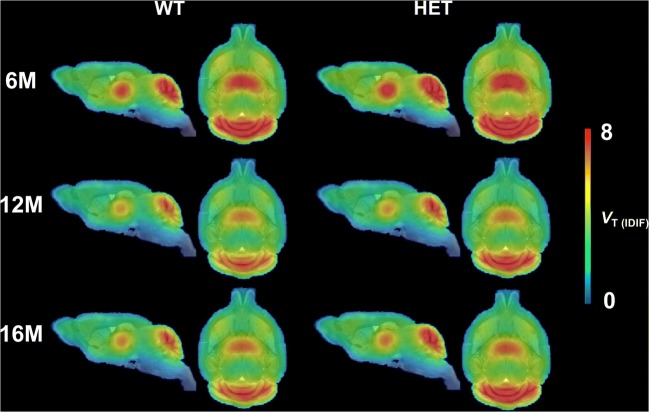
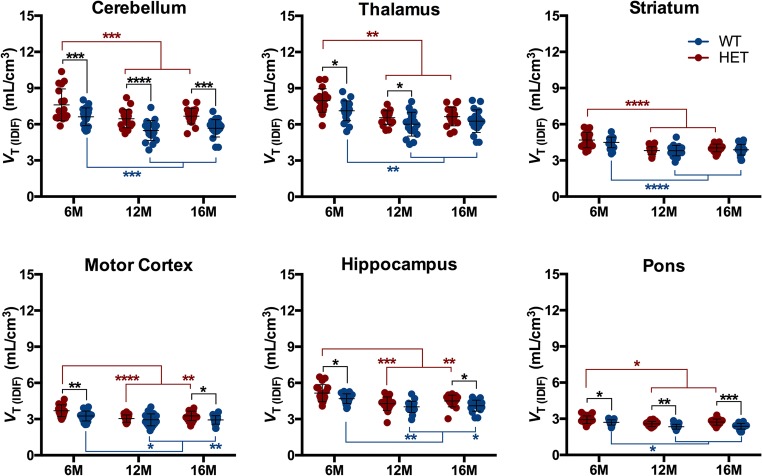
Mean parametric VT (IDIF) Logan maps for WT and HET mice at each investigated time point are shown in Fig. 1. HET mice were characterized by elevated brain [11C]ITDM VT (IDIF) values compared to WT littermates (Fig. 2). At 6 months of age, HET mice displayed higher [11C]ITDM VT (IDIF) values than WT littermates in all investigated brain regions (e.g. cerebellum: WT = 6.61 ± 0.74 mL/cm3; HET = 7.61 ± 1.32 mL/cm3; + 15.1%, p < 0.0001), with the exception of striatum (WT = 4.49 ± 0.43 mL/cm3; HET = 4.69 ± 0.63 mL/cm3; + 4.4%, p > 0.05). At 12 months of age, the [11C]ITDM VT (IDIF) values in HET mice were elevated compared to WT littermates only in cerebellum, thalamus, and pons: e.g., cerebellar VT (IDIF) values were 5.48 ± 0.93 mL/cm3 for WT mice and 6.61 ± 0.74 mL/cm3 for HET littermates (+ 17.9%, p < 0.001). Finally, [11C]ITDM PET imaging at 16 months of age revealed that VT (IDIF) values for HET mice were higher in cerebellum, motor cortex, hippocampus, and pons compared to WT littermates (e.g. cerebellum: VT (IDIF) values of 5.66 ± 0.72 mL/cm3 for WT mice and 6.66 ± 0.65 mL/cm3 for HET littermates (+ 17.6%, p < 0.0001) (Fig. 2).
Fig. 1.
Average VT (IDIF) (Logan) parametric maps of [11C]ITDM in WT and HET mice at each time point. Parametric maps are overlaid onto MRI mouse brain template for anatomical localization. WT: n = 19–21; HET: n = 18–19. WT = wild-type, HET = heterozygous, M = months
Fig. 2.
Regional [11C]ITDMVT (IDIF)quantification. HET mice demonstrated higher [11C]ITDM VT (IDIF) values compared to WT littermates. Both genotypes displayed an age-related temporal decline. Black stars indicate significance between genotypes, red stars within HET, and blue stars within WT. WT: n = 19–21; HET: n = 18–19. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. WT = wild-type, HET = heterozygous, M = months
A significant temporal decline in [11C]ITDM VT (IDIF) values was observed at 12 months of age in all investigated brain regions independently of the genotype. Accordingly, cerebellar VT (IDIF) values between 6 and 12 months of age were reduced in both WT (− 17.2%, p < 0.001) and HET (− 15.1%, p < 0.001) mice. No further change was observed between 12 and 16 months of age (Fig. 2).
Voxel-wise analysis of the parametric maps confirmed the extensive higher [11C]ITDM binding in HET mice. For instance, in line with the regional analysis, motor cortex displayed pronounced clusters of greater [11C]ITDM binding in HET mice at 6 months of age, which were not present at 12 months, but could be detected again at 16 months of age (Fig. 3). Intriguingly, voxel-wise analysis detected clusters of lower [11C]ITDM binding in striatum of HET mice at 12 and 16 months of age (Fig. 3), which could not be detected with the regional analysis.
Fig. 3.
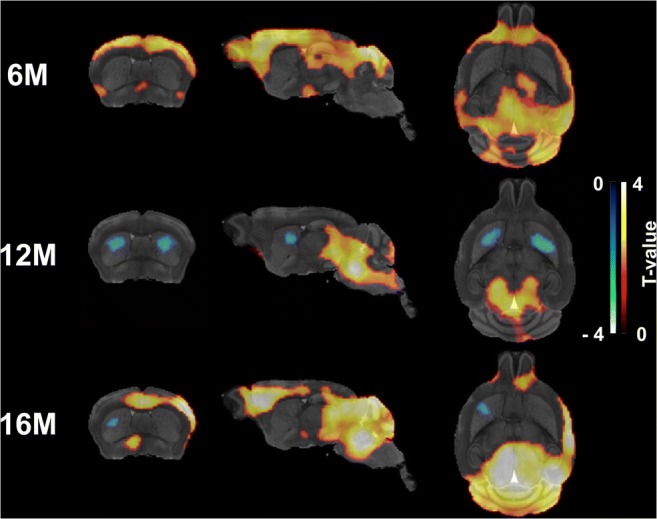
Voxel-wise differences in [11C]ITDM binding between WT and HET mice. Clusters of increased [11C]ITDM uptake in HET mice compared to WT littermates (HET > WT) are shown as hot scale, while clusters of reduced uptake in HET mice (WT > HET) as cold scale. WT: n = 19–21; HET: n = 18–19. M = months
In Vitro mGluR1 Levels Were Greater in HD Mice
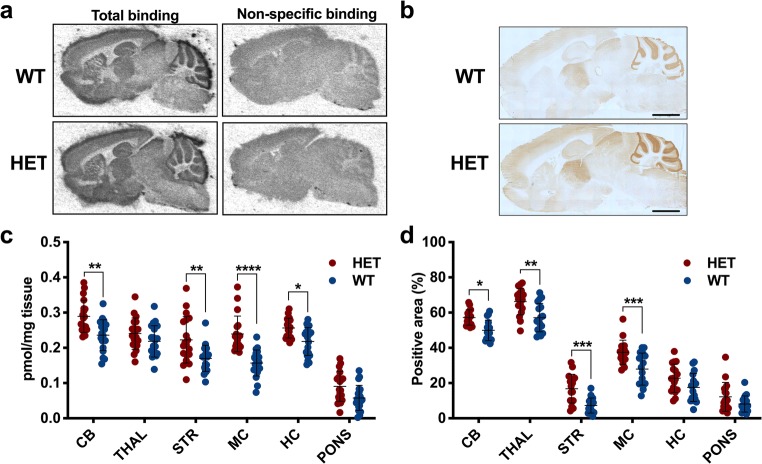
Post-mortem quantification of mGluR1 was achieved using both [3H]ITDM autoradiography and mGluR1 immunohistochemistry in the same animals (WT, n = 19; HET, n = 18). Representative [3H]ITDM autoradiograms and mGluR1 immunoreactivity for WT and HET mice are shown in Fig. 4a and b, respectively. [3H]ITDM-specific binding was greater in cerebellum, striatum, motor cortex, and hippocampus of HET mice compared to WT littermates (Fig. 4c) (e.g. in cerebellum: 0.24 ± 0.04 pmol/mg for WT; 0.29 ± 0.04 pmol/mg for HET; + 21.7%, p = 0.010).
Fig. 4.
Post-mortemevaluation of mGluR1 at 16 months of age. Representative a [3H]ITDM autoradiograms and b mGluR1 immunohistochemistry for WT and HET mice. c [3H]ITDM specific binding as well as d mGluR1 immunoreactivity were higher in HET mice compared to WT littermates. WT: n = 17–19; HET: n = 17–18. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Scale bar = 2 mm. WT = wild-type, HET = heterozygous, CB = cerebellum, THAL = thalamus, STR = striatum, MC = motor cortex, HC = hippocampus
mGluR1 immunoreactivity, measured as percentage of positive area after thresholding, was significantly higher in cerebellum, thalamus, striatum, and motor cortex of HET mice compared to WT littermates (Fig. 4d) (e.g. in cerebellum: 49.9 ± 5.8% for WT; 57.2 ± 3.3% for HET; + 14.6%, p = 0.021).
Reference Region-Based Analysis Biased the In Vivo mGluR1 Quantification
In order to determine the effect of reference region-based quantification, we applied this approach to the time point in which we measured the largest genotypic difference (16 months of age). As shown in Fig. 5, relative quantification using pons as reference region resulted in contradictory results with HET mice displaying significantly lower values in thalamus (− 13.2%, p < 0.001) and striatum (− 24.9%, p < 0.01) compared to WT littermates.
Fig. 5.
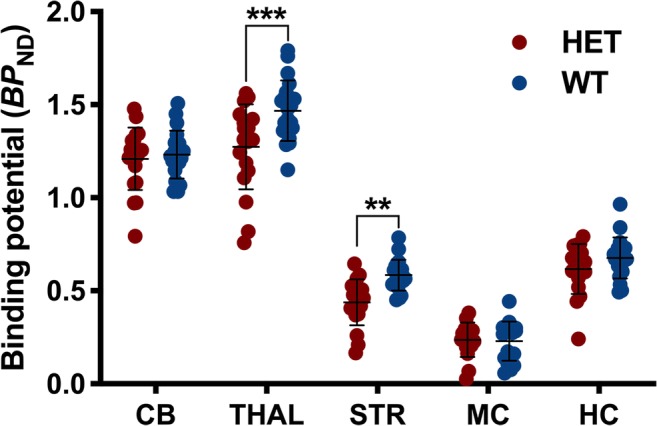
Effect of reference region-based analysis on the in vivo [11C]ITDM quantification. Relative quantification using pons as reference region resulted in contradictory results with higher binding potential in WT mice compared to HET littermates. Data at 16 months of age are reported. **p < 0.01, ***p < 0.001. WT = wild-type, HET = heterozygous, CB = cerebellum, THAL = thalamus, STR = striatum, MC = motor cortex, HC = hippocampus
Discussion
To our knowledge, this is the first study to characterize temporal mGluR1 availability in HD. We used PET imaging with the radioligand [11C]ITDM to investigate changes in mGluR1 availability in HD mice during disease progression. Longitudinal PET imaging demonstrated an overall higher cerebral [11C]ITDM binding in HET mice compared to WT littermates at different stages of disease progression with the exception of striatum. The most pronounced difference was found in cerebellum, with higher mGluR1 levels ranging from 15.1% to 17.9% during HD progression. Of note, only a genotype-independent temporal decline in [11C]ITDM VT (IDIF) was observed, indicating no disease-related effect. The observed decline is in agreement with previous investigation of the mGluR1 protein levels in the mouse cerebellum, in which a significant decline associated with age was demonstrated [34].
Only one report has previously examined in vivo mGluR1 availability in a mouse model of HD [29]. In contrast to our study, the authors reported a significant decrease in [11C]ITDM binding in the R6/2 model of HD; however, these results were obtained using a reference region-based (pons) kinetic modelling for quantification of tracer binding. Our group recently found that the pons is not a suitable reference region for [11C]ITDM as its binding in this region can be blocked and displaced both in vivo and in vitro [19]. Thus, application of reference tissue models can lead to incorrect results.
Post-mortem analysis showed a significant increase in [3H]ITDM-specific binding as well as greater mGluR1 immunoreactivity. This consistently supports the increase in mGluR1 levels of our in vivo [11C]ITDM PET imaging-based study. No post-mortem clinical investigation of mGluR1 has been performed yet, and only sparse evidence exists on the role of mGluR1 in HD at the preclinical level. Previous autoradiography studies investigating changes in the level of group I mGluRs were inconclusive, reporting only small trends, in both the R6/2 [35] and YAC128 [36] transgenic models of HD. Interestingly, we previously showed mGluR5 protein levels to be decreased in Q175 HD mice [17], while here, we demonstrated an increase in mGluR1 levels. Because of the opposing direction of mGluR1 and mGluR5 expression levels, previous studies on Group I mGluRs may have failed to detect differences due to the lack of subtype specificity.
One potential limitation of the current study was the lack of post-mortem validation at 6 and 12 months of age using satellite cohorts of animals. While such analysis could have provided a further indication of the temporal decline in mGluR1 levels measured in vivo, it was not performed as the aim of the post-mortem work was to confirm in vitro the changes measured in vivo in the same animals.
Interestingly, the voxel-wise PET analysis detected a selective decrease of in vivo striatal mGluR1 availability in HET mice primarily at 12 months with a small cluster still present at 16 months. In humans, mHTT accumulation originates in the striatum and progresses to other brain regions [37]. Since striatal medium spiny neurons are particularly vulnerable to mHTT-induced increased levels of intracellular Ca2+, the process of receptor desensitization and internalization [38] may be enhanced in order to compensate for the several glutamatergic inputs from the basal ganglia, thalamus, and cortex [39]. This would represent a protective process as previously postulated [10] and could explain the focal cluster of decreased in vivo mGluR1 availability observed in striatum with the voxel-based analysis. Conversely, post-mortem analyses showed increased striatal mGluR1 levels. This in vivo and in vitro restricted discrepancy may be a further indication that striatal mGluR1 is undergoing internalization in order to reduce the intracellular toxic levels of Ca2+.
A complex interplay exists between HTT and mGluR1/5. HTT and HTT-associated protein 1 have been shown to regulate the levels of intracellular Ca2+ by interacting with IP3R1 [40]. On one hand, mHTT enhances this signalling pathway, resulting in toxic levels of intracellular Ca2+ and intensified activation of protein kinase C (PKC), which desensitize the mGluR1/5 receptors [40, 41]. On the other hand, PKC also increases other downstream pathways including extracellular regulated kinase (ERK) and protein kinase B (also known as Akt) [42], which may promote cell survival and facilitate clearance of mHTT species. In addition, mHTT impairs the functionality of the Rab8/optineurin complex [43]. Since Rab8 modulates the trafficking and signalling of mGluR1 [44], the observed mGluR1 increase may represent a combination of an adaptive response to promote cell survival and impairment in regulation of mGluR1 surface localization. In this perspective, mGluR1 has the potential to represent a therapeutic target analogously to mGluR5 as recently reported [12, 45, 46]. Nonetheless, due to the intricate mGluR1/5 downstream signalling pathways, it is challenging to interpret the pathological effect of these changes. Further research is needed to gain a better understanding of the potential therapeutic significance of the measured mGluR1 changes during the course of disease in this HD model and to validate these findings clinically.
Conclusion
We report quantification of in vivo mGluR1 availability using [11C]ITDM PET imaging during disease course in the Q175DN mouse model of HD. These findings demonstrated higher [11C]ITDM binding in extra-striatal brain regions during disease progression in HD mice, further corroborating HD as a complex whole brain disorder.
Electronic supplementary material
(DOCX 154 kb)
Acknowledgements
The authors thank Philippe Joye, Caroline Berghmans, Eleni Van der Hallen, and Annemie Van Eetveldt of the Molecular Imaging Center Antwerp (MICA) for their valuable technical support.
Authors’ Contribution
DB, JV, LW, SStr, LM, CD, MS, LL, IMS, and SSta designed the study. ŠK, KC, and LW synthesized the radioligand. DB, JV, and AM acquired and processed the data. DB and JV analysed the data. DB, JV, LW, LM, CD, MS, LL, IMS, and SSta contributed to the interpretation of the data. DB, JV, and SStr wrote the manuscript. All authors assisted in reviewing the manuscript and approved its final form.
Funding Information
DB has a PhD fellowship from the Research Foundation Flanders (FWO, 11W2516N/11W2518N). Antwerp University, Belgium founded the work through a partial assistant professor position for JV and LW, and a full professor position for SStr and SSta. LW and SStr are also supported by Antwerp University Hospital, Belgium through a departmental position.
Compliance with Ethical Standards
Conflict of Interest
The Antwerp University group received financial support from CHDI Foundation, Inc., a non-profit biomedical research organization exclusively dedicated to collaboratively developing therapeutics that will substantially improve the lives of HD-affected individuals. LM, CD, LL, MS, IMS are employees of CHDI foundation, Inc. The authors declare no other potential conflicts of interest relevant to this work.
Ethical Approval
All applicable institutional and/or national guidelines for the care and use of animals were followed. Experiments were performed according to the European Committee Guidelines (decree 2010/63/CEE) and the Animal Welfare Act (7 USC 2131), and they were approved by the Ethical Committee for Animal Testing (ECD 2017–27) at the University of Antwerp (Belgium).
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferraguti F, Crepaldi L, Nicoletti F. Metabotropic glutamate 1 receptor: Current concepts and perspectives. Pharmacol Rev. 2008;60(4):536–581. doi: 10.1124/pr.108.000166. [DOI] [PubMed] [Google Scholar]
- 2.Francesconi A, Duvoisin RM. Role of the second and third intracellular loops of metabotropic glutamate receptors in mediating dual signal transduction activation. J Biol Chem. 1998;273(10):5615–5624. doi: 10.1074/jbc.273.10.5615. [DOI] [PubMed] [Google Scholar]
- 3.Ribeiro FM, Paquet M, Cregan SP, Ferguson SS. Group I metabotropic glutamate receptor signalling and its implication in neurological disease. CNS Neurol Disord Drug Targets. 2010;9(5):574–595. doi: 10.2174/187152710793361612. [DOI] [PubMed] [Google Scholar]
- 4.Ribeiro FM, Pires RG, Ferguson SS. Huntington's disease and group I metabotropic glutamate receptors. Mol Neurobiol. 2011;43(1):1–11. doi: 10.1007/s12035-010-8153-1. [DOI] [PubMed] [Google Scholar]
- 5.Group THsDCR A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's disease collaborative research group. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 6.Ross CA, Tabrizi SJ. Huntington's disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10(1):83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 7.Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, Warner JH, Scahill RI, Leavitt BR, Stout JC, Paulsen JS, Reilmann R, Unschuld PG, Wexler A, Margolis RL, Tabrizi SJ. Huntington disease: Natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol. 2014;10(4):204–216. doi: 10.1038/nrneurol.2014.24. [DOI] [PubMed] [Google Scholar]
- 8.Abe T, Sugihara H, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. Molecular characterization of a novel metabotropic glutamate receptor mGluR5 coupled to inositol phosphate/Ca2+ signal transduction. J Biol Chem. 1992;267(19):13361–13368. [PubMed] [Google Scholar]
- 9.Fotuhi M, Sharp AH, Glatt CE, Hwang PM, von Krosigk M, Snyder SH, Dawson TM. Differential localization of phosphoinositide-linked metabotropic glutamate receptor (mGluR1) and the inositol 1,4,5-trisphosphate receptor in rat brain. J Neurosci. 1993;13(5):2001–2012. doi: 10.1523/JNEUROSCI.13-05-02001.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribeiro FM, Paquet M, Ferreira LT, Cregan T, Swan P, Cregan SP, Ferguson SS. Metabotropic glutamate receptor-mediated cell signaling pathways are altered in a mouse model of Huntington's disease. J Neurosci. 2010;30(1):316–324. doi: 10.1523/JNEUROSCI.4974-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulyas B, Sovago J, Gomez-Mancilla B, Jia Z, Szigeti C, Gulya K, Schumacher M, Maguire RP, Gasparini F, Halldin C. Decrease of mGluR5 receptor density goes parallel with changes in enkephalin and substance P immunoreactivity in Huntington's disease: A preliminary investigation in the postmortem human brain. Brain Struct Funct. 2015;220(5):3043–3051. doi: 10.1007/s00429-014-0812-y. [DOI] [PubMed] [Google Scholar]
- 12.Abd-Elrahman KS, Hamilton A, Hutchinson SR, Liu F, Russell RC, Ferguson SSG (2017) mGluR5 antagonism increases autophagy and prevents disease progression in the zQ175 mouse model of Huntington's disease. Sci Signal 10(510). 10.1126/scisignal.aan6387 [DOI] [PubMed]
- 13.Ribeiro FM, Devries RA, Hamilton A, Guimaraes IM, Cregan SP, Pires RG, Ferguson SS. Metabotropic glutamate receptor 5 knockout promotes motor and biochemical alterations in a mouse model of Huntington's disease. Hum Mol Genet. 2014;23(8):2030–2042. doi: 10.1093/hmg/ddt598. [DOI] [PubMed] [Google Scholar]
- 14.Doria JG, de Souza JM, Andrade JN, Rodrigues HA, Guimaraes IM, Carvalho TG, Guatimosim C, Dobransky T, Ribeiro FM. The mGluR5 positive allosteric modulator, CDPPB, ameliorates pathology and phenotypic signs of a mouse model of Huntington's disease. Neurobiol Dis. 2015;73:163–173. doi: 10.1016/j.nbd.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 15.Doria JG, de Souza JM, Silva FR, Olmo IG, Carvalho TG, Alves-Silva J, Ferreira-Vieira TH, Santos JT, Xavier CQS, Silva NC, Maciel EMA, Conn PJ, Ribeiro FM. The mGluR5 positive allosteric modulator VU0409551 improves synaptic plasticity and memory of a mouse model of Huntington's disease. J Neurochem. 2018;147(2):222–239. doi: 10.1111/jnc.14555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menalled LB, Kudwa AE, Miller S, Fitzpatrick J, Watson-Johnson J, Keating N, Ruiz M, Mushlin R, Alosio W, McConnell K, Connor D, Murphy C, Oakeshott S, Kwan M, Beltran J, Ghavami A, Brunner D, Park LC, Ramboz S, Howland D. Comprehensive behavioral and molecular characterization of a new knock-in mouse model of Huntington's disease: zQ175. PLoS One. 2012;7(12):e49838. doi: 10.1371/journal.pone.0049838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertoglio D, Kosten L, Verhaeghe J, Thomae D, Wyffels L, Stroobants S, Wityak J, Dominguez C et al (2018) Longitudinal characterization of mGluR5 using (11)C-ABP688 PET imaging in the Q175 mouse model of Huntington's disease. J Nucl Med. 10.2967/jnumed.118.210658 [DOI] [PubMed]
- 18.Fujinaga M, Yamasaki T, Maeda J, Yui J, Xie L, Nagai Y, Nengaki N, Hatori A, Kumata K, Kawamura K, Zhang MR. Development of N-[4-[6-(isopropylamino)pyrimidin-4-yl]-1,3-thiazol-2-yl]-N-methyl-4-[11C]methylb enzamide for positron emission tomography imaging of metabotropic glutamate 1 receptor in monkey brain. J Med Chem. 2012;55(24):11042–11051. doi: 10.1021/jm301597s. [DOI] [PubMed] [Google Scholar]
- 19.Bertoglio D, Verhaeghe J, Korat S, Miranda A, Wyffels L, Stroobants S, Mrzljak L, Dominguez C et al (2019) in vitro and in vivo assessment of suitable reference region and kinetic modelling for the mGluR1 radioligand [11C]ITDM in mice. Mol Imaging Biol. 10.1007/s11307-019-01435-1 [DOI] [PMC free article] [PubMed]
- 20.Cudalbu C, McLin VA, Lei H, Duarte JM, Rougemont AL, Oldani G, Terraz S, Toso C, Gruetter R. The C57BL/6J mouse exhibits sporadic congenital portosystemic shunts. PLoS One. 2013;8(7):e69782. doi: 10.1371/journal.pone.0069782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng Q, Wu B, Jiang M, Jin J, Hou Z, Zheng J, Zhang J, Duan W. Characterization of behavioral, Neuropathological, brain metabolic and key molecular changes in zQ175 knock-in mouse model of Huntington's disease. PLoS One. 2016;11(2):e0148839. doi: 10.1371/journal.pone.0148839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heikkinen T, Lehtimaki K, Vartiainen N, Puolivali J, Hendricks SJ, Glaser JR, Bradaia A, Wadel K, Touller C, Kontkanen O, Yrjanheikki JM, Buisson B, Howland D, Beaumont V, Munoz-Sanjuan I, Park LC. Characterization of neurophysiological and behavioral changes, MRI brain volumetry and 1H MRS in zQ175 knock-in mouse model of Huntington's disease. PLoS One. 2012;7(12):e50717. doi: 10.1371/journal.pone.0050717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haggkvist J, Toth M, Tari L, Varnas K, Svedberg M, Forsberg A, Nag S, Dominguez C et al (2016) Longitudinal microPET imaging of the zQ175 mouse model of Huntington's disease shows in vivo changes of molecular targets in the striatum and the cerebral cortex. J Nucl Med. 10.2967/jnumed.116.180497 [DOI] [PubMed]
- 24.Bertoglio D, Verhaeghe J, Kosten L, Thomae D, Van der Linden A, Stroobants S, Wityak J, Dominguez C, Mrzljak L, Staelens S. MR-based spatial normalization improves [18F]MNI-659 PET regional quantification and detectability of disease effect in the Q175 mouse model of Huntington's disease. PLoS One. 2018;13(10):e0206613. doi: 10.1371/journal.pone.0206613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verhaeghe J, Bertoglio D, Kosten L, Thomae D, Verhoye M, Van Der Linden A, Wyffels L, Stroobants S, Wityak J, Dominguez C, Mrzljak L, Staelens S. Noninvasive relative quantification of [(11)C]ABP688 PET imaging in mice versus an input function measured over an Arteriovenous shunt. Front Neurol. 2018;9:516. doi: 10.3389/fneur.2018.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson GA, Badea A, Brandenburg J, Cofer G, Fubara B, Liu S, Nissanov J. Waxholm space: an image-based reference for coordinating mouse brain research. Neuroimage. 2010;53(2):365–372. doi: 10.1016/j.neuroimage.2010.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Julien-Dolbec C, Tropres I, Montigon O, Reutenauer H, Ziegler A, Decorps M, Payen JF. Regional response of cerebral blood volume to graded hypoxic hypoxia in rat brain. Br J Anaesth. 2002;89(2):287–293. doi: 10.1093/bja/aef182. [DOI] [PubMed] [Google Scholar]
- 28.Logan J, Fowler JS, Volkow ND, Wolf AP, Dewey SL, Schlyer DJ, MacGregor RR, Hitzemann R, Bendriem B, Gatley SJ, et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(−)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab. 1990;10(5):740–747. doi: 10.1038/jcbfm.1990.127. [DOI] [PubMed] [Google Scholar]
- 29.Yamasaki T, Fujinaga M, Yui J, Ikoma Y, Hatori A, Xie L, Wakizaka H, Kumata K, Nengaki N, Kawamura K, Zhang MR. Noninvasive quantification of metabotropic glutamate receptor type 1 with [(1)(1)C]ITDM: A small-animal PET study. J Cereb Blood Flow Metab. 2014;34(4):606–612. doi: 10.1038/jcbfm.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, Holden J, Houle S, Huang SC, Ichise M, Iida H, Ito H, Kimura Y, Koeppe RA, Knudsen GM, Knuuti J, Lammertsma AA, Laruelle M, Logan J, Maguire RP, Mintun MA, Morris ED, Parsey R, Price JC, Slifstein M, Sossi V, Suhara T, Votaw JR, Wong DF, Carson RE. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27(9):1533–1539. doi: 10.1038/sj.jcbfm.9600493. [DOI] [PubMed] [Google Scholar]
- 31.Wu YJ, Carson RE. Noise reduction in the simplified reference tissue model for neuroreceptor functional imaging. J Cereb Blood Flow Metab. 2002;22(12):1440–1452. doi: 10.1097/01.WCB.0000033967.83623.34. [DOI] [PubMed] [Google Scholar]
- 32.Lammertsma AA, Hume SP. Simplified reference tissue model for PET receptor studies. Neuroimage. 1996;4(3 Pt 1):153–158. doi: 10.1006/nimg.1996.0066. [DOI] [PubMed] [Google Scholar]
- 33.Paxinos G, Franklin K (2003) The mouse brain in stereotaxic coordinates. Academic press; 2 edition (December 8, 2003),
- 34.Simonyi A, Xia J, Igbavboa U, Wood WG, Sun GY. Age differences in the expression of metabotropic glutamate receptor 1 and inositol 1,4,5-trisphosphate receptor in mouse cerebellum. Neurosci Lett. 1998;244(1):29–32. doi: 10.1016/s0304-3940(98)00127-x. [DOI] [PubMed] [Google Scholar]
- 35.Cha JH, Kosinski CM, Kerner JA, Alsdorf SA, Mangiarini L, Davies SW, Penney JB, Bates GP, Young AB. Altered brain neurotransmitter receptors in transgenic mice expressing a portion of an abnormal human Huntington disease gene. Proc Natl Acad Sci U S A. 1998;95(11):6480–6485. doi: 10.1073/pnas.95.11.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benn CL, Slow EJ, Farrell LA, Graham R, Deng Y, Hayden MR, Cha JH. Glutamate receptor abnormalities in the YAC128 transgenic mouse model of Huntington's disease. Neuroscience. 2007;147(2):354–372. doi: 10.1016/j.neuroscience.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carty N, Berson N, Tillack K, Thiede C, Scholz D, Kottig K, Sedaghat Y, Gabrysiak C, Yohrling G, von der Kammer H, Ebneth A, Mack V, Munoz-Sanjuan I, Kwak S. Characterization of HTT inclusion size, location, and timing in the zQ175 mouse model of Huntington's disease: an in vivo high-content imaging study. PLoS One. 2015;10(4):e0123527. doi: 10.1371/journal.pone.0123527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferguson SS. Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol Rev. 2001;53(1):1–24. [PubMed] [Google Scholar]
- 39.Joel D, Weiner I. The organization of the basal ganglia-thalamocortical circuits: Open interconnected rather than closed segregated. Neuroscience. 1994;63(2):363–379. doi: 10.1016/0306-4522(94)90536-3. [DOI] [PubMed] [Google Scholar]
- 40.Tang TS, Tu H, Chan EY, Maximov A, Wang Z, Wellington CL, Hayden MR, Bezprozvanny I. Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron. 2003;39(2):227–239. doi: 10.1016/S0896-6273(03)00366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang TS, Slow E, Lupu V, Stavrovskaya IG, Sugimori M, Llinas R, Kristal BS, Hayden MR, Bezprozvanny I. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington's disease. Proc Natl Acad Sci U S A. 2005;102(7):2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choe ES, Wang JQ. Group I metabotropic glutamate receptors control phosphorylation of CREB, Elk-1 and ERK via a CaMKII-dependent pathway in rat striatum. Neurosci Lett. 2001;313(3):129–132. doi: 10.1016/S0304-3940(01)02258-3. [DOI] [PubMed] [Google Scholar]
- 43.del Toro D, Alberch J, Lazaro-Dieguez F, Martin-Ibanez R, Xifro X, Egea G, Canals JM. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol Biol Cell. 2009;20(5):1478–1492. doi: 10.1091/mbc.E08-07-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esseltine JL, Ribeiro FM, Ferguson SS. Rab8 modulates metabotropic glutamate receptor subtype 1 intracellular trafficking and signaling in a protein kinase C-dependent manner. J Neurosci. 2012;32(47):16933–16942a. doi: 10.1523/JNEUROSCI.0625-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abd-Elrahman KS, Ferguson SSG. Modulation of mTOR and CREB pathways following mGluR5 blockade contribute to improved Huntington's pathology in zQ175 mice. Mol Brain. 2019;12(1):35. doi: 10.1186/s13041-019-0456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribeiro FM, Hamilton A, Doria JG, Guimaraes IM, Cregan SP, Ferguson SS. Metabotropic glutamate receptor 5 as a potential therapeutic target in Huntington's disease. Expert Opin Ther Targets. 2014;18(11):1293–1304. doi: 10.1517/14728222.2014.948419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 154 kb)